Abstract
Malaria and filariasis are transmitted in the Southwest Pacific region by Anopheles punctulatus sibling species including An. punctulatus, An. koliensis, the An. farauti complex 1–8 (includes An. hinesorum [An. farauti 2], An. torresiensis [An. farauti 3]). Distinguishing these species from each other requires molecular diagnostic methods. We developed a multiplex polymerase chain reaction (PCR)–based assay specific for known species-specific nucleotide differences in the internal transcribed spacer 2 region and identified the five species most frequently implicated in transmitting disease (An. punctulatus, An. koliensis, An. farauti 1, An. hinesorum, and An. farauti 4). A set of 340 individual mosquitoes obtained from seven Papua New Guinea provinces representing a variety of habitats were analyzed by using this multiplex assay. Concordance between molecular and morphological diagnosis was 56.4% for An. punctulatus, 85.3% for An. koliensis, and 88.9% for An. farauti. Among 158 mosquitoes morphologically designated as An. farauti, 33 were re-classified by PCR as An. punctulatus, 4 as An. koliensis, 26 as An. farauti 1, 49 as An. hinesorum, and 46 as An. farauti 4. Misclassification results from variable coloration of the proboscis and overlap of An. punctulatus, An. koliensis, the An. farauti 4. This multiplex technology enables further mosquito strain identification and simultaneous detection of microbial pathogens.
Introduction
Mosquitoes belonging to the Anopheles punctulatus group are sibling species found in the Southwest Pacific region ranging from the Weber Line and Moluccas (former Spice Islands) to Vanuatu, including New Guinea and islands of the Bismarck Archipelago, the Solomon Islands and northern Australia.1 The group includes 13 sibling species (An. punctulatus, An. species near punctulatus, morphologically indistinguishable An. farauti 1–8 [Farauti complex; former An. farauti 2 and 3, now An. hinesorum and An. torresiensis, respectively], An. koliensis, An. clowi, and An. rennellensis).1–5 Studies by Bryan and Foley and others initially described species diversity within the Farauti complex by cross-mating,6,7 and allozyme polymorphisms.5 Beebe and others, and Cooper and others have extended molecular characterization of the Punctulatus group by DNA sequence analyses of ribosomal RNA loci.4,8–12 Additional diversity has been suggested among isolates of An. koliensis between collection sites in East Sepik and Madang Provinces in Papua New Guinea.13
Although members of the Punctulatus group have been characterized as unspecialized in regard to blood feeding behaviors and breeding habitats,14 individual species within the group are not distributed ubiquitously throughout the region. In previous surveys, An. farauti s.s and An. hinesorum have been characterized as most widely distributed, throughout Papua New Guinea, northern Australia, the Solomon Islands, and Vanuatu. Anopheles farauti s.s. is found most often within 1–2 km of the coast. In contrast An. hinesorum is most commonly found in areas between 10 and 100 km from the coast. Anopheles koliensis and An. punctulatus are also widely distributed throughout lowlands in Papua New Guinea and the Solomon Islands. Anopheles torresiensis has been found in northern Australia and southwestern Papua New Guinea where climate has been characterized as monsoonal. Anopheles farauti 4 is found primarily in northern Papua New Guinea throughout the Sepik and Ramu River plains. Less widely distributed species include An. farauti 5 and 6, found in the Papua New Guinea highlands; An. farauti 7, found in the Solomon Islands and Vanuatu; and An. near species punctulatus, found in inland habitats of southern and northern Papua New Guinea.4,14–16 Anopheles farauti s.l. larvae are found most frequently in coastal streams and brackish pools or swamps. Anopheles punctulatus larvae are found in sunlit rainwater pools made in tire tracks or drainage pools. Anopheles koliensis larvae are found in temporary grassland or forest edge pools more than 2 km from the coast.17
In Papua New Guinea, studies have most often implicated An. farauti s.s., An. hinesorum, An. farauti 4, An. koliensis, and An. punctulatus as the primary vectors of malaria and filarial parasites.13,18,19 However, given the heterogeneous distribution of An. punctulatus species complex members across Papua New Guinea, in addition to what is known about the variable breeding and biting preferences among species3,17,20 it will be important to understand if all members of the Punctulatus group transmit parasites, or if there is further heterogeneity among species in their competence to transmit parasites.
Assessment and continuous monitoring of the vector species composition in disease-endemic regions is a necessary component of vector biology and consistent with the integrated vector management strategy of the World Health Organization.21 Although DNA probe hybridization and polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) methods have been developed for performing species-level classification of Punctulatus group sibling species,11–13 currently, morphology remains the most common method of species identification. Morphological traits used to distinguish species (proboscis coloration) are unreliable because individual mosquitoes within single species display polymorphism, such that similar proboscis coloration is observed among species.1,4,13,22 This finding emphasizes that molecular diagnostic strategies will be required tools to enhance the capacity of entomologists for assessing vector capacity, insecticide resistance, and distribution patterns of insect species complexes in many infectious disease scenarios. Significant technical advances can now be incorporated into new diagnostic strategies to enable more efficient surveillance of these important human disease vectors.
We describe a high-throughput multiplex strategy for molecular identification of the Punctulatus group sibling species that serve as primary vectors of malaria and filarial parasites in Papua New Guinea based on species-specific sequence differences in the internal transcribed spacer 2 (ITS2) region of ribosomal DNA surveyed in this study and previous studies.10 This species identification tool applies technology used previously to perform multiplex analysis of Plasmodium and Wuchereria bancrofti infection in human blood samples.23,24
Materials and Methods
Mosquitoes.
Representative mosquitoes were obtained by the Entomology Unit of the Papua New Guinea Institute of Medical Research (PNGIMR) through on-going studies and activities associated with distribution of long-lasting insecticide-treated bed nets; collection sites from seven provinces in Papua New Guinea (Table 1). Mosquito capture methods included landing catches, Centers for Disease Control light traps,20 and larval collections. Samples collected at the larval stage were allowed to mature in the laboratory before morphological identification could be completed. Sample locations were defined by global positioning system and represented a range of altitudes, habitats, and varied distance from the coast to capture a variety of species (Table 1). Samples collected from Madang Province (Bilbil), Manus Province (Lorengau), and Morobe Province (Godowa) represented collections within < 1 km of the coast. Sampling from West Sepik Province (Pitapena: 202 km from coast) and Western Highlands Province (Brimba and Singoropa: 120 km from coast) represented collections from the most inland locations.
Table 1.
Mosquito collection site data, Papua New Guinea
| Site | Province | Altitude, meters | Latitude | Longitude | Approximate distance to coast, km |
|---|---|---|---|---|---|
| Dreikikir | East Sepik | 341 | −3.5445 | 142.77123 | 30 |
| Nale | East Sepik | 104 | −3.7725 | 143.07056 | 30 |
| Peneng | East Sepik | 254 | −3.5435 | 142.65018 | 30 |
| Bilbil | Madang | 4 | −5.2885 | 145.76371 | < 0.5 |
| Dimer | Madang | 327 | −4.8007 | 145.62395 | 7 |
| Hudini | Madang | 27 | −5.2890 | 145.74413 | 2 |
| Kokofine | Madang | 128 | −5.6903 | 145.48009 | 40 |
| Naru | Madang | 154 | −5.4067 | 145.59528 | 15 |
| Sausi | Madang | 160 | −5.6999 | 145.53802 | 35 |
| Yagaum | Madang | 180 | −5.2904 | 145.79873 | 2 |
| Lorengau | Manus | 68 | −2.0348 | 147.26136 | 1 |
| Galawo | Morobe | 1119 | −6.9314 | 146.60930 | 40 |
| Godowa | Morobe | 2 | −6.5899 | 147.82233 | < 0.5 |
| Ramu | Morobe | 444 | −6.0900 | 145.98100 | 65 |
| Pitapena | West Sepik | 183 | −4.6297 | 141.10485 | 202 |
| Kuru | Western | 43 | −8.8718 | 143.06471 | 20 |
| Brimba | WHP* | 1484 | −5.5883 | 144.68255 | 120 |
| Singoropa | WHP* | 1357 | −5.5420 | 144.65368 | 120 |
WHP = Western Highlands Province.
Species morphology.
Mosquitoes were morphologically identified using methods previously described in which members of the An. punctulatus group were classified as An. punctulatus, An. koliensis, or An. farauti s.l. by morphological characteristics.4,22 A set of 340 mosquitoes morphologically identified as members of the An. punctulatus group were then kept in coded vials containing silica gel until DNA extraction could be completed.
DNA extraction.
Genomic DNA was extracted from single whole mosquitoes (n = 340) by using either a QIAamp 96 Kit or an individual spin blood and tissue kit (QIAGEN, Valencia, CA, recommended protocol) or a modification of the method of Bender and others.25 In the modified method, individual whole mosquitoes were thoroughly ground by vortexing each mosquito with a copper pellet (BB) in a 1.5-mL microfuge tube containing 100 μL of grinding buffer (0.1 M NaCl, 0.2 M sucrose, 0.1 M Tris-HCl, pH 9.1–9.2, 0.05 M EDTA, and 0.5% sodium dodecyl sulfate). Samples were incubated at 65°C for 30 minutes; 8 M potassium acetate was then added (final concentration = 1 M) to each tube and incubated on ice for 30 minutes. Samples were centrifuged at 13,500 rpm (15 minutes) and the supernatant was transferred to a new sterile tube; 100 μL of 100% ethanol was added for precipitation of mosquito DNA. Tubes were incubated at room temperature (5 minutes) and then centrifuged at 13,500 rpm (15 minutes). Supernatants were removed; 100 μL of ice-cold 70% ethanol was added to each sample, mixed, and centrifuged at 13,500 rpm (5 minutes). Supernatants were removed and samples were allowed to dry overnight; precipitated DNA was resuspended in 30–100 μL of sterile deionized H2O or Tris-acetate buffer.
PCR amplification and DNA sequence analysis.
PCR amplifications (25 μL) of the ITS2 locus were performed in a solution containing 67 mM Tris-HCl, pH 8.8, 6.7 mM MgSO4, 16.6 mM (NH4)2SO4, 10 mM 2-mercaptoethanol, 100 μM dATP, dGTP, dCTP, and dTTP, 2.5 units of thermostable DNA polymerase, and upstream and downstream primers described by Beebe and Saul.12 The thermocycling program was 95°C for 2 minutes (1×), 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 minute (35×), 72°C for 4 minutes (1×) and was performed by using a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad, Hercules, CA) for all 340 mosquito samples. To evaluate overall amplification efficiency, PCR products were separated by electrophoresis on 2% agarose gels (1× Tris-Borate-EDTA buffer), stained with SYBR® Gold (Invitrogen, Carlsbad, CA), and visualized on a Storm 860 using ImageQuant, 5.2 software (Molecular Dynamics, Sunnyvale, CA). TOPO TA cloning (Invitrogen) of the amplified ITS2 region was performed on a subset (n = 90) of the 340 mosquitoes identified, followed by single-pass bidirectional plasmid sequencing (Beckman Coulter Genomics, Danvers, MA) to identify motifs in the ITS2 sequence for further design and development of species-specific DNA probes. DNA sequence data was analyzed by using Geneious version 5.0.3.26 ClustalW227 (default settings: gap open penalty = 10, gap extend penalty = 6.66) was used to generate the ITS2 species consensus sequence alignment as shown in Figure 1.
Figure 1.

Internal transcribed spacer 2 (ITS2) consensus alignment for Anopheles punctulatus (AP), An. farauti s.s. (AF1), An. hinesorum (AH), An. farauti 4 (AF4), and An. koliensis (AK) from Papua New Guinea. Consensus sequence for each member was created by using ITS2 sequence from multiple representatives from each species: An. punctulatus (n = 27) (Genbank accession no. HM584428–HM584454), An. farauti s.s. (n = 15) (HM584365–HM584379), An. hinesorum (n = 12) (HM584380–HM584391), An farauti 4 (n = 16) (HM584392–HM584406, HM584427), and An. koliensis (n = 20) (HM584407–HM584426). Locations of ligase detection reaction classification and reporter probes are shaded in gray and black, respectively.
Molecular species identification.
Anopheles punctulatus, An. koliensis, An. farauti s.s., An. hinesorum, and An. farauti 4 species were differentiated in a high throughput manner targeting unique species-specific polymorphisms in the ITS2 ribosomal DNA. After PCR amplification, products were added to a multiplex ligase detection reaction (LDR) in which species-specific upstream classification probes (containing anti-TAG sequences specific to Luminex® microsphere sets) ligate to downstream sequence reporter probes (3′-biotinylated) when appropriate target template sequences are available (Table 2). The LDRs were conducted in a solution (15 μL) containing 20 mM Tris-HCl buffer, pH 7.6, 25 mM potassium acetate, 10 mM magnesium acetate, 1 mM NAD+, 10mM dithiothrietol, 0.1% Triton X-100, 10 nM (200 pmol) of each LDR probe, 1 μL of each PCR product, and 2 units of Taq DNA ligase (New England Biolabs, Beverly, MA). The LDRs were initially heated at 95°C for 1 minute, followed by 32 thermal cycles at 95°C for 15 seconds and 58°C for 2 minutes. Labeling and detection of LDR fluorescent microsphere assay (FMA) products was completed as described23 by using a Bio-Plex Array Reader (Bio-Rad).
Table 2.
Internal transcribed spacer 2 DNA ligation detection reaction primers for Papua New Guinea Anopheles species identification*
| Species-specific primers† | Primer sequence‡ | FlexMAP™ microsphere§ |
|---|---|---|
| AF1 TAG | 5′-tacaaatcatcaatcactttaatcGCG CCT GCC CCC AGM GA-3′ | 11 |
| AF1 Biotin | 5′-Phos-TCT CGC GAC CCA CAT GCA C-3′-Biotin | |
| AH TAG | 5′-tcatcaatcaatctttttcacttt TCG CGC GCC AAC CCA TG-3′ | 59 |
| AH Biotin | 5′-Phos-CAC ACT GCC GCG CAA CC/3′-Biotin | |
| AP TAG | 5′-ctttaatctcaatcaatacaaatcAGC GCG GGG CGK CCG T-3′ | 1 |
| AP Biotin | 5′-Phos-GTT ACC GGC CCC TGC AC-3′-Biotin | |
| AF4 TAG | 5′-tcataatctcaacaatctttctttCGC ACG CGG CCT CGG CGG GAC TT-3′ | 68 |
| AK TAG | 5′-ctatctatctaactatctatatcaCTC TGT GTG GGA GGG AGT GCG TT-3′ | 78 |
| AF4 and AK Biotin | 5′-Phos-CGG TGC GTT YAC YCG ACT AC-3′-Biotin |
AF1 = An. farauti s.s; AH = An. hinesorum; AP = An. punctulatus; AF4 = An. farauti 4; AK = An. koliensis.
Species-specific TAG and Biotin primers are based on GenBank accession nos. AF1 (AF055984, EF042696, and EF619441, HM584365–HM584379), AH (AF033213, HM584380–HM584391), AP (AF033220, HM584428–HM584454), AF4 (AF033214, HM584392–HM584427), and AK (AF033219, HM584407–HM584426)
Nucleotides in lower case (24 bases) represent TAG sequences added to the 5′ end of each species-specific ligase detection reaction primer. Single letter nucleotide codes: M = A or C; K = G or T; Y = T or C.
Luminex® microsphere sets are synthesized to exhibit unique fluorescence and each microsphere set is coupled to different anti-TAG sequences, which are complementary to the species-specific TAG sequence primers.
Individual Luminex FlexMap™ microsphere sets cost US $25.00/vial. Interrogation of each species-specific target sequence requires 250 microspheres (1,000 assays/vial); 2.5 cents/target/mosquito.
A subset (n = 117) of the original 340 mosquitoes was also characterized by using an ITS2 RFLP assay.12 This subset was chosen at random and the PNGIMR Entomology Unit conducted RFLP analysis in a blinded manner. A portion (n = 13) of this subset of mosquitoes had also been evaluated by DNA sequence analysis as described above.
Statistical analysis.
Specificity, sensitivity, and positive and negative predictive value calculations were performed by using standard methods (http://statpages.org/ctab2x2.html).
Results
ITS2 sequence analysis and post–PCR-LDR-FMA assay design.
Previous reports of ITS2 polymorphism among Punctulatus group sibling species10 demonstrated that there is significant polymorphism among species. Moreover, previous analysis showed polymorphism within An. koliensis isolates in northern Papua New Guinea.13 Therefore, to identify conserved regions within ITS2 PCR products specific for LDR-FMA interrogation, we performed DNA sequence analysis of multiple alleles for each species of interest. After alignment of 27 alleles of An. punctulatus, 15 of An. farauti s.s., 12 of An. hinesorum, 16 of An. farauti 4, and 20 of An. koliensis, conserved species-specific sequence polymorphisms were identified. Locations in which species-specific classification and conserved sequence LDR reporter probes hybridized with template PCR products are shown in ITS2 consensus alignment for An. punctulatus, An. farauti s.s., An. hinesorum, An. farauti 4, and An. koliensis (Figure 1). Consensus sequences for each sibling species were created from ITS2 sequences after alignment of multiple representatives from each species: An. punctulatus, n = 27 (98.9% similarity within species); An. farauti s.s., n = 15 (99.3% similarity); An. hinesorum, n = 12 (99.6% similarity); An. farauti 4, n = 16 (99.5% similarity); and An. koliensis, n = 20 (98.3% similarity). These consensus sequences were compared with previously published sequences10 and found to be greater than 97.5% similar: AP consensus versus GenBank accession no. AF033220 = 98.7%, AF1 consensus (An.farauti s.s.) versus GenBank accession no. AF055984 = 98.2%, AH consensus versus GenBank accession no. AF033213 = 98.1%, AF4 consensus versus GenBank accession no. AF033215 = 97.5%, and AK consensus versus GenBank accession no. AF033219 = 98.4%.
Specificity of the Punctulatus group post–PCR-LDR-FMA.
The specificity of the assay was then demonstrated by using probes identified in Figure 1 and Table 2 to differentiate PCR products amplified from previously sequenced species-specific controls. Because members of the Punctulatus group are known to carry Plasmodium19,28,29 and Wuchereria parasites13 and human blood meals,3,28 we included six additional controls containing Plasmodium falciparum, P. vivax, P. malariae, P. ovale, Wuchereria bancrofti, and human genomic DNA.
Results of the specificity test (Figure 2) show by strong species-specific fluorescent signals (An. punctulatus median fluorescence intensity [MFI] > 5,000; An. koliensis > 11,000; An. farauti s.s. > 20,000; An. hinesorum > 21,000; An. farauti 4 > 5,000) that each LDR-FMA probe set detected only the species expected, and that all background signals were below MFIs of 900. This finding demonstrated that the LDR probes designed to target species-specific polymorphisms described here and in previously published sequences10 detected only the species present and no others. Additionally, fluorescent signals from the parasite and human samples were below a background of 900 MFI, demonstrating that the presence of parasites in a human blood meal would not obscure Anopheles species identification.
Figure 2.
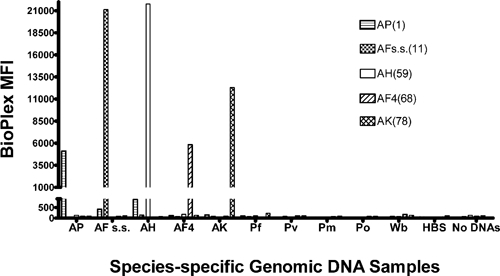
Detection of species-specific DNAs by the Anopheles species multiplex polymerase chain reaction–ligase detection reaction–fluorescent microsphere assay (PCR-LDR-FMA) for mosquitoes from Papua New Guinea. Data represent a summary of 12 individual PCR LDR-FMA detecting An. koliensis (AK), An. farauti s.s. (AF), An. hinesorum (AH), An. farauti 4 (AF4), An. koliensis (AK), and Plasmodium falciparum (Pf), P. vivax (Pv), P. malariae (Pm), P. ovale (Po), Wuchereria bancrofti (Wb), human (HBS) genomic DNAs, and negative control (no DNA). Whereas genomic DNAs were added individually into PCRs for each individual mosquito species, LDR and FMA included oligonucleotide probes representing all five Anopheles species. Numbers in parentheses next to species designations in the legend ((1), (11), (59), (68), (78)) identify Luminex FlexMap™ microspheres (Luminex Corp., Austin, TX). No DNAs indicates a blank sample to which no genomic DNAs were added.
Evaluation of morphological and molecular identification in field survey collections.
Vector surveys completed previously in malaria and filariasis endemic communities have shown the Anopheles mosquito populations to be primarily composed of An. farauti s.s., An. hinesorum, An. farauti 4, An. koliensis, and An. punctulatus.3,13,17,19 Given this background, among the 340 mosquitoes subjected to ITS2 LDR-FMA analysis, morphological assessment identified 67 as An. punctulatus, 115 as An. koliensis, and 158 as An. farauti s.l. Amplification of the ITS2 sequence produced a PCR product of the expected size range (653–783 basepairs) from all 340 insects. ITS2 LDR-FMA observed mono-specific species identification for each individual mosquito.
The overall comparison of morphology and molecular species classification for the 340 mosquitoes sampled are summarized in Table 3. First, where morphology classifies mosquitoes into only 3 categories (An. punctulatus, An. koliensis, and An. farauti s.l.), the ITS2-based multiplex assay performs species-level classification for An. punctulatus, An. koliensis, and three members of the Farauti complex (An. farauti s.s., An. hinesorum, and An. farauti 4).
Table 3.
Concordance assessment between morphology and internal transcribed spacer 2 classification for Anopheles punctulatus sibling species, Papua New Guinea*
| Morphological ID | No. | LDR-FMA Species ID | ||||
|---|---|---|---|---|---|---|
| AP | AK | AF | AH | AF4 | ||
| AP | 67 | 57 | 10 | – | – | – |
| AK | 115 | 11 | 81 | 15 | 3 | 5 |
| AF s.l. | 158 | 33 | 4 | 26 | 49 | 46 |
LDR-FMA = ligase detection reaction–fluorescent microsphere assay; ID = identification; AP = An. punctulatus s.s.; AK = An. koliensis; AF = An. farauti s.s./An. farauti 1; AH = An. hinesorum/An. farauti 2; AF4 = An. farauti 4; AF s.l. = An .farauti s.l. (specific species morphologically unidentifiable).
Comparison of morphology to molecular classification by ITS2 LDR-FMA shows the following results (Table 3). Of the 67 mosquitoes morphologically identified as An. punctulatus, the ITS2 assay identified 57 mosquitoes as An. punctulatus (85% concordance). Given the specificity of LDR-FMA for hybridization to species-specific sequence polymorphisms, results suggest that 15% of these mosquitoes should be re-classified as An. koliensis (n = 10). Of the 115 mosquitoes morphologically identified as An. koliensis, the ITS2 assay identified 81 as An. koliensis (70% concordance); 34 mosquitoes were re-classified (30%) as An. punctulatus (n = 11), An. farauti s.s. (n = 15), An. hinesorum (n = 3), and An. farauti 4 (n = 5). Of the 158 mosquitoes identified as An. farauti s.l. by morphology, ITS2 LDR-FMA identified 121 as one of three Farauti complex sibling species (26 An. farauti s.s., 49 An. hinesorum, and 46 An. farauti 4) (76.5% concordance); 37 mosquitoes were re-classified (23.5%) as An. punctulatus (n = 33) and An. koliensis (n = 4). Overall concordance between morphology and molecular identification was 76.2%. The most common source of discordance (33 of 81 misclassified, 40.7%) was observed for mosquitoes identified as An. farauti s.l. by morphology and An. punctulatus by ITS2 LDR-FMA.
Results from comparison of ITS2 LDR-FMA probe classification to morphology are also shown in Table 3. Of the 101 mosquitoes identified as An. punctulatus by ITS2 LDR-FMA, 57 were morphologically identified as An. punctulatus (56.4% concordance); 11 were identified as An. koliensis and 33 as An. farauti s.l. Ninety-five mosquitoes were identified as An. koliensis by LDR-FMA, of these 81 had been morphologically identified as An. koliensis (85% concordance), 10 as An. punctulatus, and 4 as An. farauti s.l. Of the 41 mosquitoes identified as An. farauti s.s. by LDR-FMA, 26 were morphologically identified as An. farauti s.l. and 15 as An. koliensis. Of the 52 mosquitoes identified as An. hinesorum by LDR-FMA, 49 were morphologically identified as a member of An. farauti s.l. and 3 as An. koliensis. Of the 51 mosquitoes identified as An. farauti 4 by LDR-FMA, 46 were morphologically identified as a member of An. farauti s.l. and 5 as An. koliensis.
Overall sensitivities and specificities of Anopheles spp. identification in Papua New Guinea are shown in Table 4. Values differ based on the direction of these comparisons (morphology to molecular, left; molecular to morphology, right).
Table 4.
Sensitivity and specificity of Anopheles spp. identification, Papua New Guinea*
| Characteristic | Morphology | LDR-FMA | ||||
|---|---|---|---|---|---|---|
| AP | AK | AF | AP | AK | AF | |
| Sensitivity | 0.56 | 0.85 | 0.840 | 0.85 | 0.7 | 0.77 |
| Specificity | 0.96 | 0.86 | 0.81 | 0.84 | 0.94 | 0.87 |
LDR-FMA = ligase detection reaction–fluorescent microsphere assay; AP = An. punctulatus s.s.; AK = An. koliensis; AF = An. farauti s.s./An. farauti 1.
Comparisons between ITS2 RFLP and ITS2 LDR-FMA techniques were made for a subset (n = 117) of the 340 mosquitoes. ITS2 RFLP species determination was completed by the PNGIMR Entomology Unit with no prior knowledge of the ITS2 LDR-FMA results. A 98.3% concordance was observed between ITS2 RFLP and LDR-FMA species identification methods (Table 5). Overall concordance between ITS2 RFLP and morphology was 76%, which was similar to that of ITS2 LDR-FMA and morphology. Coincidentally, ITS2 DNA sequence was also obtained for 13 of the 117 mosquitoes evaluated by LDR-FMA and RFLP. All 13 (An. punctulatus = 5, An. farauti s.l. = 2, and An. koliensis = 6) of these mosquitoes were identified by ITS2 RFLP, LDR-FMA and reference sequence comparison with 100% concordance.
Table 5.
Concordance assessment of molecular internal transcribed spacer 2 classification methods (RFLP vs. LDR-FMA) for Anopheles punctulatus sibling species, Papua New Guinea*
| RFLP | LDR-FMA Species ID | ||||
|---|---|---|---|---|---|
| AP | AK | AF | AH | AF4 | |
| AP | 37 | – | 1 | – | – |
| AK | – | 39 | – | – | – |
| AF | – | – | 23 | – | – |
| AH | – | – | – | 3 | – |
| AF4 | 1 | – | – | – | 13 |
RFLP = restriction fragment length polymorphism; LDR-FMA = ligase detection reaction–fluorescent microsphere assay; ID = identification; AP = An. punctulatus s.s.; AK = An. koliensis; AF = An. farauti s.s./An. farauti 1; AH = An. hinesorum/An. farauti 2; AF4 = An. farauti 4.
Discussion
DNA sequence analysis of the ITS2 region of the ribosomal RNA genes of An. punctulatus group sibling species has identified numerous sequence polymorphisms (single and multiple nucleotide polymorphisms; insertions and deletions; repeat variants or types). Beebe and others have used these molecular polymorphisms to perform phylogenetic analyses to demonstrate sequence differences that routinely distinguish morphologically identical sibling species, particularly those within the Farauti complex.10 This work has been the platform for developing both post-PCR DNA probe and RFLP strategies for describing species relationships and ancestry, to perform species identification in field surveys, and to begin evaluation of strain distribution between sites within the Southwest Pacific region. New technology incorporating fluorescent microspheres coupled to an array of variable sequence oligonucleotides (Luminex FlexMAP™ microspheres) enables liquid-phase microarray analysis that significantly expands capacity for developing multiplex DNA sequence-based diagnostic strategies. Given further results illustrating An. punctulatus and An. bancrofti group ITS2 sequence polymorphisms within individual species,8,30 we wanted to make multiple within and among species sequence comparisons before developing species-specific DNA probes for application through this technology. Our analysis of more than 100 ITS2 alleles from An. punctulatus (n = 27), An. koliensis (n = 20), An. farauti s.s. (n = 28; includes 13 previously published sequences8 [Genbank accession nos. AF104314–AF104326]), An. hinesorum (n = 12), and An. farauti 4 (n = 16) showed considerable ITS2 sequence divergence between species (from 65.0% to 92.9% pairwise percent identity; includes previously published sequences10 for An. torresiensis [Genbank accession no. AF033214], An. farauti 5 [AF033216], An. farauti 6 [AF033217], An. farauti 7 [AF033218], An. near punctulatus [AF033221]), further comparisons showed high conservation of ITS2 sequence (≥ 99.0% pairwise percent identity) within species. These comparisons enabled identification of sequence regions that varied significantly among species that did not vary within species. Probes based on these sequences have now been shown to differentiate the five Punctulatus group sibling species (Figure 2) implicated in transmitting malaria and filarial parasites in Papua New Guinea.
Using the multiplex molecular diagnostic assay, we analyzed 340 mosquitoes obtained from 18 locations (7 Provinces in Papua New Guinea) that had been morphologically identified as members of the Punctulatus group. Because morphology can only differentiate members of the An. punctulatus complex into three species categories (An. farauti s.l., An. koliensis, and An. punctulatus) it was clear that the ITS2 multiplex assay would provide more definitive identification of individual mosquitoes than classical morphometric methods, and this was borne out in further identification of An. farauti s.s., An. hinesorum, and An. farauti 4. Comparison of the two methods was performed by classifying mosquitoes into the three generalized species. Among the 101 individual insects identified by the An. punctulatus probe, 44 mosquitoes were classified as either An. koliensis or An. farauti s.l. by morphology (DNA sensitivity versus morphology, morphology positive predictive value versus DNA = 0.564). Species identification between the An. koliensis probe and morphology was more highly concordant (DNA sensitivity versus morphology = 0.853). However, 34 of 115 insects identified as An. koliensis by morphology were observed to hybridize with An. punctulatus and An. farauti probes (morphology sensitivity versus DNA = 0.704). Similarly, 37 of 158 insects identified by morphology as An. farauti s.l. were observed to hybridize to An. punctulatus and An. koliensis probes (morphology sensitivity versus DNA = 0.766). When evaluated in aggregate, DNA probes detecting sibling species of the Farauti complex were observed to hybridize with 23 mosquitoes identified by morphology as An. koliensis. However, none of the insects identified by morphology as An. punctulatus hybridized with any of the Farauti complex probes (DNA sensitivity versus morphology = 0.840). To compare this new LDR-FMA method with the currently established molecular method of identification, ITS2 RFLP, we tested a subset of 117 mosquito samples and found concordance rates between ITS2 RFLP and ITS2 LDR-FMA to be above 98%. Similar to ITS2 LDR-FMA, RFLP species differentiation was 76% concordant with morphological species identification.
Given the variability of proboscis coloration and overlapping geographic distribution of morphologically similar Punctulatus group sibling species previously documented by Cooper and others4 and Beebe and Cooper,14 we anticipated that we would observe discordance between the two classification schemes (ITS2 LDR-FMA and morphological identification). At 10 of the 18 collection sites, 100% concordance was observed between DNA probe and morphologic classification of An. punctulatus; in four of these sites An. punctulatus was the only species collected. In the 12 sites where An. koliensis was identified by morphology, DNA probe classification was 100% concordant in only one site (Hudini). In the 11 sites where An. farauti s.l. was collected, 100% concordance between DNA probe and morphology was observed in two sites (Sausi and Bilbil). In all 13 sites where more than one species was present, discordance was observed between DNA probe and morphology, and in two sites where only one species was identified by morphology, different species were identified by DNA probes. Because morphological classifications and LDR-FMA probe hybridization discordance was highest between An. koliensis and An. farauti s.l., further research is needed to characterize the relationships of these species more clearly. Finally, observed discordance between ITS2 LDR-FMA DNA probes and morphology was observed in collection sites from all 7 of the provinces included in our survey.
We have described and demonstrated a new multiplex DNA-based assay designed to identify DNA sequence polymorphisms in the primary disease vectors of the Punctulatus group in Papua New Guinea. This method is based on technology that facilitates multiplex analysis, automation, and uniformity of PCR-based mosquito species diagnosis because procedures from sample processing and DNA extraction to entry of results into database files can now be handled continuously in 96-well plate format. Costs associated with the post-PCR fluorescent microsphere assay for the five mosquito species surveyed total to 12.5 cents per individual mosquito. The LDR-FMA multiplex avoids use of ethidium bromide (biohazardous waste) used to visualize RFLP patterns. Also, although the RFLP method reliably differentiates sibling species within the Punctulatus group (Table 5), the MspI RFLP analysis will not identify strain-specific single nucleotide or small insertion/deletion polymorphisms (this study and Beebe and others10) observed within species that appear to be associated with different geographic locations. The LDR-FMA assay design allows for modifications and additions, which could identify these polymorphisms. Moreover, because the same LDR-FMA multiplex strategy is used for diagnosis of Plasmodium species and Wuchereria bancrofti,23,24 it is foreseeable that a single post-PCR assay would be able to perform Anopheles species identification and diagnosis of important human parasites simultaneously from a single tube.
Understanding habitat preference, vector competence, biting habits, and distribution of Punctulatus group sibling species and strains is paramount to understanding mosquito ecology and designing disease and vector control plans. Given the overall complexity of vector-borne disease transmission associated with the Punctulatus group, it is critical to develop diagnostic surveillance strategies that can monitor markers associated with infection by efficient methods. In comparison with RFLP analysis, the LDR-FMA multiplex strategy has greater capacity for high throughput analysis of known genetic targets and expansion to assess additional polymorphism in a single post-PCR assay. Therefore, this new diagnostic approach provides significant potential to improve research studies and to monitor and evaluate operational control programs to reduce disease transmission by these important vectors.
ACKNOWLEDGMENTS
We are grateful to the PNGIMR Entomology Unit for aid in mosquito collections and morphological identification. We dedicate this work to our colleague Henry Dagoro who passed away on August 4, 2010. Over the past 30 years, he contributed careful analysis and significant energy to the study of the An. punctulatus group sibling species as a senior member of the PNGIMR Entomology Laboratory. He patiently mentored numerous students and scientists from around the world on the behaviors and local habitats characterizing these important human disease vectors.
Footnotes
Financial support: This study was supported by grants from the National Institutes of Health (AI065717) and the Fogarty International Center (TW007872, TW007377, and TW007735). Mosquito collection was also supported by the Global Fund to Fight Aids, Tuberculosis and Malaria Round 3 malaria grant to Papua New Guinea.
Authors' addresses: Cara Henry-Halldin, Allison M. Zimmerman, and Peter A. Zimmerman, Center for Global Health and Diseases, Case Western Reserve University School of Medicine, Wolstein Research Building, Cleveland, OH, E-mails: cnh@case.edu, amz23@case.edu, paz@case.edu. Lisa Reimer, Edward Thomsen, Gussy Koimbu, John B. Keven, and Henry Dagoro (deceased), Papua New Guinea Institute of Medical Research-Madang, Madang, Papua New Guinea, E-mails: lisa.reimer@case.edu, edward.thomsen@case.edu, gkoimbu@gmail.com, jo.bosco@live.com. Manuel W. Hetzel, Ivo Mueller, and Peter Siba, Papua New Guinea Institute of Medical Research-Goroka, Goroka, Papua New Guinea, E-mails: manuel.hetzel@pngimr.org.pg, ivomueller@fastmail.fm, pmaxsiba@gmail.com
References
- 1.Rozeboom LE, Knight KL. The punctulatus complex of Anopheles (Diptera: Culicidae) J Parasitol. 1946;32:95–131. [PubMed] [Google Scholar]
- 2.Bryan JH. Morphological studies on the Anopheles punctulatus Dönitz complex. Trans R Entomol Soc Lond. 1974;125:413–435. [Google Scholar]
- 3.Charlwood JD, Dagoro H, Paru R. Blood-feeding and resting behaviour in the Anopheles punctulatus Dönitz complex (Diptera: Culicidae) from coastal Papua New Guinea. Bull Entomol Res. 1985;75:463–476. [Google Scholar]
- 4.Cooper RD, Waterson DG, Frances SP, Beebe NW, Sweeney AW. Speciation and distribution of the members of the Anopheles punctulatus (Diptera: Culicidae) group in Papua New Guinea. J Med Entomol. 2002;39:16–27. doi: 10.1603/0022-2585-39.1.16. [DOI] [PubMed] [Google Scholar]
- 5.Foley DH, Paru R, Dagoro H, Bryan JH. Allozyme analysis reveals six species within the Anopheles punctulatus complex of mosquitoes in Papua New Guinea. Med Vet Entomol. 1993;7:37–37. doi: 10.1111/j.1365-2915.1993.tb00649.x. [DOI] [PubMed] [Google Scholar]
- 6.Bryan J. Studies on the Anopheles punctulatus complex. 3. Mating behaviour for the F1 hybrid adults from crosses between Anopheles farauti no. 1 and Anopheles farauti no. 2. Trans R Soc Trop Med Hyg. 1973;67:85–91. doi: 10.1016/0035-9203(73)90324-6. [DOI] [PubMed] [Google Scholar]
- 7.Bryan JH. Studies on the Anopheles punctulatus complex. 1. Identification by proboscis morphological criteria and by cross-mating experiments. Trans R Soc Trop Med Hyg. 1973;67:64–69. doi: 10.1016/0035-9203(73)90322-2. [DOI] [PubMed] [Google Scholar]
- 8.Beebe NW, Cooper RD, Foley DH, Ellis JT. Populations of the south-west Pacific malaria vector Anopheles farauti s.s. revealed by ribosomal DNA transcribed spacer polymorphisms. Heredity. 2000;84:244–253. doi: 10.1046/j.1365-2540.2000.00665.x. [DOI] [PubMed] [Google Scholar]
- 9.Beebe NW, Cooper RD, Morrison DA, Ellis JT. A phylogenetic study of the Anopheles punctulatus group of malaria vectors comparing rDNA sequence alignments derived from the mitochondrial and nuclear small ribosomal subunits. Mol Phylogenet Evol. 2000;17:430–436. doi: 10.1006/mpev.2000.0853. [DOI] [PubMed] [Google Scholar]
- 10.Beebe NW, Ellis JT, Cooper RD, Saul A. DNA sequence analysis of the ribosomal DNA ITS2 region for the Anopheles punctulatus group of mosquitoes. Insect Mol Biol. 1999;8:381–390. doi: 10.1046/j.1365-2583.1999.83127.x. [DOI] [PubMed] [Google Scholar]
- 11.Beebe NW, Foley DH, Saul A, Cooper L, Bryan JH, Burkot TR. DNA probes for identifying the members of the Anopheles punctulatus complex in Papua New Guinea. Am J Trop Med Hyg. 1994;50:229. doi: 10.4269/ajtmh.1994.50.229. [DOI] [PubMed] [Google Scholar]
- 12.Beebe NW, Saul A. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 1995;53:478. doi: 10.4269/ajtmh.1995.53.478. [DOI] [PubMed] [Google Scholar]
- 13.Benet A, Mai A, Bockarie F, Lagog M, Zimmerman P, Alpers MP, Reeder JC, Bockarie MJ. Polymerase chain reaction diagnosis and the changing pattern of vector ecology and malaria transmission dynamics in Papua New Guinea. Am J Trop Med Hyg. 2004;71:277. [PubMed] [Google Scholar]
- 14.Beebe NW, Cooper RD. Distribution and evolution of the Anopheles punctulatus group (Diptera: Culicidae) in Australia and Papua New Guinea. Int J Parasitol. 2002;32:563–574. doi: 10.1016/s0020-7519(01)00359-9. [DOI] [PubMed] [Google Scholar]
- 15.Cooper RD, Waterson DG, Kupo M, Foley DH, Beebe NW, Sweeney AW. Anopheline mosquitoes of the Western province of Papua New Guinea. J Am Mosq Control Assoc. 1997;13:5–12. [PubMed] [Google Scholar]
- 16.Sweeney AW, Cooper RD, Frances SP. Distribution of the sibling species of Anopheles farauti in the Cape York Peninsula, northern Queensland, Australia. J Am Mosq Control Assoc. 1990;6:425–429. [PubMed] [Google Scholar]
- 17.Charlwood JD, Graves PM, Alpers MP. The ecology of Anopheles punctulatus group of mosquitoes from Papua New Guinea: a review of recent work. P N G Med J. 1986;26:19–26. [PubMed] [Google Scholar]
- 18.Cooper RD, Frances SP. Malaria vectors on Buka and Bougainville Islands, Papua New Guinea. J Am Mosq Control Assoc. 2002;18:100–106. [PubMed] [Google Scholar]
- 19.Cooper RD, Waterson DG, Frances SP, Beebe NW, Pluess B, Sweeney AW. Malaria vectors of Papua New Guinea. Int J Parasitol. 2009;39:1495–1501. doi: 10.1016/j.ijpara.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Bockarie MJ, Alexander N, Bockarie F, Ibam E, Barnish G, Alpers M. The late biting habit of parous Anopheles mosquitoes and pre-bedtime exposure of humans to infective female mosquitoes. Trans R Soc Trop Med Hyg. 1996;90:23–25. doi: 10.1016/s0035-9203(96)90465-4. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization . Global Strategic Framework for Integrated Vector Management. Geneva: World Health Organization; 2004. [Google Scholar]
- 22.Belkin J. The Mosquitoes of the South Pacific (Diptera: Culicidae) Berkeley, CA: University of California Press; 1962. [Google Scholar]
- 23.McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, Zimmerman PA. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am J Trop Med Hyg. 2006;74:413–421. [PMC free article] [PubMed] [Google Scholar]
- 24.Mehlotra R, Gray LR, Blood Zikursh MJ, Kloos Z, Henry-Halldin CN, Tisch DJ, Thomsen E, Reimer L, Kastens W, Baea M, Baea K, Baisor M, Tarongka N, Kazura JW. Molecular-based assay for simultaneous detection of four Plasmodium spp. and Wuchereria bancrofti infections. Am J Trop Med Hyg. 2010;82:1030–1033. doi: 10.4269/ajtmh.2010.09-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender W, Spierer P, Hogness DS, Chambon P. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- 26.Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious Pro Version 5.0. 2009. http://www.geneious.com Available at.
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Burkot TR, Graves PM, Paru R, Wirtz RA, Heywood PF. Human malaria transmission studies in the Anopheles punctulatus complex in Papua New Guinea: sporozoite rates, inoculation rates, and sporozoite densities. Am J Trop Med Hyg. 1988;39:135. doi: 10.4269/ajtmh.1988.39.135. [DOI] [PubMed] [Google Scholar]
- 29.Burkot TR, Molineaux L, Graves PM, Paru R, Battistutta D, Dagoro H, Barnes A, Wirtz RA, Garner P. The prevalence of naturally acquired multiple infections of Wuchereria bancrofti and human malarias in anophelines. Parasitology. 1990;100:369. doi: 10.1017/s003118200007863x. [DOI] [PubMed] [Google Scholar]
- 30.Beebe NW, Maung J, van den Hurk AF, Ellis JT, Cooper RD. Ribosomal DNA spacer genotypes of the Anopheles bancroftii group (Diptera: Culicidae) from Australia and Papua New Guinea. Insect Mol Biol. 2001;10:407–413. doi: 10.1046/j.0962-1075.2001.00278.x. [DOI] [PubMed] [Google Scholar]


