Abstract
Pluripotent human embryonic stem cells (hESCs) provide an unprecedented opportunity for the study of human tissue development, and the development of cell-based therapies for human disease. To realize these potential advances, however, methods for monitoring expression of intracellular proteins in live hESCs without altering cellular properties are needed. Molecular beacons are single-stranded oligonucleotides that have been employed to assay gene expression. To test their potential for high-throughput isolation of hESCs, we developed a dual fluorescence resonance energy transfer (FRET) molecular beacon system using fluorescence-activated cell sorting (FACS) with Oct4 as a target. We demonstrate that Oct4 can be detected by FRET using confocal microscopy, that this can be applied in a high-throughput manner to the identification and isolation of Oct4-expressing hESCs by FACS, that FRET-positive hESCs demonstrate pluripotency in culture and in vivo, and that hESCs transfected with molecular beacons demonstrate normal growth rates and oligonucleotide extinction over time. These studies demonstrate that FRET-based FACS using molecular beacons provides a useful tool for isolating Oct4-expressing pluripotent hESCs, and may also be adapted to selecting differentiating hESCs at specific developmental time points determined by transcription factor expression without functional or genomic alteration. As such, it provides an important new method for high-throughput isolation of hESC-derived tissue-specific precursors for analytic and therapeutic purposes.
Introduction
Pluripotent human embryonic stem cells (hESCs) have an unlimited capacity for self-renewal and the ability to differentiate in culture and in vivo into tissues derived from all 3 embryonic germ layers. To date, most hESC lines have been characterized by their expression of cell surface antigens [1]. These studies have identified a battery of glycolipids and glycoproteins that are found on a high percentage of undifferentiated hESCs, including the stage-specific antigens, SSEA-3 and SSEA-4, and the keratin sulfate-related antigens, Tra-1–60 and Tra-1–81, among others [2]. These antigens are commonly used to assess the pluripotency of hESCs, for within days upon the induction of differentiation their expression dramatically decreases [3]. It also has been appreciated that low levels of spontaneous differentiation occur within hESC cultures grown under proliferation conditions, and that cells within proliferating colonies can express early markers of specific embryonic germ layers [4]. As such, the presence of these cells may bias the study of mechanisms of pluripotency in proliferating hESC colonies. Nuclear transcription factors such as Oct4 and Nanog have been implicated in pathways regulating pluripotency [5,6]; however, expression of these proteins is more difficult to assess in live cells. Virally transduced reporters have been shown to be specific and efficient for this purpose [7]; however, these have the potential to alter cell behavior, especially when randomly integrated into the cell genome.
Molecular beacons are single-stranded oligonucleotides that have been employed to assay gene expression in vitro, as in real-time polymerase chain reaction (PCR), and in single cells using microscopy [8]. These consist of short sequences capable of forming stem-loop structures bearing a fluorescent reporter group at one end and a fluorescent quencher at the opposite end [8]. In the absence of a target sequence, the oligonucleotide self-anneals, forming a stem that brings the reporter and quencher in close proximity, thereby quenching fluorescence. In the presence of a target sequence, the oligonucleotide anneals to the target, separating the reporter and quencher, thereby allowing fluorescence. To test the potential of this technology for identifying and isolating live pluripotent hESCs in a high-throughput manner, we developed a fluorescence-activated cell sorting (FACS)-based, dual fluorescence resonance energy transfer (FRET) molecular beacon system that utilizes pairs of molecular beacons containing donor and acceptor fluorescent groups. FRET results when the 2 fluorescent groups are brought into proximity by both beacons annealing to a target sequence, thus increasing specificity by requiring recognition by both oligonucleotides. The probes are synthesized using O-methylated nucleotides, which are not recognized by ribonucleases and avoid activating the RNA interference system [9]. FACS allows for excitation of the donor group, detection of emission from the acceptor group, and high-throughput sorting of cells expressing the target nuclear protein based on FRET. Using this approach, we developed a high-throughput method for isolating live hESCs based on expression of intracellular proteins, without altering the functional or genomic characteristics of the cells.
Materials and Methods
Molecular beacon design
RNAfold was used to generate a map of the Oct4 mRNA secondary structure using a minimum free energy algorithm (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi; [10]). On the basis of this map, accessible regions were evaluated using Beacon Designer (Premier Biosoft), and 2 oligoncleotide sequences spanning ribonucleotides 488–541 were chosen based on published parameters [11]. The beacons were synthesized using 2-O-methyl ribonucleotides, purified by high-performance liquid chromatography, and molecular weights confirmed by mass spectrometry (SynGen, Inc.) with the following final sequences: Donor, 5′-6FAM-GCUCUUCUGCUUCAGGAGCUUAGAGC-BHQ1-3′; Acceptor, 5′-BHQ2-ACCCUGCCUGUGUAUAUCCCAGGGU-5ROX-3′ (where underlined, ribonucleotides were added to facilitate annealing of the stem domains).
hESC culture and transfection
All work with hESCs was approved by the UCSF Human Gamete, Embryo and Stem Cell Research Committee. The H9 hESC line (WiCell) was maintained as previously described [12] on irradiated mouse embryonic fibroblast (MEF) feeder cells in a medium comprised of knockout Dulbecco's Modified Eagle's Medium (DMEM) (Invitrogen) supplemented with 20% knockout serum replacer (Invitrogen), 2 mM glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, and 15 ng/mL recombinant human basic fibroblast growth factor (R&D Systems). Cells were passaged 1:2 or 1:3 every 3 days by incubating in the growth medium containing 1 mg/mL collagenase IV (Sigma-Aldrich) and dispersing cells onto fresh feeder cell layers. All cultures were routinely screened for mycoplasma, and normal karyotype was monitored by array comparative genomic hybridization using published methods [13]. Transfection with molecular beacons was accomplished by dissociating hESC colonies as described for passaging, and electroporating 5 μg donor beacon and/or 1 μg acceptor beacon in a volume of 0.5 mL using a 4 mm cuvette with a Bio-Rad Gene Pulser II set at 320 V and 200 μF (time constant ∼5 s). After electroporation, cells were transferred to 1 mL growth medium and allowed to recover for 48 h on feeder cells. Differentiation was accomplished by human embryoid body (hEB) formation in suspension as previously described [12]. Briefly, colonies of hESCs were dissociated into small clusters by exposure to collagenase IV (Sigma-Aldrich), and then allowed to differentiate in a medium comprised of knockout DMEM (Invitrogen) supplemented with 20% defined fetal bovine serum (Hyclone), 2 mM glutamine, 0.1 mM nonessential amino acids, and 0.1 mM β-mercaptoethanol. After 1 week in suspension, hEBs were attached to gelatin-coated 12-well culture plates and allowed to differentiate for an additional 14 days (21 days total). To examine mixed cultures of undifferentiated and partially differentiating hESCs, adherent hESC colonies were transferred to differentiation medium and allowed to differentiate spontaneously for 5 days as previously described [12].
Confocal microscopy
At 48 h after electroporation, hESCs transfected with molecular beacons were scraped from wells in cold phosphate-buffered saline, fixed for 15 min at room temperature in 4% paraformaldehyde, resuspended in cold phosphate-buffered saline, and mounted onto microscope slides with cover slips. Cells were analyzed using a Zeiss Confocal Laser-Scanning Microscope LSM 510 META with NLO. Samples were excited with 488 or 584 nm lasers. 6FAM and 5ROX emissions were detected with LP505 and LP615 filters, respectively. FRET was determined by emission detection at 610 nm using the LP615 filter following excitation with the 488 nm laser.
FACS analysis
hESCs transfected with molecular beacons were scraped into cold phosphate-buffered saline, dissociated in 1 mg/mL collagenase IV for 15 min at 37°C, and analyzed or sorted on a FACSAria (BD Biosciences) using previously described methods [14,15]. Cells were stained with 1 μg/mL propidium iodide to distinguish between live and dead cells. Samples were excited with a 488 nm laser for detection of 6FAM using a 515–545 nm filter, or of FRET using a 600–620 nm filter. For detection of 5ROX, a 560 nm laser and 600–620 nm filter were used. In all experiments, the positive gates were determined by analyzing untransfected cells. Data were analyzed using FlowJo 8 (TreeStar, Inc.) on a Macintosh platform.
Immunocytochemistry and reverse transcription-PCR
hESCs sorted on the basis of FRET were plated on cover slips, fixed with 4% paraformaldehyde, and then stained for Oct4 expression and counterstained with DAPI as previously described [16]. hEBs were plated on cover slips for differentiation, fixed with 4% paraformaldehyde at 21 days after transfer to differentiation medium, and then permeabilized, fixed, and stained for β-III-tubulin, smooth muscle actin, and α-fetoprotein expression as previously described [12]. Primary antibodies used were mouse anti-human Oct4 (Santa Cruz Biotechnology sc-5279), mouse anti-human α-fetoprotein (Sigma A8452), mouse anti-human β-III-tubulin (R&D Systems MAB1195), or mouse anti-human smooth muscle actin (R&D Systems MAB1420). Goat anti-mouse Alexa-Fluor 594 (1:500; Invitrogen A20185) was used as secondary antibody for all studies. Stained cells were analyzed by immunofluorescence microscopy using a Nikon Microphot-FXA fluorescence/phase microscope and QImaging Retiga 2000R digital camera (Diagnostic Instruments) with MetaMorph software (Molecular Devices).
Reverse transcription (RT)-PCR analysis was performed as previously described [17]. Total RNA was isolated from sorted hESCs using the RNeasy Micro Kit (Qiagen) according to the manufacturer's instructions, and 0.2 μg was used as template for RT using Superscript II reverse transcriptase (Invitrogen). Linear amplification was achieved by 25 cycles with PlatinumTaq polymerase (Invitrogen). PCR products were separated on 1% agarose and detected with ethidium bromide. Quantitation was achieved by densitometric analysis of digitized gel images using NIH Image. PCR amplification of the housekeeping gene, GAPDH, was performed as a control. Primer sets used for RT-PCR were as follows: human Oct4, 5′-GACAACAATGAGAACCTTCAGGAGA-3′ and 5′-CTGGCGCCGGTTACAGAACCA-3′; human Nanog, 5′-GACTGAGCTGGTTGCCTCAT-3′ and 5′-TTTCTTCAGGCCCACAAATC-3′; GAPDH, 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′ and 5′-CATGTAGGCCATGAGGTCCACCAC-3′.
Teratoma formation
These experiments were performed with approval from the UCSF Institutional Animal Care and Use Committee. Teratomas were formed and analyzed as previously described [12,18]. Sorted or unsorted hESCs, or MEFs, were mixed with an equal volume of 1 mg/mL Phaseolus vulgaris lectin (PHA-P; Sigma L1668), pelleted, and incubated in the growth medium overnight at 37°C, 5% CO2 in a 0.4 μm MILLICELL (Millipore). Two cell pellets (105 cells per pellet) were grafted under the right kidney capsule of an 8-week-old male CB17 SCID-Beige mouse for each cell type. The left kidney was used as an untransplanted control. Transplanted cells formed teratomas in the recipients and were analyzed 10 weeks after grafting. Left and right kidneys were removed separately, with adherent teratomas removed en bloc with right kidneys, and weighed. Ratios of the right kidney+teratoma:left kidney were calculated. Teratomas were fixed in 10% buffered formalin and embedded in paraffin, and 5 μm sections were stained with hematoxylin and eosin to identify tissue structures using a Zeiss Axiovision microscope equipped with an MBF Bioscience CX9000 camera and StereoVision microbrightfield software.
Cell proliferation and molecular beacon extinction
To determine the rate of cell growth, transfected hESCs were sorted 48 h after transfection based on FRET emission at 610 nm. Approximately 15,000 sorted cells and an equivalent number of untransfected hESCs were placed back into the growth medium on feeder cells, and total hESC numbers were counted every 24 h. Cell cycle length was calculated as 24/[1 + ([N2 − 2N1]/2N1)], where N1 is number of cells counted on a given day and N2 is number of cells counted 24 h later. This assumes that all cells have the same cycle length, that cell cycle progression occurs at a constant rate, and that all cells are actively cycling [19]. To determine molecular beacon loss over time, FRET-based sorted cells were placed back into the growth medium on feeder cells, passaged every 3 days as described above, and re-analyzed by flow cytometry for FRET at 7, 14, and 21 days. The percentage of cells exhibiting FRET, as assessed by emission at 610 nm with excitation at 488 nm, was recorded.
Results
Dual FRET molecular beacons can be used to detect Oct4 expression in single cells
We chose Oct4 (also known as Pou5F1) as the target gene for these studies since its expression is downregulated coincidently with stem cell differentiation, and its functional role in maintaining pluripotency has ascribed to Oct4 its designation as a pluripotency marker [5]. The Oct4 gene encodes a nuclear protein that belongs to a family of transcription factors containing the POU DNA-binding domain, and its expression can be detected in embryonic as well as some multipotent adult stem cells [5].
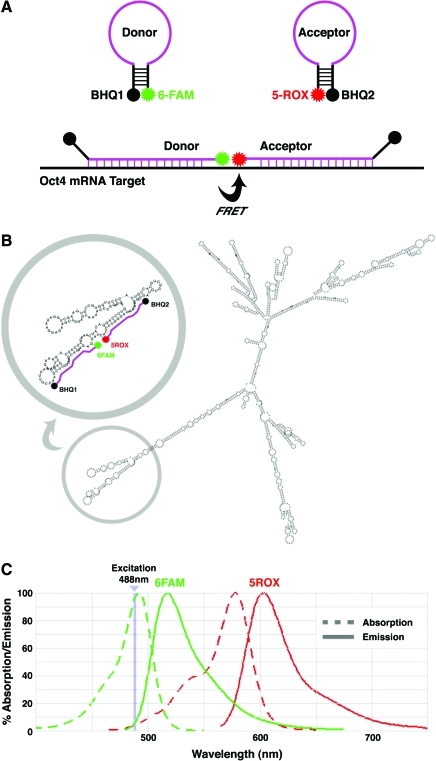
We used 6FAM, which has an absorption maximum at 490 nm and emission maximum at 520 nm, as the donor fluorophore, and 5ROX, with an absorption maximum at 580 nm and emission maximum at 610 nm, as the acceptor fluorophore. BHQ1 was used to quench 6FAM in the donor beacon and BHQ2 to quench 5ROX in the acceptor beacon when in solution as short hairpins (Fig. 1A). The oligonucleotide sequence for each beacon was determined based on computational modeling of the Oct4 mRNA secondary structure using a minimum free energy algorithm [10] (Fig. 1B). When annealed to the target mRNA in vivo, excitation of 6FAM at 488 nm emits in the spectral range of 515–560 nm, overlapping with the absorption range of 5ROX (540–578 nm) (Fig. 1C). Excitation of 5ROX by 6FAM emission would result in 5ROX emission at 600–620 nm (Fig. 1C).
FIG. 1.
Molecular beacon strategy for Oct4 detection. (A) Schematic model for dual fluorescence resonance energy transfer (FRET) molecular beacon system. 6FAM and 5ROX fluorophores were chosen for their spectral overlap (Ex 490 nm/Em 520 nm and Ex 580 nm/Em 610 nm, respectively). BHQ1 and BHQ2 were used to quench 6FAM and 5ROX emissions, respectively, in solution. (B) Prediction of Oct4 mRNA secondary structure using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Gray circle highlights region amenable to molecular beacon design. (C) Absorption/emission spectra for 6FAM and 5ROX demonstrating overlap between 6FAM emission and 5ROX absorption spectra with >30% response at 540–560 nm. Color images available online at www.liebertonline.com/scd.
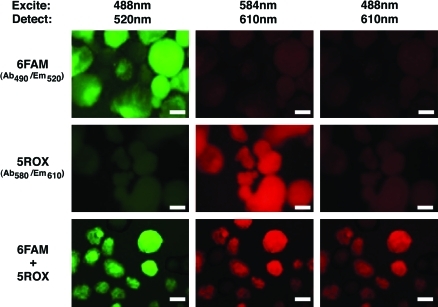
To determine whether FRET would occur in hESCs, either donor or acceptor or both beacons were introduced into proliferating hESCs by electroporation, and then suspension cultures of aggregated cells were analyzed by confocal microscopy (Fig. 2). Emission at 520 nm was detected in cells transfected with the 6FAM-labeled donor beacon subjected to excitation at 488 nm. Similarly, emission at 610 nm was detected in cells transfected with the 5ROX-labeled acceptor beacon subjected to excitation at 584 nm. However, only cells transfected with both donor and acceptor beacons demonstrated emission at 610 nm when subjected to excitation at 488 nm, suggesting that FRET had occurred between the 2 beacons in proliferating, undifferentiated hESCs expressing Oct4.
FIG. 2.
Oct4 expression is detected in human embryonic stem cells (hESCs) by fluorescence resonance energy transfer (FRET) using confocal microscopy. Typical confocal micrographs of aggregated hESCs in suspension transfected with 6FAM (donor)- and 5ROX (acceptor)-labeled, Oct4-specific molecular beacons separately and in combination are shown. The same field is shown across each row, with each panel produced using the indicated excitation wavelength/detection filter combination. Individual beacons fluoresce at appropriate wavelengths (6FAM, upper left; 5ROX, middle middle), with absence of overlapping emissions (6FAM, middle left; 5ROX, upper middle). Only cells transfected with both beacons fluoresce at the acceptor's wavelength (610 nm) when excited within the donor's absorption spectrum (488 nm) (6FAM+5ROX, right lower). Scale bar: 100 μm. Color images available online at www.liebertonline.com/scd.
FACS-based dual FRET can detect and isolate Oct4-expressing hESCs
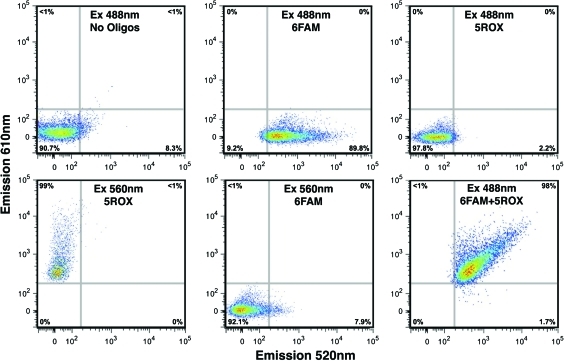
To determine whether the system could be used in a high-throughput, FACS-based method to isolate Oct4-expressing hESCs, we examined fluorescence from each beacon, and assayed for FRET, by flow cytometry (Fig. 3). Emission at 520 nm was again seen in the majority of donor beacon-transfected cells excited at 488 nm, whereas emission at 610 nm was observed in the majority of acceptor beacon-transfected cells excited at 560 nm. Only cells transfected with both donor and acceptor beacons demonstrated emission at both 520 and 610 nm when excited at 488 nm, establishing that FRET occurred between the 2 beacons and was sufficient to allow cell sorting (Fig. 3).
FIG. 3.
Oct4-expressing hESCs can be identified and isolated by FRET-based fluorescence-activated cell sorting (FACS). Typical FACS histograms of proliferating hESCs transfected with 6FAM (donor)- and 5ROX (acceptor)-labeled, Oct4-specific molecular beacons separately and in combination are shown. Flow cytometry detects fluorescence of individual beacons at appropriate wavelengths (6FAM, upper middle; 5ROX, lower left), with absence of overlapping emissions (6FAM, lower middle; 5ROX, upper right). Flow cytometry detects fluorescence at the acceptor's wavelength (610 nm) with excitation of the donor (488 nm) only in cells transfected with both beacons (6FAM+5ROX, lower right). Color images available online at www.liebertonline.com/scd.
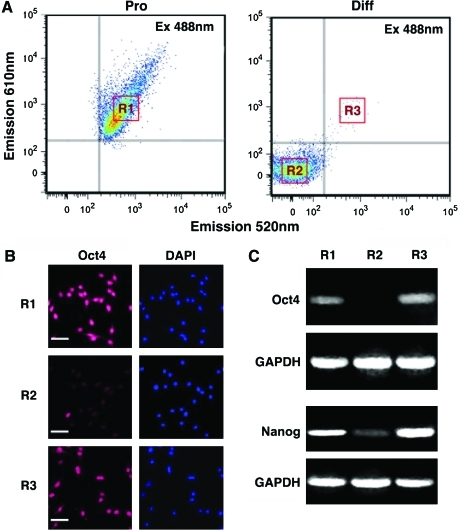
hESCs isolated by high-throughput FACS-based FRET specifically express Oct4
To validate specific expression of Oct4 in sorted cells, we cultured hESC colonies for 7 days under growth or differentiation conditions, and then sorted FRET-positive cells from both proliferating and differentiating cultures, as well as FRET-negative cells from differentiating cultures (Fig. 4A). FRET-positive cells from both proliferating and differentiating cultures demonstrated Oct4 expression by both immunocytochemistry (Fig. 4B) and RT-PCR analysis (Fig. 4C), whereas FRET-negative cells did not (Fig. 4B, C). In addition, RT-PCR showed that FRET-positive cells expressed significantly higher levels of another pluripotency marker, Nanog.
FIG. 4.
hESCs isolated by FRET-based FACS specifically express Oct4. (A) Typical FACS histograms of proliferating (Pro) and differentiating (Diff) hESCs transfected with 6FAM- and 5ROX-labeled, Oct4-specific molecular beacons as in Figure 3 are shown. R1 and R3 indicate FRET-positive proliferating and differentiating hESCs, respectively; R2 indicates FRET-negative differentiating hESCs. (B) Immunocytochemical analysis demonstrated that FACS-isolated FRET-positive hESCs from proliferating (R1) and differentiating (R3) cultures expressed Oct4 (red), whereas FRET-negative hESCs (R2) did not. Nuclei were counterstained with DAPI. Scale bar: 100 μm. (C) Reverse transcription–polymerase chain reaction confirmed that FACS-isolated FRET-positive hESCs (R1, R3) expressed Oct4, as well as Nanog, whereas FRET-negative hESCs (R2) did not. GAPDH was used as a control. Color images available online at www.liebertonline.com/scd.
hESCs isolated by FACS-based FRET detection of Oct4 demonstrate functional pluripotency
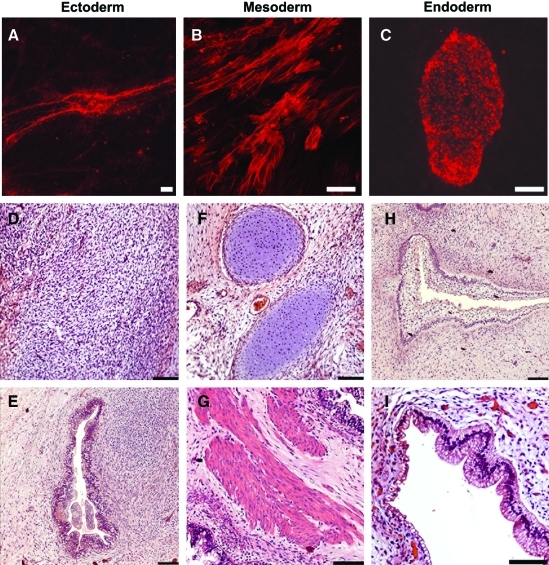
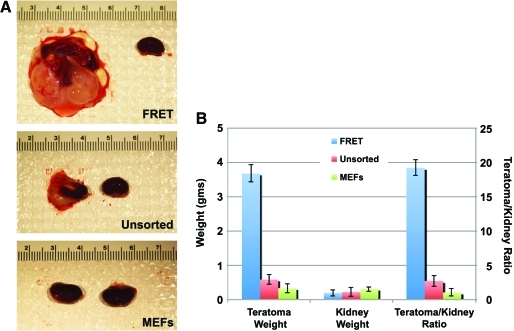
To determine whether hESCs transfected with molecular beacons and sorted on the basis of Oct4 expression retained the defining characteristics of pluripotent stem cells, we differentiated sorted cells in culture and in vivo (Fig. 5). FRET-positive cells isolated from partially differentiated cultures (as shown in Fig. 4A) and subsequently re-cultured were able to generate cells expressing β-III-tubulin (ectoderm), smooth muscle actin (mesoderm), and α-fetoprotein (endoderm) when grown under differentiation conditions (Fig. 5A–C). FRET-positive cells also gave rise to tissues derived from all 3 embryonic germ layers in an in vivo teratoma formation assay (Fig. 5D–I). Interestingly, FRET-positive cells gave rise to significantly larger teratomas than unsorted, partially differentiated hESCs, or MEF controls (Fig. 6), suggesting an enrichment for pluripotent hESCs through FRET-based selection of Oct4-expressing cells.
FIG. 5.
Oct4-expressing hESCs isolated by FRET-based FACS demonstrate pluripotency in culture and in vivo. (A–C) Immunocytochemistry demonstrated that FRET-positive hESCs differentiate in culture into cells arising from all 3 embryonic lineages, including cells expressing βIII-tubulin (A; ectoderm), smooth muscle actin (B; mesoderm), and α-fetoprotein (C; endoderm). Scale bars: 100 μm. Immunohistochemical analysis of teratomas showed that FRET-positive hESCs gave rise to tissues derived from the 3 embryonic germ layers, including ectoderm (D, E), mesoderm (F, G), and endoderm (H, I). (D) Bed of eosinophilic neural precursors. (E) Nascent neural tube structure. (F) Pools of cartilage surrounded by loose mesenchyme. (G) Skeletal muscle surrounded by dense mesenchyme. (H) Primitive glandular structure. (I) Glandular intestinal structure. Scale bars: 100 μm. Color images available online at www.liebertonline.com/scd.
FIG. 6.
FRET-based FACS of Oct4-expressing hESCs achieves enrichment of pluripotency. (A) Typical teratomas produced from 105 FRET-positive hESCs (FRET), unsorted hESCs, or mouse embryonic fibroblasts (MEFs) are shown. For each panel, the ungrafted control kidney is shown to the right of the grafted kidney. (B) The ratio of teratoma+kidney weight:contralateral kidney weight is significantly higher for FRET-positive hESCs than for unsorted hESCs, suggesting enrichment for pluripotent hESCs. As expected, MEFs did not produce teratomas. Weight axis (left) applies to columns for teratoma and kidney weights; teratoma/kidney ratio axis (right) applies to columns for teratoma/kidney ratios. Data shown represent mean ± standard error of the mean (SEM) (n = 2). Color images available online at www.liebertonline.com/scd.
hESCs isolated by FACS-based FRET demonstrate normal growth and shed molecular beacons over time
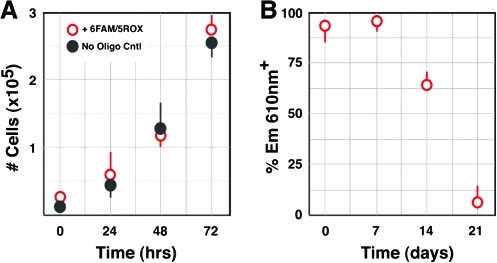
Since the presumed advantages of using molecular beacons include the ability to detect expression of intracellular proteins in living cells in a reversible manner without genomic or functional alteration, we tested the proliferation rate of transfected cells, as well as the extinction of the FRET signal over time (Fig. 7). hESCs transfected with both donor and acceptor beacons were sorted based on emission at 610 nm with excitation at 488 nm as above, placed back into culture under proliferation conditions, and cell numbers were counted over time. Transfected cells grew at rates similar to untransfected cells, with a cell cycle length of ∼15 h (Fig. 7A), similar to that previously reported for the H9 hESC line [20]. We also analyzed shedding of molecular beacons from hESC cultures by evaluating FRET over time in sorted cultures maintained under growth conditions. While the majority of cells initially sorted based on FRET continued to demonstrate FRET at 7 days in culture, the percentage of cells emitting at 610 nm with excitation at 488 nm declined to ∼5% by 21 days (Fig. 7B). This suggested that the molecular beacons were shed over time in proliferating cultures.
FIG. 7.
Normal growth rates and molecular beacon extinction over time in hESCs. (A) Cells numbers were counted every 24 h over a 72 h period. Data shown represent mean ± SEM (n = 4). No difference was detected between growth rates for cells transfected with molecular beacons and demonstrating FRET by FACS compared to untransfected cells, with a calculated cell cycle length of ∼15 h. (B) Cells transfected with both molecular beacons were sorted by FRET-based FACS and placed back into culture under proliferation conditions. At the indicated time intervals, cultures were re-analyzed by flow cytometry for FRET-based evidence of the presence of both beacons. Data shown represent mean ± SEM (n = 3). Molecular beacons persisted, as evidenced by FRET, for at least 7 days in culture, but were shed over time, as indicated by extinction of FRET signal. Color images available online at www.liebertonline.com/scd.
Discussion
Molecular beacons have paved the way for technologies such as real-time PCR and microarray analysis, and have been used to detect gene expression in single cells. Compared to viral reporter systems, molecular beacons do not genetically modify cells and avoid having to produce clonal cell lines. Therefore, they can easily be deployed in any cell line. In these studies, we have shown that molecular beacons can be used to identify and sort hESCs in a high-throughput manner based on expression of Oct4. We also show that they are eventually shed by transfected cells. This demonstrates the usefulness of this system for selecting pluripotent hESCs from a heterogeneous culture for further study. In addition, the pitfalls of various detection methods for examining Oct4 expression recently have been discussed [21]. These include the detection of Oct4 pseudogenes by PCR, detection of a cytoplasmic isoform, Oct4B, by immunoblot analysis, and cross-reactivity of available antibodies in immunochemistry and FACS analyses [21]. Molecular beacons allow for the design of oligonucleotides that avoid these pitfalls, provide an approach that assays transcript expression, and use FRET to assure specificity.
In designing the molecular beacons, careful consideration should be given to both the site of binding on the mRNA as well as the choice of fluorophores. We designed several pairs of beacons aligning to what appeared to be exposed regions of the Oct4 mRNA (Fig. 1B); however, the set described here, which was separated by 5 nucleotides with predicted melting temperatures of 56.6°C and 58.7°C, was the only set that demonstrated FRET. In addition, we also tested Cy3 and Cy5 as donor and acceptor fluorescent groups, since others have used these fluorophores for single-cell FRET analysis by microscopy. With greater separation between their donor emission/acceptor absorption spectra, we hypothesized that they would be less likely to give false-positive FRET results. However, these beacons failed to produce FRET when analyzed by flow cytometry. One possible explanation is that the abbreviated time spent passing through the laser/filter unit of a cell sorter requires greater overlap between donor emission/acceptor absorption spectra to achieve FRET, than during analysis by microscopy, which allows a longer period for donor excitation/emission and acceptor absorption/emission to occur.
The dual FRET molecular beacon system presented here provides a useful new tool for isolating pluripotent hESCs in a high-throughput manner, and may also be adapted to selecting cells as they achieve specific gene expression signatures [22]. This approach is especially desirable because of its high-throughput capability, the normal hESC behavior exhibited despite molecular beacon introduction, and the short lifespan of molecular beacons within cells. As such, it provides an important new tool for isolating hESC-derived, tissue-specific precursors for both analysis and therapeutic purposes.
Acknowledgments
The authors thank William Hyun and Tara Rambaldo (UCSF Helen Diller Family Comprehensive Cancer Center Laboratory for Cell Analysis) for technical assistance with FACS, and Marc Diamond (UCSF) and members of the Bernstein laboratory for helpful discussions. This work was supported by a Comprehensive Research Grant (RC1-00104) from the California Institute for Regenerative Medicine and a Public Health Service Grant (HL085377) from NHLBI to H.S.B. F.W.K. was supported by a National Service Research Award (HL007544) from NHLBI.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Adewumi O. Aflatoonian B. Ahrlund-Richter L. Amit M. Andrews PW. Beighton G. Bello PA. Benvenisty N. Berry LS. Bevan S. Blum B. Brooking J. Chen KG. Choo AB. Churchill GA. Corbel M. Damjanov I. Draper JS. Dvorak P. Emanuelsson K. Fleck RA. Ford A. Gertow K. Gertsenstein M. Gokhale PJ. Hamilton RS. Hampl A. Healy LE. Hovatta O. Hyllner J. Imreh MP. Itskovitz-Eldor J. Jackson J. Johnson JL. Jones M. Kee K. King BL. Knowles BB. Lako M. Lebrin F. Mallon BS. Manning D. Mayshar Y. McKay RD. Michalska AE. Mikkola M. Mileikovsky M. Minger SL. Moore HD. Mummery CL. Nagy A. Nakatsuji N. O'Brien CM. Oh SK. Olsson C. Otonkoski T. Park KY. Passier R. Patel H. Patel M. Pedersen R. Pera MF. Piekarczyk MS. Pera RA. Reubinoff BE. Robins AJ. Rossant J. Rugg-Gunn P. Schulz TC. Semb H. Sherrer ES. Siemen H. Stacey GN. Stojkovic M. Suemori H. Szatkiewicz J. Turetsky T. Tuuri T. van den Brink S. Vintersten K. Vuoristo S. Ward D. Weaver TA. Young LA. Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter MK. Rosler ES. Fisk GJ. Brandenberger R. Ares X. Miura T. Lucero M. Rao MS. Properties of four human embryonic stem cell lines maintained in a feeder-free culture system. Dev Dyn. 2004;229:243–258. doi: 10.1002/dvdy.10431. [DOI] [PubMed] [Google Scholar]
- 3.Draper JS. Pigott C. Thomson JA. Andrews PW. Surface antigens of human embryonic stem cells: changes upon differentiation in culture. J Anat. 2002;200:249–258. doi: 10.1046/j.1469-7580.2002.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heng BC. Liu H. Rufaihah AJ. Cao T. Human embryonic stem cell (hES) colonies display a higher degree of spontaneous differentiation when passaged at lower densities. In Vitro Cell Dev Biol Anim. 2006;42:54–57. doi: 10.1290/0510071.1. [DOI] [PubMed] [Google Scholar]
- 5.Pan GJ. Chang ZY. Scholer HR. Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 6.Gokhale PJ. Andrews PW. New insights into the control of stem cell pluripotency. Cell Stem Cell. 2008;2:4–5. doi: 10.1016/j.stem.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Cao F. Drukker M. Lin S. Sheikh AY. Xie X. Li Z. Connolly AJ. Weissman IL. Wu JC. Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation. Cloning Stem Cells. 2007;9:107–117. doi: 10.1089/clo.2006.0E16. [DOI] [PubMed] [Google Scholar]
- 8.Santangelo P. Nitin N. Bao G. Nanostructured probes for RNA detection in living cells. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 9.Meister G. Landthaler M. Dorsett Y. Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber AR. Lorenz R. Bernhart SH. Neubock R. Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi S. Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 12.King FW. Ritner C. Liszewski W. Kwan HC. Pedersen A. Leavitt AD. Bernstein HS. Subpopulations of human embryonic stem cells with distinct tissue-specific fates can be selected from pluripotent cultures. Stem Cells Dev. 2009;18:1441–1450. doi: 10.1089/scd.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholas CR. Gaur M. Wang S. Pera RA. Leavitt AD. A method for single-cell sorting and expansion of genetically modified human embryonic stem cells. Stem Cells Dev. 2007;16:109–117. doi: 10.1089/scd.2006.0059. [DOI] [PubMed] [Google Scholar]
- 14.Epting CL. Lopez JE. Pedersen A. Brown C. Spitz P. Ursell PC. Bernstein HS. Stem cell antigen-1 regulates the tempo of muscle repair through effects on proliferation of alpha7 integrin-expressing myoblasts. Exp Cell Res. 2008;314:1125–1135. doi: 10.1016/j.yexcr.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epting CL. Lopez JE. Shen X. Liu L. Bristow J. Bernstein HS. Stem cell antigen-1 is necessary for cell-cycle withdrawal and myoblast differentiation in C2C12 cells. J Cell Sci. 2004;117:6185–6195. doi: 10.1242/jcs.01548. [DOI] [PubMed] [Google Scholar]
- 16.Epting CL. King FW. Pedersen A. Zaman J. Ritner C. Bernstein HS. Stem cell antigen-1 localizes to lipid microdomains and associates with insulin degrading enzyme in skeletal myoblasts. J Cell Physiol. 2008;217:250–260. doi: 10.1002/jcp.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X. Collier JM. Hlaing M. Zhang L. Delshad EH. Bristow J. Bernstein HS. Genome-wide examination of myoblast cell cycle withdrawal during differentiation. Dev Dyn. 2003;226:128–138. doi: 10.1002/dvdy.10200. [DOI] [PubMed] [Google Scholar]
- 18.Ritner C. Bernstein HS. Fate mapping of human embryonic stem cells by teratoma formation. www.jove.com/index/details.stp?id=2036; J Vis Exp. 2010;42:2036. doi: 10.3791/2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexiades MR. Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 20.Becker KA. Ghule PN. Therrien JA. Lian JB. Stein JL. van Wijnen AJ. Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 21.Liedtke S. Stephan M. Kogler G. Oct4 expression revisited: potential pitfalls for data misinterpretation in stem cell research. Biol Chem. 2008;389:845–850. doi: 10.1515/BC.2008.098. [DOI] [PubMed] [Google Scholar]
- 22.Wong SSY. Bernstein HS. Cardiac regeneration using human embryonic stem cells: producing cells for future therapy. Regen Med. 2010;5 doi: 10.2217/rme.10.52. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]