Abstract
An estimated 884 million people worldwide do not have access to an improved drinking water source, and the microbial quality of these sources is often unknown. In this study, a combined tangential flow, hollow fiber ultrafiltration (UF), and real-time PCR method was applied to large volume (100 L) groundwater (N = 4), surface water (N = 9), and finished (i.e., receiving treatment) drinking water (N = 6) samples for the evaluation of human enteric viruses and bacterial indicators. Human enteric viruses including norovirus GI and GII, adenovirus, and polyomavirus were detected in five different samples including one groundwater, three surface water, and one drinking water sample. Total coliforms and Escherichia coli assessed for each sample before and after UF revealed a lack of correlation between bacterial indicators and the presence of human enteric viruses.
Introduction
Clean water is essential for life. Throughout the world, millions of people do not have access to microbiologically safe water for drinking, cooking, and other essential purposes. It is estimated that 884 million people—one-sixth of the world's population—do not have access to improved sources of drinking water.1,2 Improved drinking water sources include piped household water connections, public standpipes, boreholes, protected dug wells, protected springs, and rainwater collection.3 However, access to improved water does not automatically equate to microbiologically safe drinking water. Health risks associated with drinking water include infectious diseases predominantly caused by human and animal enteric (e.g., fecally derived) pathogens, including bacteria (i.e., Vibrio cholerae, Escherichia coli, Campylobacter, and Salmonella), protozoa (i.e., Cryptosporidium, Entamoeba histolytica, and Cyclospora caryentensis), and viruses (i.e., norovirus, enterovirus, hepatitis A and E virus, and rotavirus).4–6 In addition, intestinal helminthes (i.e., guinea worm and liver flukes) are associated with infectious diseases related to contaminated drinking water, causing an estimated 9,400 deaths every year.4 Poor water quality, limited sanitation, and inadequate hygiene can cause gastrointestinal illnesses, resulting in an estimated 3.5 billion diarrheal episodes per year and causing 1.87 million childhood deaths per year worldwide from diarrhea.7,8 Of these deaths, 90% are children from developing countries and account for nearly 20% of the 10 million total deaths in children under 5 years each year.8,9
Many areas in developing countries lack access to safe drinking water. However, the majority (around 70%) of the global population without improved drinking water sources reside in rural areas.2 Rural communities are typically located far away from urban centers where the capacity to provide a centralized drinking water system is dramatically reduced; thus, rural populations commonly obtain water on an individual or household basis from nearby surface and groundwater sources where the microbial quality is often unknown.5,10 When improved drinking water supply infrastructure (e.g., boreholes with hand pumps) is available in rural areas, infrastructure sustainability is often limited by inadequate financial resources for operation and maintenance costs as well as the inability to obtain spare parts and necessary technical expertise.10,11 Along with technical and financial difficulties of supplying drinking water to rural communities, there is often a lack of health risk perception and understanding of basic hygiene practices.12
Potential alternatives to the lack of widespread drinking water infrastructure in rural areas are small water enterprises (SWE), including water tankers and water kiosks that make water resources available at the community and household levels. The presence of SWEs in developing regions including Southeast Asia and sub-Saharan Africa has become of increasing interest to non-governmental organizations, governments, and global institutions as a viable and sustainable option for the provision of potable drinking water to vulnerable populations.13 Access to improved drinking water sources is of particular interest for sub-Saharan African countries, where 37% of the population is still without improved water.2 In addition, Africa has the highest distribution of deaths caused by diarrhea compared with low- and middle-income countries in four other World Health Organization (WHO) regions, accounting for 41% of all diarrhea deaths occurring in the developing world.8
Ghana has experienced an increase in SWEs in the rural southern regions of Greater Accra and in the southern region of Lake Volta. Ghana is classified as a country with medium human development and ranks 152 of 182 countries included in the Human Development Index.14 In Ghana, 36% of the rural population does not have access to improved drinking water sources, and 89% lack adequate sanitation.9,15 Because of the limited access to safe drinking water combined with the lack of improved sanitation, enteric diseases are one of the leading causes of morbidity and mortality among children in Ghana. It is estimated that 13% of the children under 5 deaths in Ghana are attributable to diarrheal disease.15 Thus, the implementation of alternative strategies (e.g., SWEs) for the provision of safe drinking water to rural communities in Ghana is greatly needed.
The monitoring and evaluation of strategies designed to improve drinking water quality in developing countries, including Ghana, is insufficient and often lacking. To adequately assess microbial drinking water quality, a system-wide approach should be taken. Water quality should be assessed not only after the treatment intervention but also at the source, within the distribution system, if present, and within individual households.4,16 Bacterial indicators including total coliforms, E. coli, and fecal enterococci are predominantly relied on for the assessment of microbial water quality. Total coliforms can provide basic information on water quality, but they are not an index of fecal pollution; however, E. coli and enterococci are both indicators of fecal contamination.17 The predominant indicator of choice for fecal pollution is E. coli.4 Indicator microorganisms are intended to act as sentinels for the potential presence of human pathogens of fecal origin, including enteric viruses and protozoa. However, previous studies have shown that these bacteria, especially total coliforms, are poorly correlated with the presence of human pathogenic bacteria, enteric viruses, and protozoa.18–20 The lack of correlation between fecal indicator bacteria and the presence or absence of enteric viruses and protozoa is primarily caused by the inherent characteristics of these groups of microorganisms. Enteric viruses and protozoa are more resistant to disinfection (i.e., chlorination) processes used for drinking water treatment and degradation from environmental stressors compared with bacteria.16 Additional microbial parameters used to assess drinking water quality include thermo-tolerant fecal coliforms, heterotrophic plate count, hydrogen sulfide-producing bacteria (i.e., Salmonella, Citrobacter, and Proteus), and F-specific RNA bacteriophages.17 Although traditional microbial indicators have proved to be useful for assessment of drinking water quality in the past, direct detection of pathogens through advanced sampling and detection techniques (i.e., large-volume concentration and molecular analyses) can play an important role by providing a more representative evaluation of microbial water quality.
Studies evaluating improved drinking water sources and drinking water interventions in developing counties have historically relied on fecal indicator bacteria for assessment of microbial water quality.21–26 For the present study, the primary objective was to provide a more representative assessment of available water sources in southern Ghana, with a focus on recovery and detection of enteric viruses. The microbial quality of treated drinking water vended from water kiosks (i.e., SWEs) and surface and groundwater sources in six rural villages within Southern Ghana was assessed using an optimized tangential flow, hollow fiber ultrafiltration (UF) method for the concentration and recovery of indicator bacteria and viruses from 100-L water samples. Molecular methods including real-time polymerase chain reaction (PCR) and reverse transcription PCR (RT-PCR) were then applied for the detection of human enteric viruses, including human norovirus (HuNoV), enterovirus (EV), human adenovirus (HuAdV), human polyomavirus (HuPyV), and hepatitis E virus (HEV). To date, no studies have reported the use of UF for the recovery of microorganisms from 100-L water samples for evaluating the microbial quality of community water resources in a developing country.
Materials and Methods
Location and sample collection.
Water samples were collected between July 28 and August 8, 2009 from groundwater (GW; N = 4), surface water (SW; N = 9), and finished (i.e., receiving treatment) drinking water (DW; N = 6) sites. These samples were collected in six separate villages (A, B, C, D, E, and F) in Southern Ghana and within the city of Accra. Villages A, B, C, D, and E had access to treated drinking water through a community-based water kiosk in addition to other household water sources, including untreated GW and SW. Village F did not have a community water kiosk; thus, this community used primarily GW and SW for drinking and household water supplies. Figure 1 shows the location of each village in relation to Accra. Additional sampling sites were determined in each village by observing and inquiring to residents about primary household water sources. At each site, 100 L of water were collected using sterilized 25-L plastic water storage containers with a removable plastic funnel in addition to 20-L buckets when necessary (i.e., primarily for SW sites). Turbidity, pH, total dissolved solids, dissolved oxygen, chloride, nitrate, and ammonium were collected for each water sample using an YSI Multiparameter Water Quality Sonde (Model #6820 V2) and Data System (Model #650MDS; YSI Incorporated, Yellow Springs, OH). Samples were transported to the field laboratory in Accra, Ghana and processed on the same day as collection.
Figure 1.
Map of sampling area in Southern Ghana.
Preparation of water samples.
At the field laboratory in Accra, Ghana, the chemical surfactant sodium polyphosphate (NaPP; Sigma, St. Louis, MO) was added to each 25-L container to achieve a final concentration of 0.01%. Each water sample was then transferred to a 20-L bucket containing a disposable low-density polyethylene (LPDE) bucket liner (U.S. Plastics Corporation, Lima, OH). The 20-L bucket containing a portion of the 100-L water sample was continuously filled with the sample during UF until the entire sample was concentrated down to 200–300 mL.
UF setup.
The UF setup was conducted as previously reported with modifications.27 High-performance, platinum-cured LS/36 and LS/24 silicon tubing (Masterflex; Cole-Parmer Instrument Co., Vernon Hills, IL) was used in each experiment and then reused after disinfection. Disinfection consisted of submersion in 50 ppm chlorine bleach (i.e., hypochlorous acid) solution followed by a 5-min rinse with reverse osmosis (RO) purified water. After rinsing with RO water, the tubing was soaked in a 3 M excess sodium thiosulfate (Sigma) solution for neutralization of remaining chlorine followed by a final 5-min rinse with RO water. Polypropylene NS4 quick-disconnect couplings (Colder Products Company, St. Paul, MN), screw clamps, brass fittings, rubber stoppers (Fisher Scientific, Waltham, MA), and 25-L water collection containers were disinfected in the same way before use in the UF setup and between each water sample. RO water for rinsing and preparing chlorine and sodium thiosulfate solutions was prepared from Accra City drinking water using a Countertop 4-Stage Reverse Osmosis 75/90 GPD System with Ultraviolet Light for disinfection (APEC Water Systems, City of Industry, CA). Baxter Exceltra Plus 210 (Baxter International, Deerfield, IL) dialysis filters were used during UF. The Exceltra Plus 210 filter is composed of cellulose triacetate hollow fiber membranes with a molecular weight cut-off of 70,000 daltons and surface area of 2.1 m2. New filters and LPDE bucket liners were used for each sample. A Cole-Parmer Model 7524-40 peristaltic pump and Masterflex Model 77800-52 pump heads were used for processing all samples.
UF procedure.
Before filtration, ultrafilters were blocked with 0.1% NaPP. The peristaltic pump was set to pump at 1,700 mL/min under 10 lb/in2 of pressure, with an average flow of 900 mL/min and 800 mL/min for the cross-flow rate and permeate rate, respectively. Filtration was performed until approximately 200–300 mL of concentrated sample remained in the ultrafiltration system. After sample concentration, the ultrafilter was eluted as described previously with modifications.27 Briefly, a solution containing 0.1% Tween 80 (Sigma), 0.01% NaPP, and 0.001% Antifoam A (Sigma) was prepared with RO water and added to the concentrate at a solution-to-concentrate ratio of 1:9. During elution, both permeate ports were closed, and backpressure was removed by releasing the screw clamp. The concentrate and elution buffer mixture was then recirculated for 5 minutes at a flow rate of 1,700 mL/min and then collected. The elution step was performed on all samples except SW (N = 9) and Accra City DW (N = 1) samples. UF concentrates were stored at 4°C and shipped back on ice to The Johns Hopkins Bloomberg School of Public Health laboratories in Baltimore, MD for additional sample processing and analysis.
Indicator bacteria analysis.
Bacterial analyses for total coliforms and E. coli were completed before and after UF using the IDEXX Quanti-tray system (IDEXX Laboratories, Westbrook, ME). Samples of 101 mL were collected from GW and DW samples, and 10.1 mL samples were collected from SW samples, respectively, before filtration to analyze for the presence of total coliforms and E. coli. After filtration, 10.1 mL samples were collected from GW and DW UF concentrates, and 1.1 mL samples were collected from SW UF concentrates. For detection and enumeration of total coliforms and E. coli, a Colilert Quanti-tray system was used to determine the most probable number (MPN) in each sample before and after UF. For GW and DW samples, 100 and 1 mL volumes were analyzed before UF, and 10 mL and 100 μL volumes were analyzed after UF. For SW samples, 10 mL and 100 μL volumes were analyzed before UF, and 1 mL and 100 μL volumes were analyzed after UF. Prepared samples in Quanti-trays were incubated at 37°C for 24 hours. Sample volumes of less than 100 mL (i.e., 10 mL, 1 mL, or 0.1 mL) were added to 0.1% peptone (Invitrogen, Carlsbad, CA) to bring the total volume to 100 mL. A negative control containing 100 mL 0.1% peptone was analyzed by Colilert for each batch of samples.
UF concentrate sample processing.
A secondary concentration step was applied to a portion of the UF concentrates for the molecular analysis of human enteric viruses. Briefly, using Centricon Plus-70 (Millipore, Billerica, MA) centrifugal filtration devices with a molecular weight cut-off of 30,000 daltons or 100,000 daltons, 70 mL of UF concentrates were further concentrated. Before beginning secondary concentration, 70 mL SW samples were pre-clarified by centrifugation at 5,000 × g for 5 minutes at 4°C. The supernatant was separated from the pellet, and the entire supernatant volume was then applied to a Centricon Plus-70 filter unit. SW pre-clarification pellets were resuspended as needed in 500 μL diethyl pyrocarbonate (DEPC)-treated water (Quality Biologicals, Gaithersburg, MD) and archived at −80°C. The final Centricon concentrate volumes ranged from 0.18 to 0.84 mL, 0.16 to 3.8 mL, and 0.18 to 9.3 mL for GW, SW, and DW, respectively. Two hundred microliters of each pellet were processed separately during total viral nucleic acid (NA) extraction as described below.
For the analysis of human enteric viruses, total viral NA was extracted from GW, SW, and DW secondary concentrates and SW pre-clarification pellets using QIAamp MinElute Virus Spin kit (Qiagen, Valencia, CA). Eluted viral NA was portioned out and archived at −80°C until analysis. During total viral NA extraction, a negative control extraction containing 200 μL DEPC-treated water was also processed to verify that no cross-contamination occurred.
Real-time PCR and identification of inhibition.
Amplification of viral DNA and RNA targets was performed using an ABI Prism 7300 Sequence Detection System (Applied Biosystems, Carlsbad, CA). Total viral NA extracted from UF secondary concentrates was analyzed for HEV (GI–GIV), HuAdV (Types A–F), HuNoV (GI and GII), EV, and HPyV, including JC virus and BK virus, by real-time PCR or RT-PCR. All assays were performed in a 96-well plate format. The sequences and sources of the primers and probes used in this present study are shown in Table 1.28–32 Each assay was validated using positive controls and negative controls consisting of non-target NA and DEPC-treated water.
Table 1.
Primers and probes used in this study
| Microorganism | GenBank accession number | Primer or probe name | Primer/probe final concentration (nM) | Probe label | Sequence* | Product size (bp) | Product region | Ref. |
|---|---|---|---|---|---|---|---|---|
| Hepatitis E virus (GI–GIV) | M73218 | JVHEVF | 250 | 5′ GGTGGTTTCTGGGGTGAC 3′ | 70 | ORF3 region | 30 | |
| JVHEVR | 250 | 5′ AGGGGTTGGTTGGATGAA 3′ | ||||||
| JVHEVP | 100 | FAM† | 5′ TGATTCTCAGCCCTTCGC 3′ | |||||
| Human norovirus GI | M87661 | COG1F | 1,000 | 5′ CGYTGGATGCGNTTYCATGA 3′ | 85 | ORF1–ORF2 junction | 31 | |
| COG1R | 1,000 | 5′ CTTAGACGCCATCATCATTYAC 3′ | ||||||
| RING 1A | 100 | FAM† | 5′ AGATYGCGATCYCCTGTCCA 3′ | |||||
| RING 1B | 100 | FAM† | 5′ AGATCGCGGTCTCCTGTCCA 3′ | |||||
| Human norovirus GII | AF145896 | COG2F | 1,000 | 5′ CARGARBCNATGTTYAGRTGGATGAG 3′ | 88 | ORF1–ORF2 junction | 31 | |
| COG2R | 1,000 | 5′ TCGACGCCATCTTCATTCACA 3′ | ||||||
| RING2-TP | 200 | FAM† | 5′ TGGGAGGGCGATCGCAATCT 3′ | |||||
| Human adenovirus (Type A–F) | AC_000008 | JTVXF | 400 | 5′ GGACGCCTCGGAGTACCTGAG 3′ | 96 | Hexon region | 29 | |
| JTVXR | 400 | 5′ ACIGTGGGGTTTCTGAACTTGTT 3′ | ||||||
| JTVXP | 150 | FAM† | 5′ CTGGTGCAGTTCGCCCGTGCCA 3′ | |||||
| Human polyomavirus (JC and BK) | AB092584 | SM2 | 500 | 5′ AGTCTTTAGGGTCTTCTACCTTT 3′ | 173 (JC); 176 (BK) | Partial T antigen | 28 | |
| P6 | 500 | 5′ GGTGCCAACCTATGGAACAG 3′ | ||||||
| KGJ3 | 400 | FAM‡ | 5′ TCATCACTGGCAAACAT 3′ | |||||
| Human enteroviruses | AJ293918 | EV1R | 700 | 5′ TGTCACCATAAGCAGCCA 3′ | 143 | 5′ untranslated region (UTR) | 32 | |
| EV1F | 700 | 5′ CCCTGAATGCGGCTAAT 3′ | ||||||
| EV probe | 120 | FAM† | 5′ ACGGACACCCAAAGTAGTCGGTTC 3′ | |||||
| Hepatitis G virus (internal standard) | U44402 | HepG-F | 400 | 5′ CGGCCAAAAGGTGGTGGATG 3′ | 185 | 5′ UTR | 33, 34 | |
| HepG-R | 400 | 5′ CGACGAGCCTGACGTCGGG 3′ | ||||||
| HepG probe | 200 | FAM† | 5′ AGGTCCCTCTGGCGCTTGTGGCGAG 3′ |
Mixed bases in degenerate primers and probes are as follows: Y = C, T; R = A, G; B = C,T, G; N = A, C, T, G.
The FAM (6-carboxyfluorescein) quencher is BHQ-1 (black hole quencher).
The FAM quencher is a minor groove binder non-fluorescent quencher (MGBNFQ).
For viral RNA amplification, each 25-μL reaction contained 12.5 μL of 2× master mix (QuantiTect Probe RT-PCR Kit [Qiagen]), 5U RNAse Inhibitor (Applied Biosystems), custom primers (Invitrogen), and dual-labeled TaqMan probes (Biosearch Technologies, Novato, CA) at final concentrations listed in Table 1 for each assay, 5 μL of prepared sample, and DEPC-treated water for the remaining volume. Real-time RT-PCR amplification for three of the assays (HuNoV GI and GII and EV) was performed under the following conditions: reverse transcription for 30 minutes at 50°C, then denaturation for 15 minutes at 95°C followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 60 seconds. Real-time RT-PCR amplification for HEV RNA was the same as for the above three assays for HuNoV and HuEV, except the annealing and extension were performed separately at 55°C for 30 seconds and 72°C for 30 seconds, respectively.
For viral DNA amplification, each 25-μL reaction contained 10 μL of 2× master mix (QuantiTect Probe PCR Kit [Qiagen]), custom primers (Invitrogen), and dual-labeled TaqMan probes (Biosearch Technologies) at concentrations listed in Table 1 for each assay, 5 μL of prepared sample, and DEPC-treated water for the remaining volume. Real-time PCR amplification for HuAdV was performed under the following conditions: denaturation for 15 minutes at 95°C followed by 40 cycles of denaturation at 94°C for 15 seconds and annealing/extension at 60°C for 60 seconds. Real-time PCR amplification for HuPyV was performed under the following conditions: denaturation for 15 minutes at 95°C followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 55°C for 15 seconds, and extension at 60°C for 60 seconds. Dilutions of sample NA extracts were prepared in DEPC-treated water. All PCR analyses were performed using a positive control for each target and DEPC-treated water as the negative control with each thermocycler run to ensure reagent and cycling efficiency.
An internal standard for the identification of inhibition in real time PCR and RT-PCR assays was prepared using Hepatitis G virus (HGV) Armored RNA (Asuragen, Austin, TX). RNA was extracted from 140 μL HGV Armored RNA using QIAamp Viral RNA Mini Spin kit (Qiagen) following the manufacturer's spin protocol. HGV RNA was eluted from the Qiagen spin column by performing a double elution using 2 × 40 μL DEPC-treated water supplemented with 0.01% 500 U/μL RNAse Inhibitor (Applied Biosystems). The extracted RNA was then amplified by real-time RT-PCR using an ABI Prism 7300 Sequence Detection System. Real-time RT-PCR conditions were the same as HuNoV and EV amplification. Primers and probe for the HGV assay are shown in Table 1.33,34 Each batch of samples assayed for inhibition included a negative control of HGV master mix containing no HGV RNA and at least three positive control (PC) reactions containing only HGV RNA and no sample. For controls, 5 μL DEPC-treated water was added to bring the reaction volume to 25 μL.
Back-volume calculations and statistical analysis.
For indicator bacteria, the percent recovery efficiency (%RE) was calculated as follows. Correlation analyses were performed using the Pearson product–moment correlation.
Strong correlations were defined as a correlation with a Pearson's r value of ≥ 0.6 or ≤ −0.6. Back-volume values were calculated for molecular data to estimate the volume of pre-concentrated sample analyzed during a given assay.
Results
Water quality.
A total of 19 100-L GW (N = 4), SW (N = 9), and DW (N = 6) samples were collected. Water quality parameters were obtained for these samples at the time of collection or before UF at the field laboratory. Table 2 lists the maximum contaminant levels (MCL) and guidelines for each water quality parameter as proposed by the United States Environmental Protection Agency (USEPA) and WHO, respectively.4,35 Table 3 displays average values with lower and upper points for each parameter by source water type. Nitrate and turbidity are the only two parameters set using a health-based standard. The remaining parameters can have cosmetic (e.g., skin or tooth discoloration) or aesthetic (e.g., taste, odor, or color) issues associated with them if present at an elevated level.35 Figures 2 and 3 display turbidity and nitrate levels for each water source type, respectively, as well as the corresponding USEPA MCL or WHO guideline. Overall, median turbidity levels for each water source type exceeded the USEPA MCL of ≤ 0.3 NTU (Nephelometric Turbidity Unit). One GW, one DW (Accra City drinking water), and five of nine SW samples were above the WHO guideline of 5 NTU for acceptable appearance. Nitrates were detected in all water sources with highest levels (14–224 mg/L-N) present in GW sources. Finished DW samples were all below the USEPA MCL and WHO guidelines; however, two DW samples for water kiosks had elevated (5–9.9 mg/L-N) nitrate levels.
Table 2.
| Water parameter | US EPA (MCL) | WHO guideline | Health-based standard | |
|---|---|---|---|---|
| US EPA | WHO | |||
| Turbidity (NTU) | ≤ 0.3* | < 0.1† | Yes | No |
| Nitrate (mg/L) | 10 | 50‡ | Yes | Yes |
| pH | 6.5–8.5 | 6.5–8.0 | No | No |
| TDS (mg/L) | 500 | 600–1,000§ | No | No |
| DO (mg/L) | – | – | – | – |
| Ammonium (mg/L) | – | 1.5¶ | No | No |
| Chloride (mg/L) | 250 | 250 | No | No |
US EPA = US Environmental Protection Agency; WHO = World Health Organization; MCL = maximum contaminant level; TDS = total dissolved solids; DO = dissolved oxygen.
Ninety-five percent of samples in a 1-month period with no one sample exceeding 1 NTU (Nephelometric Turbidity Unit).
Median recommended value; however, 5 NTU is acceptable appearance.
Set as 50 mg/L as nitrate, which is equivalent to 10 mg/L nitrate as nitrogen.
Less than 600 mg/L is best, and above 1,000 mg/L has objectionable taste.
Based on taste threshold.
Table 3.
Water quality data from 100 L GW, SW, and DW samples
| Sample type | n | Water quality parameters average values (lower to upper) | ||||||
|---|---|---|---|---|---|---|---|---|
| pH | TDS (mg/L) | Turbidity (NTU) | DO (mg/L) | Chloride (mg/L) | Nitrate (mg/L N) | Ammonium (mg/L N) | ||
| GW | 4 | 6.11 (5.55–6.40) | 370 (335–409) | 3.45 (< 0.1–11.3) | 7.94 (6.92–10.05) | 78.0 (51.4–106) | 112 (14.1–224) | 0.376 (0.299–0.465) |
| SW | 9 | 6.84 (5.76–7.22) | 193 (42–420) | 8.04 (1.4–18.5) | 6.07 (5.61–8.14) | 33.5 (1.16–74.8) | 26.8 (2.96–179) | 0.212 (0.091–0.347) |
| DW | 6 | 6.92 (6.19–7.31) | 226 (58–421) | 1.78 (< 0.1–2.7) | 6.99 (5.09–8.99) | 36.5 (0.99–77.2) | 3.54 (1.50–7.70) | 0.262 (0.181–0.419) |
GW = groundwater; SW = surface water; DW = finished drinking water; TDS = total dissolved solids; DO = dissolved oxygen.
Figure 2.
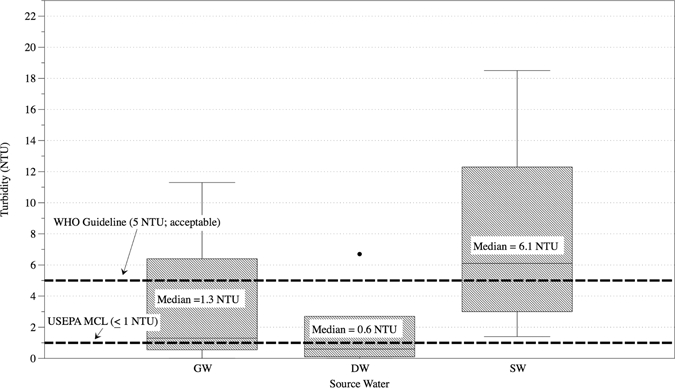
Measured turbidity levels by source water type. GW = groundwater; DW = treated drinking water; SW = surface water; MCL = maximum contaminant level.
Figure 3.
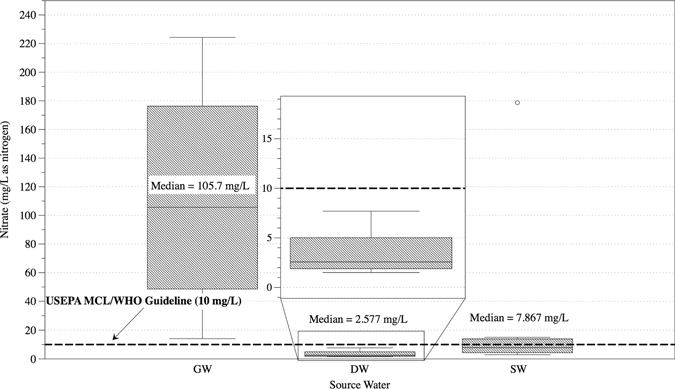
Measured nitrate levels by source water type. GW = groundwater; DW = treated drinking water; SW = surface water; MCL = maximum contaminant level.
Detection of bacterial indicators.
Total coliforms were detected in 16 of 19 samples, including three GW, four DW, and all SW samples. E. coli was detected in 13 of 19 samples, including three GW samples. None of the village water kiosk DW samples were positive for E. coli. However, Accra municipal drinking water was positive for E. coli. The calculation of absolute recovery efficiencies for total coliforms and E. coli after UF was often not possible because of values of too numerous to count (TNTC) or no positive detection before UF. Recovery efficiency for E. coli in GW (N = 3) and SW (N = 9) ranged from 34% to 67% and < 1% to > 120%, respectively. Two samples, one GW and one DW, negative for total coliforms before UF tested positive for total coliforms after UF. The average UF concentrate volumes were 338, 374, and 231 mL for GW, SW, and DW, respectively. The reduction of total coliforms and E. coli after treatment at village water kiosks was also analyzed. Figure 4 displays the reduction of both total coliforms and E. coli in the finished drinking water from the water kiosks by village. As indicated, E. coli was not detected in any water kiosk DW samples nor was E. coli detected in any water kiosk DW UF concentrate.
Figure 4.
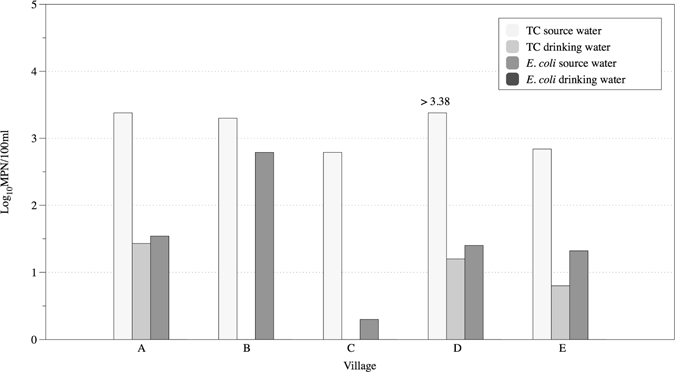
Reduction of total coliforms and E. coli in water sources after treatment at water kiosks. TC = total coliform.
Molecular analysis.
Total viral NA extracted from the secondary concentrates and pellets of each 100-L UF concentrate was analyzed for HuAdV (Type A–F), EV, HuPyV (JC virus and BK virus), HuNoV (GI and GII), and HEV (GI–GIV); 5-μL GW, SW, DW, and SW pellet viral NA extracts were analyzed undiluted and in 10- or 100-fold dilutions. Each assay included a negative control with DEPC-treated water as well as a positive control for each target NA. Table 4 displays results of human enteric virus analysis for GW, DW, and SW NA extracts. SW water results are reported incorporating the pellet and supernatant NA extracts as one sample therefore, if target NA was detected in the pellet or supernatant NA extracts, the sample was considered positive. Human enteric viruses including HuNoV GI and GII, HuAdV, and HuPyV were detected in five different samples, including one GW, three SW, and one DW sample (Table 4). Of these samples, four of five tested positive for E. coli before UF (Figure 5). The one DW sample positive for two human enteric viruses did not test positive for E. coli before or after UF; however, total coliforms were present in both instances.
Table 4.
Detection of human enteric viruses in total viral NA extracts by real-time PCR and RT-PCR
| Source water | n | Number of positive samples for virus target by real-time PCR or RT-PCR | |||||
|---|---|---|---|---|---|---|---|
| EV | HuNoV | HuAdV (Type A–F) | HuPyV (JC or BK) | HEV | |||
| GI | GII | ||||||
| GW | 4 | ND | 1 | ND | ND | ND | ND |
| DW* | 6 | ND | ND | 1 | ND | 1 | ND |
| SW | 9 | ND | ND | 1 | 2 | ND | ND |
ND = not detected; GW = groundwater; DW = treated drinking water; SW = surface water; EV = human enterovirus; HuNoV = human norovirus; HuAdV = human adenovirus; HuPyV = human polyomavirus; HEV = hepatitis E virus.
One water kiosk DW sample tested positive for both HuNoV GII and HuPyV.
Figure 5.
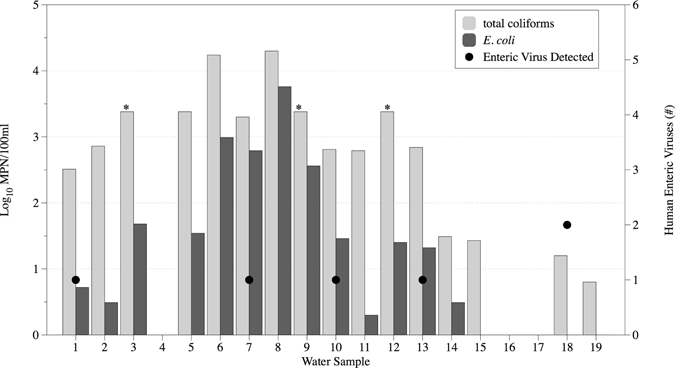
Bacterial indicator and human enteric virus levels in 100-L groundwater (samples 1–4), surface water (samples 5–3) and drinking water (samples 14–19). *Total coliform log10MPN/100 mL is greater than 3.38, but samples were too numerous to count (> 2,419.6 MPN/100 mL).
Evaluation of inhibition.
Inhibition was evaluated in viral NA extracts from GW, SW, DW, and SW pellets using a HGV RNA internal standard. Five-microliter viral NA extracts were analyzed undiluted and in 10- or 100-fold dilutions to determine the level of inhibition. Sample inhibition occurred when the sample HGV RNA cycle threshold (Ct) value deviated from the average Ct value of the PC HGV RNA by one Ct value. Inhibition was detected in 11 of 19 samples (Table 5). Analysis for additional target viral NA was subsequently determined based on the dilutions where inhibition did not occur. For example, if inhibition was detected in the undiluted but not at 10-fold dilutions, then target NA was analyzed in 10- and 100-fold but not undiluted samples.
Table 5.
Inhibition analysis on 100-L surface water, groundwater, and drinking water UF concentrate total NA extracts from secondary concentration method
| Source water | n | Number of samples inhibited during real-time RT-PCR* |
|---|---|---|
| HGV | ||
| GW | 4 | 1 |
| DW | 6 | 3 |
| SW | 9 | 7 |
GW = groundwater; DW = treated drinking water; SW = surface water and pellet; HGV = Hepatitis G virus internal standard.
Five microliters of each nucleic acid extract were analyzed at undiluted, 10-fold, and/or 100-fold for inhibition; results indicate inhibition in at least one dilution.
Back-volume calculations.
To determine the volume of initial sample analyzed, back calculations from the total viral NA extracts were completed for each GW, SW, and DW sample. Table 6 shows the average volume per sample processing step as it relates to the initial sample volume. GW, SW, and DW samples were grouped separately for clarity. The average sample volumes analyzed in a 5-μL real-time PCR or RT-PCR reactions were 699, 629, and 1,323 mL for GW, SW, and DW, respectively. These volumes decreased by 10- or 100-fold as the sample was diluted to overcome the effects of inhibition.
Table 6.
Back-volume calculations to determine average final sample volume in UF concentrates, secondary concentrates, and total viral NA extracts
| Sample type | n | Total sample volume (L) | Average concentrate in mL (range) | Total sample (mL)/average concentration (mL) | Average total secondary concentrate (mL)* | Average total sample (mL)/secondary concentrate (L) | Average total viral NA extract (μL)† | Average total sample in 5 μL NA extract (mL) |
|---|---|---|---|---|---|---|---|---|
| GW | 4 | 100 | 338 (218–495) | 328 | 0.42 | 55 | 93.6 | 699 |
| DW | 6 | 100 | 229 (86–434) | 632 | 2.67 | 17 | 92.2 | 1,323 |
| SW | 9 | 100 | 232 (143–419) | 493 | 1.05 | 33 | 95.0 | 629 |
NA = nucleic acid; GW = groundwater; DW = treated drinking water; SW = surface water.
Total secondary concentrate from 70 mL UF concentrate.
Total viral NA extract from 200-μL secondary concentrate except in two GW and one DW secondary concentrates, for which 150-μL volumes were used.
Discussion
Access to improved drinking water sources is currently unavailable to an estimated 13% of the world's population, and access to microbiologically safe drinking water sources is almost certainly unavailable to an even greater portion of the population.2 The world's population residing in developing countries and more specifically, those living in rural areas of these countries often use multiple types of water sources for drinking and other household purposes (e.g., cleaning, bathing, and cooking). Some of these sources may be considered improved water sources; however, steps are rarely taken to conduct a representative evaluation of their microbial water quality. In the instances that improved water sources are evaluated, fecal indicator bacteria are primarily relied on to provide information about the presence of pathogenic bacteria, enteric viruses, and protozoa. The primary objective of the present study was to provide a broader assessment of the microbial quality of drinking water sources available to rural communities in southern Ghana. To achieve this, a combined tangential flow UF and real-time PCR methodology was applied for the assessment of human enteric viruses in 100-L GW, DW, and SW samples.
During the collection of GW, DW, and SW samples, water quality parameters were taken at each site. Currently, health-based USEPA MCLs and WHO guidelines for physical parameters exist only for nitrate and turbidity levels in water sources intended for human consumption. As expected, treated drinking water samples from water kiosks had the lowest turbidity levels. This is primarily because of the use of a coagulant before treatment as well as a series of filtration techniques during treatment. Even with this treatment, three of five water kiosk samples were above the USEPA MCL of ≤ 0.3 NTU. Turbidity is primarily regulated as an indicator of filtration effectiveness—higher turbidity levels could correspond to presence of pathogenic microorganisms.35 The water kiosk sample with the highest turbidity (2.7 NTU) had no E. coli present before or after UF; however, total coliforms were present, and two human enteric viruses (HuNoV and HuPyV) were detected. However, this relationship between presence of pathogens and elevated turbidity does not always hold true. Nitrate levels were exceeded in seven (four GW and three SW) samples. An additional six samples had elevated (5–9.9 mg/L-N) levels of nitrate, including two water kiosk samples. The presence of nitrates in drinking water sources—GW and SW sources are included in this category for Ghana—is indicative of agricultural contamination (i.e., surface run-off of inorganic nitrogenous fertilizers) and human and animal fecal pollution (i.e., mismanagement of wastewater and inappropriate placement of pit latrines).4,36
The analysis of bacterial indicators in GW, DW, and SW samples before and after UF provided general information on microbial water quality at each source and enabled the %RE for bacteria to be assessed. As indicated, RE of total coliforms and E. coli could not be assessed for every sample. However, RE was calculated for all SW samples with a range of less than 1% to greater than 120%. Reasons for variability in %RE in the present study are not immediately apparent, because the recovery efficiency did not have a strong correlation with levels of turbidity or any other water quality parameter (data not shown). However, poor recovery of bacterial indicators does not automatically imply low RE for enteric viruses as well.
Molecular methods applied in this study focused on the detection of both RNA and DNA human enteric viruses. The human enteric viruses that were targeted in the present study (HuNoV, EV, HuAdV, and HEV) were primarily selected based on their widespread association with acute gastrointestinal illnesses as recognized by both the USEPA and WHO.4,37 Of these enteric viruses, HEV infections have predominantly been an issue in developing countries because of fecal contamination of drinking water supplies and inadequate sanitation.38 Of the four HEV genotypes, genotype 2 has been reported as endemic in most regions of Africa and primarily affects young adults.39 An additional target, HuPyV, was also included, based on recent research suggesting that HuPyV could be used as a reliable viral indicator of human fecal contamination.28,40–44 In this study, each type of water source was positive for a human enteric virus. The only human enteric viruses not detected in UF concentrates were EV and HEV. One GW source was positive for detection of an enteric virus. The presence of viruses in GW has been associated with unsanitary wellhead conditions, local sources of fecal contamination in the immediate area around the well (e.g., pit latrines), improper disinfection after construction and repairs, substandard well construction, and groundwater aquifers under the influence of surface water (i.e., alluvial and sand gravel aquifers).45,46
Enteric viruses were also detected in a treated DW source from a community-based water kiosk. The water kiosks operating in the study area of southern Ghana use a multi-barrier system—similar to conventional drinking water treatment plants—for the treatment of source water. These steps include pretreatment for removal of suspended solids, rapid sand filtration, activated carbon filtration, successive filtration through 5- and 1-μm pore size microfilter membranes, and ultraviolet disinfection. The presence of viruses in the finished DW is potentially the result of multiple failures during the treatment process, including ineffective pretreatment (i.e., coagulation) and insufficient disinfection by UV. Because enteric viruses are small (0.03–0.1 μm), the filtration selected in the treatment process would not normally be very effective in viral removal due to the larger nominal filter pore sizes (i.e., 1–5 μm). Three separate SW sources were also positive for human enteric viruses, two of which were source waters for community water kiosks. The lack of widespread detection of enteric viruses in SW sources during this evaluation could be the result of low viral recovery, low viral concentrations in source water, or sample inhibition during real-time PCR and RT-PCR.
This study reports detection of human enteric viral RNA and DNA in GW, SW, and treated DW; however, the actual public health implications are unclear. Unlike cell culture systems, real-time PCR and RT-PCR do not detect infectious virion particles, and insufficient evidence exists regarding the stability of viral NA when inactivation occurs in the environment because of various stressors including ultraviolet light and shifts in temperature and pH levels. However, the detection of viral NA should not be considered detection of non-infectious particles, because, although infectivity cannot be determined, the potential for that microorganism to be infectious before NA extraction cannot be excluded.47,48
In conjunction with molecular analysis of human enteric viruses, a simple method for the identification of sample inhibition during real-time PCR and RT-PCR assays was also applied. This method uses a commercially available HGV RNA internal standard. Despite using a sample processing method that was optimized for increased elimination of sample inhibitors, one of four, three of six, and seven of nine GW, DW, and SW viral NA extracts, respectively, were inhibited. The use of an RNA or DNA internal standard during PCR and RT-PCR has been a common method reported for evaluating sample inhibition in complex environmental and clinical matrices.32,49–51 However, this step is often missing during evaluation of environmental water samples. Identification of inhibition is imperative when reporting results that could potentially impact public health, especially if false negatives are reported. If inhibition analysis is not included, the estimated risk of exposure to a particular pathogen in drinking water could be an underestimate, because it may likely be based on a decreased value or result caused by sample inhibitors.
Back-volume calculations for each sample were also provided in this study. These calculations were done to determine the estimated original sample volume that is being analyzed during molecular analyses. Not unexpectedly (because of greater SW turbidity), two times the volume of original sample was analyzed for DW compared with SW. Interestingly, a similar difference was seen between DW and GW as well. The ability to relate a Ct value output to a certain volume of water is important when characterizing the level of human enteric viruses potentially present in the original 100-L samples. The combined tangential flow UF and real-time PCR methodology applied in this study allows for a more representative assessment of microbial water quality. In addition, the ability to analyze 100 L of water within a 300-mL concentrate provides a more complete picture of true water quality than does a smaller volume grab sample. This distinction is important to understand with respect to applying microbial risk assessment approaches for determination of health-based standards for individual microorganisms. For example, a risk estimate based on the presence of a given microorganism in 100 mL of water would likely provide less protection of public health than an estimate based on a 100-L composite sample, because large volumes are frequently required for the direct detection of pathogens because of their low-level concentration in ambient waters.
Overall, this study has shown the application of a combined tangential flow UF and real-time PCR methodology as a technique to broaden the assessment of microbial quality of groundwater, treated drinking water, and surface water sources in rural communities of southern Ghana. To date, these sampling and detection techniques have not been applied for the microbial assessment of large-volume water samples in developing countries.
ACKNOWLEDGMENTS
The authors thank Dr. Suzanne Emerson for providing Hepatitis E virus positive control stool specimens. We also thank the Ghana field team from Research International and Safe Water Network for facilitating access to the water kiosks.
Footnotes
Financial support: This study was supported by a USEPA Science to Achieve Results (STAR) Grant (R833002), the Osprey Foundation of Maryland, The Johns Hopkins Global Water Program, and The Johns Hopkins Bloomberg School of Public Health's Center for a Livable Future. K.E.G. is a Center for a Livable Future Predoctoral Fellow. The views expressed herein have not been subjected to USEPA review and therefore, do not necessarily reflect the views of the agency; no official endorsement should be inferred.
Authors' addresses: Kristen E. Gibson, Division of Agriculture, Department of Food Science, University of Arkansas and Center for Food Safety, Fayetteville, AR, E-mail: keg005@uark.edu. Melissa C. Opryszko, James Schissler, Yayi Guo, and Kellogg J. Schwab, Department of Environmental Health Sciences, Division of Environmental Health Engineering, Johns Hopkins Bloomberg School of Public Health and the Johns Hopkins Center for Water and Health, Baltimore, MD, E-mails: mopryszk@jhsph.edu, jschissl@jhsph.edu, yguo@jhsph.edu, and kschwab@jhsph.edu.
References
- 1.Mara DD. Water, sanitation and hygiene for the health of developing nations. Public Health. 2003;117:452–456. doi: 10.1016/S0033-3506(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization and UNICEF Progress on Sanitation and Drinking Water: 2010 Update. 2010. http://www.who.int/water_sanitation_health/publications/9789241563956/en/index.html Available at. Accessed September 7, 2010.
- 3.World Health Organization and UNICEF Meeting the MDG Drinking Water and Sanitation Target: The Urban and Rural Challenges of the Decade. 2004. http://www.who.int/water_sanitation_health/monitoring/jmp2006/en/index.html Available at. Accessed June 11, 2010.
- 4.World Health Organization . Guidelines for Drinking-Water Quality. Geneva, Switzerland: World Health Organization; 2008. pp. 1–668. [Google Scholar]
- 5.Ashbolt NJ. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology. 2004;198:229–238. doi: 10.1016/j.tox.2004.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds K, Mena K, Gerba C. Risk of waterborne illness via drinking water in the United States. Rev Environ Contam Toxicol. 2008;192:117–158. doi: 10.1007/978-0-387-71724-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold BF, Colford JM. Treating water with chlorine at point-of-use to improve water quality and reduce child diarrhea in developing countries: a systematic review and meta-analysis. Am J Trop Med Hyg. 2007;76:354–364. [PubMed] [Google Scholar]
- 8.Boschi-Pinto C, Velebit L, Shibuya K. Estimating child mortality due to diarrhoea in developing countries. Bull World Health Organ. 2008;86:710–717. doi: 10.2471/BLT.07.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UNICEF The State of the World's Children 2008: Child Survival. 2007. http://www.unicef.org/sowc08/ Available at. Accessed June 11, 2010.
- 10.Pronk W, Zurbrugg C, Swartz C, Pronk W. Decentralized systems for potable water and the potential of membrane technology. Water Res. 2008;43:245–265. doi: 10.1016/j.watres.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Lee EJ, Schwab KJ. Deficiencies in drinking water distribution systems in developing countries. J Water Health. 2005;3:109–127. [PubMed] [Google Scholar]
- 12.Sobsey MD. Managing Water in the Home: Accelerated Health Gains from Improved Water Supply. 2002. http://www.who.int/water_sanitation_health/dwq/wsh0207/en/ Available at. Accessed June 11, 2010.
- 13.Opryszko MC, Huang H, Soderlund K, Schwab KJ. Data gaps in evidence-based research on small water enterprises in developing countries. J Water Health. 2009;7:609–622. doi: 10.2166/wh.2009.213. [DOI] [PubMed] [Google Scholar]
- 14.United Nations Development Programme . Human Development Report 2009. Overcoming Barriers: Human Mobility and Development. 2009. http://hdr.undp.org/en/reports/global/hdr2009/ Available at. Accessed June 4, 2010. [Google Scholar]
- 15.World Health Organization Mortality Country Fact Sheet 2006: Ghana. 2006. http://www.who.int/countries/gha/gha/en/ Available at. Accessed May 31, 2010.
- 16.Medema GJ, Payment P, Dufour A, Robertson W, Waite M, Hunter P, Kirby R, Anderson Y. Assessing Microbial Safety in Drinking Water: Improving Approaches and Methods. London, United Kingdom: IWA Publishing; 2003. pp. 11–46. (Safe drinking water: an ongoing challenge). World Health Organization, Organisation for Economic Cooperation and Development. [Google Scholar]
- 17.Payment P, Waite M, Dufour A. Assessing Microbial Safety of Drinking Water: Improving Approaches and Methods. London, United Kingdom: IWA Publishing; 2003. pp. 47–78. (Introducing parameters for the assessment of drinking water quality). World Health Organization, Organisation for Economic Cooperation and Development. [Google Scholar]
- 18.Gerba C, Goyal S, LaBelle R, Cech I, Bodgan G. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am J Public Health. 1979;69:1116–1119. doi: 10.2105/ajph.69.11.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol. 2005;71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose JB, Gerba CP, Jakubowski W. Survey of potable water-supplies for Cryptospordium and Giardia. Environ Sci Technol. 1991;25:1393–1400. [Google Scholar]
- 21.Semenza JC, Roberts L, Henderson A, Bogan J, Rubin CH. Water distribution system and diarrheal disease transmission: a case study in Uzbekistan. Am J Trop Med Hyg. 1998;59:941–946. doi: 10.4269/ajtmh.1998.59.941. [DOI] [PubMed] [Google Scholar]
- 22.Crump JA, Okoth GO, Slutsker L, Ogaja DO, Keswick BH, Luby SP. Effect of point- of-use disinfection, flocculation and combined flocculation-disinfection on drinking water quality in western Kenya. J Appl Microbiol. 2004;97:225–231. doi: 10.1111/j.1365-2672.2004.02309.x. [DOI] [PubMed] [Google Scholar]
- 23.Crump JA, Otieno PO, Slutsker L, Keswick BH, Rosen DH, Hoekstra RM, Vulule JM, Luby SP. Household based treatment of drinking water with flocculant-disinfectant for preventing diarrhoea in areas with turbid source water in rural western Kenya: cluster randomized controlled trial. BMJ. 2005 doi: 10.1136/bmj.38512.618681.E0. doi:10.1136/bmj.38512.618681.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobsey MD, Handzel T, Venczel L. Chlorination and safe storage of household drinking water in developing countries to reduce waterborne disease. Water Sci Technol. 2003;47:221–228. [PubMed] [Google Scholar]
- 25.Colindres RE, Jain S, Bowen A, Domond P, Mintz E. After the flood: an evaluation of in-home drinking water treatment with combined flocculent-disinfectant following tropical storm jeanne—Gonaives, Haiti, 2004. J Water Health. 2007;5:367–374. doi: 10.2166/wh.2007.032. [DOI] [PubMed] [Google Scholar]
- 26.Cobbina SJ, Anyidoho LY, Nyame F, Hodgson IO. Water quality status of dugouts from five districts in northern Ghana: implications for sustainable water resources management in a water stressed tropical savannah environment. Environ Monit Assess. 2009 doi: 10.1007/s10661-009-1059-6. doi:10.1007/s10661-009-1059-6. [DOI] [PubMed] [Google Scholar]
- 27.Polaczyk A, Narayanan J, Cromeans T, Hahn D, Roberts J, Amburgey J, Hill V. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J Microbiol Methods. 2008;73:92–99. doi: 10.1016/j.mimet.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 28.McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol. 2009;75:3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jothikumar N, Cromeans T, Hill V, Lu X, Sobsey M, Erdman D. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl Environ Microbiol. 2005;71:3131–3136. doi: 10.1128/AEM.71.6.3131-3136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods. 2006;131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory JB, Litaker RW, Noble RT. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal positive control for detection of enteroviruses in environmental samples. Appl Environ Microbiol. 2006;72:3960–3967. doi: 10.1128/AEM.02291-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlueter V, Schmolke S, Stark K, Hess G, Ofenloch-Haehnle B, Engel AM. Reverse transcription-PCR detection of hepatitis G virus. J Clin Microbiol. 1996;34:2660–2664. doi: 10.1128/jcm.34.11.2660-2664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambertini E, Spencer S, Bertz P, Loge F, Kieke B, Borchardt M. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl Environ Microbiol. 2008;74:2990–2996. doi: 10.1128/AEM.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.United States Environmental Protection Agency National Primary Drinking Water Regulations. 2009. http://www.epa.gov/safewater/contaminants/index.html Available at. Accessed June 13, 2010.
- 36.Schmoll O, Howard G, Chilton J, Chorus I. Protecting Groundwater for Health: Managing the Quality of Drinking-Water Sources. London, United Kingdom: World Health Organization IWA Publishing; 2006. pp. 537–650. [Google Scholar]
- 37.United States Environmental Protection Agency Fact Sheet: Final Third Drinking Water Contaminant Candidate List (CCL 3) 2009. www.epa.gov/safewater/ccl/pdfs/ccl3_docs/fs_cc3_final.pdf Available at. Accessed May 24, 2010.
- 38.Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Emerson S, Purcell R. Hepatitis E virus. Rev Med Virol. 2003;13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 40.Bofill-Mas S, Pina S, Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol. 2000;66:238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hundesa A, Maluquer de Motes C, Bofill-Mas S, Albinana-Gimenez N, Girones R. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl Environ Microbiol. 2006;72:7886–7893. doi: 10.1128/AEM.01090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bofill-Mas S, Albinana-Gimenez N, Clemente-Casares P, Hundesa A, Rodriguez-Manzano J, Allard A, Calvo M, Girones R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl Environ Microbiol. 2006;72:7884–7896. doi: 10.1128/AEM.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albinana-Gimenez N, Clemente-Casares P, Bofill-Mas S, Hundesa A, Ribas F, Girones R. Distribution of human polyomaviruses, adenoviruses, and hepatitis E virus in the environment and in a drinking-water treatment plant. Environ Sci Technol. 2006;40:7416–7422. doi: 10.1021/es060343i. [DOI] [PubMed] [Google Scholar]
- 44.McQuaig SM, Scott TM, Harwood VJ, Farrah SR, Lukasik JO. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl Environ Microbiol. 2006;72:7567–7574. doi: 10.1128/AEM.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borchardt MA, Haas NL, Hunt RJ. Vulnerability of drinking-water wells in La Crosse, Wisconsin, to enteric-virus contamination from surface water contributions. Appl Environ Microbiol. 2004;70:5937–5946. doi: 10.1128/AEM.70.10.5937-5946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbaszadegan M, LeChevallier MW, Gerba CP. Occurrence of viruses in US groundwaters. J Am Water Works Assoc. 2003;95:107–120. [Google Scholar]
- 47.Duizer E, Bijkerk P, Rockx B, De Groot A, Twisk F, Koopmans M. Inactivation of caliciviruses. Appl Environ Microbiol. 2004;70:4538–4543. doi: 10.1128/AEM.70.8.4538-4543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Limsawat S, Ohgaki S. Fate of liberated viral RNA in wastewater determined by PCR. Appl Environ Microbiol. 1997;63:2932–2933. doi: 10.1128/aem.63.7.2932-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwab KJ, Estes MK, Neill FH, Atmar RL. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of norwalk virus from stool samples. J Clin Microbiol. 1997;35:511–514. doi: 10.1128/jcm.35.2.511-514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson J, Hymas W, Hillyard D. The use of armored RNA as a multi-purpose internal control for RT-PCR. J Virol Methods. 2008;150:73–76. doi: 10.1016/j.jviromet.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dreier J, Störmer M, Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J Clin Microbiol. 2005;43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


