Abstract
Background
A potential susceptibility locus for colorectal cancer on chromosome 9p24 (rs719725) was initially identified through a genome-wide association study, though replication attempts have been inconclusive.
Methods
We genotyped this locus and explored interactions with known risk factors as potential sources of heterogeneity, which may explain the previously inconsistent replication. We included Caucasians with colorectal adenoma or colorectal cancer and controls from four studies (total 3891 cases, 4490 controls): the Women’s Health Initiative (WHI); the Diet, Activity and Lifestyle Study (DALS); a Minnesota population-based case-control study (MinnCCS); and the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO). We used logistic regression to evaluate the association and test for gene-environment interactions.
Results
SNP rs719725 was statistically significantly associated with risk of colorectal cancer in WHI (OR per A allele 1.19; 95% CI 1.01–1.40; p-trend 0.04), marginally associated with adenoma risk in PLCO (OR per A allele 1.11; 95% CI 0.99–1.25; p-trend 0.07), and not associated in DALS and MinnCCS. Evaluating for gene-environment interactions yielded no consistent results across the studies. A meta-analysis of seventeen studies (including these four) gave an OR per A allele of 1.07 (95% CI 1.03–1.12; p-trend 0.001).
Conclusions
Our results suggest the A allele for SNP rs719725 at locus 9p24 is positively associated with a small increase in risk for colorectal tumors. Environmental risk factors for colorectal cancer do not appear to explain heterogeneity across studies.
Impact
If this finding is supported by further replication and functional studies, it may highlight new pathways underlying colorectal neoplasia.
Keywords: colorectal, adenoma, cancer, 9p24, rs719725
Introduction
Colorectal cancer is a major cause of morbidity and mortality in the United States. In 2010, there will be an estimated 142,520 new cases and 51,370 deaths in the US expected from colorectal cancer (1). Colorectal cancer is the third most commonly diagnosed cancer and the third leading cause of cancer-related deaths in the US when men and women are considered separately (2), and the second leading cause for both sexes combined (3). A substantial proportion of colorectal cancer is due to genetic risk factors, with estimates of heritable effects reaching as high as 35 percent (95% confidence interval (CI) 10–48%) in a study of twins from Finland, Denmark and Sweden (4). However, less than five percent of colorectal cancer is due to known highly penetrant variants inherited in an autosomal dominant manner (5). Thus, a large proportion of the remaining uncharacterized inherited susceptibility is expected to be due to numerous low-penetrance variants (6). The use of high-throughput technology, allowing the simultaneous ascertainment of hundreds of thousands of single nucleotide polymporphisms (SNPs), particularly in the context of genome-wide association studies (GWAS), has been especially useful in exploring such potential variants.
Recent GWAS have succeeded in identifying 10 genetic regions that contain SNPs associated with colorectal cancer risk (7). One of these regions, 8q24, has been replicated in multiple studies and has also been implicated in various other cancers, such as prostate, breast, and bladder cancer (8). Along with 8q24, a second locus 9p24 was identified in the Assessment of Risk for Colorectal Tumors in Canada (ARCTIC) genome-wide scan (8). Within this region, rs719725 was identified as being the most strongly associated with colorectal cancer (OR 1.14, p = 1.32 × 10−5), although with lower statistical significance and less cross-study consistency than for 8q24 (8). However, in replication studies by these researchers, only three of seven study populations showed a statistically significant association consistent with the GWAS finding, resulting in a lower strength of association (OR 1.08, p = 0.023) (8).
A subsequent study replicated the rs719725 association with colorectal cancer using a case-unaffected sibling control design from the Colon Cancer Family Registry (9). This study found a statistically significant association among population-based families (p = 0.011), but not among clinic-based families (p = 0.97), although the difference between these two was, itself, not statistically significant (p = 0.26) (9). A later study was unable to replicate rs719725 using a combined analysis of three study populations from Sheffield, U.K., Leeds, U.K., and Utah, U.S.A. (OR 1.04, 95% CI 0.91–1.19) (10). Overall results from these subsequent replications were inconsistent, demonstrating an association in some populations while not in others (8, 9, 10).
While GWAS have identified ten SNPs associated with colorectal tumor risk, the biological functionality of these markers are generally unknown. At the 9p24 locus, SNP rs719725 does not lie within a gene itself, but there are several genes nearby. These include: protein kinase NYD-SP25 isoform 3 (TPD52L3, 37 kb telomeric), which is a member of the tumor protein D52 family; interleukin 33 (IL33, 124 kb telomeric); ubiquitin-like PHD and RING finger domain-containing protein (UHRF2, 47 kb centromeric); and glycine dehydrogenase (GLDC, 167 kb centromeric) (9). Potentially, rs719725 could also be in linkage disequilibrium with a variant that modifies a long range enhancer of any of these genes, as seen for the variants in 8q24 and MYC (11). rs719725 is upstream of UHRF2 and downstream of TPD52L3, IL33, and GLDC; it lies in a haplotype block that is ~114 kb in size, which is part of a larger block that is ~407 kb in size (Haploview 4.1, release 21, CEU population) (12). The small block contains TPD52L3; the large block includes TPD52L3, IL33, and UHRF2. As with other tag SNPs, associations between rs719725 and colorectal tumors may be due to linkage disequilibrium between the genetic marker and the true susceptibility allele, or alleles, at a neighboring locus. Beyond risk identification, a recent study found rs719725 to be statistically significantly associated with time to tumor recurrence in adjuvant colorectal cancer patients (13), suggesting this SNP may also be potentially important for prognosis and survival.
In order to further investigate whether the genetic variant on 9p24 is associated with colorectal tumors, we examined the impact of rs719725 on both colorectal adenoma and cancer, allowing the capture of a broader spectrum of colorectal tumor development. Because the large majority of colorectal malignancies develop from an adenomatous polyp (14), any genetic influence on these precursor lesions is likely to have an effect on cancer as well. We examined this association in four well-characterized study populations. These studies allowed for a detailed investigation of potential interactions with environmental risk factors known to be associated with colorectal cancer risk, including obesity, physical activity, non-steroidal anti-inflammatory drug use, smoking, and diet. Such factors may contribute to heterogeneity between studies, and may help to explain the inconsistent findings previously reported.
Materials and Methods
Study samples
To investigate the full spectrum of colorectal cancer we included both colorectal adenoma and colorectal cancer from four study populations (total 3891 cases, 4490 matched controls): the Women’s Health Initiative (WHI: 656 colorectal cancer cases, 664 matched controls); the Diet, Activity and Lifestyle Study (DALS: 1461 colon cancer cases, 1813 matched controls); a Minnesota population-based case-control study (MinnCCS: 517 colorectal adenoma cases, 628 matched controls); and the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO: 1257 colorectal adenoma cases, 1385 matched controls).
The WHI Observational Study (OS) is a prospective cohort study, which enrolled 93,676 postmenopausal women aged 50–79 years living near 40 clinical centers across the US from 1993 to 1998, with continuous follow-up (15). We included invasive incident colorectal cancer cases diagnosed up through Sept 12th 2005. Controls were randomly selected and individually matched on age at screening, enrollment date, ethnicity, hysterectomy status, and prevalent colorectal cancer at baseline (16), using risk-set sampling (17).
DALS is a population-based case-control study of colon cancer (18). Cases and controls were recruited from the Kaiser Permanente Medical Care Program of Northern California, an eight-county area in Utah, and the metropolitan Twin Cities of Minnesota. Cases had been diagnosed with first primary colon cancer between October 1, 1991 and September 30, 1994. Cancers of the rectosigmoid junction or rectum were not included in this study. Controls were matched to cases by 5-year age groups and sex, and came from membership lists (Kaiser), driver license lists (Utah up to age 65, Minnesota), Health Care Financing Administration lists (Utah over age 65), or state identification lists (Minnesota).
The MinnCCS is a case-control study within the Minnesota Cancer Prevention Research Unit (19). Cases and controls, aged 30–74 years, were recruited from patients that were scheduled for colonoscopy from April 1991 to April 1994 within a large, multi-clinic gastroenterology practice in metropolitan Minneapolis. Cases were subjects found to have adenoma at the time of colonoscopy, and controls were subjects found to be without adenomatous or hyperplastic polyps at colonoscopy (20).
The PLCO is a randomized trial of ~155,000 persons aged 55–74, enrolled during 1993–2001 from 10 US centers, to evaluate the effectiveness of screening on cancer mortality (21). We conducted a nested case-control study among participants randomized to the screening arm, who underwent a 60-cm flexible sigmoidoscopy examination at study entry. Cases included participants with advanced colorectal adenoma of the distal colon or rectum (adenoma ≥ 1 cm in size, diagnosed as: villous/tubulovillous; high-grade dysplasia; or carcinoma in situ). Controls were participants who had a successful baseline screening exam that was negative for polyps in the distal colon and rectum. Controls were frequency-matched to cases on ethnicity, sex, and, for a subset, age (21).
Further details regarding the methods used for selection of subjects and data collection in these studies have been previously reported. Given the small proportion of non-Caucasians in these studies (2.9% to 16.2%) all four datasets were restricted to Caucasians only, which the numbers above reflect. All reported studies obtained informed consent and received IRB approval.
Genotyping
For the WHI and DALS studies, genotyping was performed by MALDI-TOF mass spectrometry on the Sequenom MassARRAY 7K platform using the iPLEX Gold (low-plex) reaction (16). For the PLCO and MinnCCS studies, genotyping was performed using the TaqMan assay system: PLCO used the Fludigm Biomark system (Fluidigm, South San Francisco, CA, USA) and MinnCCS used an ABI 7900HT instrument (ABI, Foster City, CA, USA) (20). These methods have been previously reported and described in further detail elsewhere (16, 20). Quality-control measures in these studies included call-rate cutoffs, blinded duplicates, and other standard methods. In all four studies, the minor allele frequency (MAF; C allele) was ~37% and controls were in Hardy-Weinberg equilibrium (HWE; p > 0.18).
Statistical analysis
We calculated odds ratios (ORs) and 95% confidence intervals (95% CIs) to estimate the association between SNP rs719725 and colorectal neoplasia. We conducted logistic regression analyses for each study separately, including study-specific covariates, such as age, sex, and study center, as appropriate. To be consistent with most previous studies, we used the minor allele C as the reference allele. We calculated risk based on the log-additive model, by including a single variable coded as 0, 1, or 2, reflecting the number of copies of the major A allele. We also evaluated risk for an unrestricted model, without any assumption of the underlying genetic model, which provides OR estimates comparing genotypes AC vs. CC and AA vs. CC. We combined the individual study-specific adjusted log-ORs using a linear mixed-effects model and then estimated the overall ORs and 95% CIs (22).
To investigate whether the association between rs719725 and cancer differed by anatomical location, we also performed stratified analyses for colon vs. rectum. Cases with tumors located at both the colon and the rectum and those with unknown location were excluded from these stratified analyses. Polytomous regression was used to evaluate statistical differences among models. Since WHI only included females, we also evaluated these associations stratified by sex.
Risk factors potentially related to colorectal cancer were evaluated for interaction with rs719725 using likelihood ratio tests. The pertinent risk factors considered were: family history of colorectal cancer (having at least one first degree relative, yes/no); cigarette smoking (never/former/current and pack-years); use of non-steroidal anti-inflammatory drugs (NSAIDs, yes/no); alcohol consumption (g/day); physical activity (hours of moderate/vigorous exercise per week); body-mass index (BMI, kg/m2); folate intake (total folate in mcg/day), calcium intake (mg/day), red meat intake (g/day); and use of postmenopausal hormones (never/former/current and duration). Dietary risk factors were adjusted for daily caloric intake by including this term in the regression models used for the likelihood ratio tests. Risk factors with a statistically significant test in at least two studies were considered to lead to potential interaction effects, and OR estimates were calculated stratified on that factor. To assess the overall effect of the interaction we also performed random-effects meta-analyses of the betas and standard errors of the interaction terms for each risk factor.
We performed a random-effects meta-analysis combining our four studies with thirteen results presented in three previously published studies on rs719725 and colorectal cancer (8, 9, 10). For this combined analysis, we included the log-additive OR estimates and 95% CIs for each study population; for a few published results we needed to transform from A allele referent to C allele referent (10), or from an unrestricted model to a log-additive model (9). Forest plots were used to display the results from individual studies, as well as the summary results. The statistical significance of between-study heterogeneity was evaluated using Cochran’s Q statistic (23). If the p-value was less than 0.10, the heterogeneity was considered statistically significant. We also quantified heterogeneity using the I2 metric. I2 takes values between 0 percent and 100 percent, with higher values indicating higher levels of heterogeneity (24). Potential for publication bias was assessed using Egger’s test and visual inspection of funnel plots (25).
Analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC), STATA version 11 (StataCorp, College Station, TX), and HaploView version 4.2 (12). Our statistical significance cutoff was a p-value of 0.05, and for marginal statistical significance was 0.10.
Results
Demographic information for each study is shown in Tables 1a and 1b. Participants in the four studies had a mean age of 57.9 to 67.1 years. By definition the WHI study population was 100% female, whereas the proportion female was 45.5% in DALS, 50.7% in MinnCCS, and 35.8% in PLCO. As cases and controls were matched on age and sex, there were no substantial differences between cases and controls on these variables. Due to restriction, all participants were self-reported Caucasian. As expected, cases were more likely to have a positive family history of CRC, except in MinnCCS. The difference in family history in MinnCCS is plausibly explained by the fact that participants were selected among those that elected to get screened for CRC: for cases, indications for screening are likely to be symptom- or disease-related, whereas among controls, screening was more likely sought by the worried well with a positive family history. Except for WHI, cases tend to be more likely to be current or former smokers. The proportion of smokers was generally similar across these populations. Rectal cancers made up 18.2% of the WHI cases and 0% of the DALS cases; rectal adenoma made up 16.3% of the MinnCCS cases and 22.3% of the PLCO cases (Table 2).
Table 1.
Demographic and lifestyle factors in four study populations.*
| Characteristic | Measure | WHI | DALS | MinnCCS | PLCO | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | ||
| Total | Count | 656 | 664 | 1461 | 1813 | 517 | 628 | 1257 | 1385 |
| Age | Mean | 67.11 | 67.09 | 65.15 | 65.16 | 57.91 | 52.77 | 63.1 | 62.65 |
| Sex | % Female | 100.0% | 100.0% | 43.7% | 46.9% | 37.1% | 61.8% | 36.6% | 36.0% |
| Ethnicity | % White | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Relative with colorectal cancer | % Yes | 21.8% | 19.2% | 17.0% | 9.7% | 16.6% | 28.3% | 12.1% | 9.6% |
| BMI | Mean | 27.89 | 27.09 | 27.66 | 26.63 | 27.29 | 26.87 | 27.91 | 27.4 |
| Cigarette smoking status | % Current | 7.0% | 5.3% | 14.5% | 13.6% | 20.8% | 16.1% | 15.1% | 6.8% |
| % Former | 44.7% | 48.9% | 44.5% | 39.4% | 46.4% | 37.1% | 50.1% | 48.1% | |
| % Never | 48.4% | 45.8% | 41.0% | 47.0% | 32.8% | 46.9% | 34.8% | 45.1% | |
| Pack-years | Mean | 14.2 | 12.0 | 0.7 | 0.5 | 19.7 | 12.8 | 2.2 | 1.6 |
| NSAIDs | % Use | 30.6% | 30.1% | 39.3% | 48.4% | 11.2% | 19.8% | 59.0% | 63.2% |
| Aspirin | % Use | 24.7% | 25.5% | 27.0% | 36.0% | 28.7% | 31.6% | 48.1% | 51.1% |
| Alcohol | Mean g/day | 5.8 | 6.1 | 12.4 | 10.0 | 10.4 | 6.5 | 15.7 | 12.6 |
| Physical activity | Mean hours/week modest/vigorous | 1.6 | 1.8 | 1.6 | 1.8 | 9.6 | 8.7 | 2.5 | 2.9 |
| Folate intake | Mean mcg/day | 1197.1 | 1231.8 | 385 | 385.1 | 317.1 | 318.4 | 576.2 | 630.7 |
| Calcium | Mean mg/day | 1192.1 | 1268.3 | 1095.1 | 1107.8 | 958.4 | 991.6 | 1164.3 | 1258.8 |
| Red meat intake | Mean g/day | 15.0 | 15.2 | 26.9 | 23.9 | 4.8 | 4.6 | 88.1 | 85.5 |
| HRT Use | % ever use | 51.6% | 57.6% | 24.0% | 30.8% | 27.6% | 52.8% | 64.6% | 70.5% |
| Duration of use | 10.3 | 10.3 | - | - | 2.3 | 2.4 | 44.1% | 43.8% | |
For duration of HRT use, WHI and MinnCCS are years of use, PLCO is percentage of participants using 6 or more years, and DALS does not report HRT use duration.
Table 2.
Numbers of colorectal cancer and adenoma stratified by site.
| Tumor Type | Tumor Site | WHI | DALS | MinnCCS | PLCO | ||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||
| Cancer | Colon | 534 | 81.4% | 1461 | 100.0% | - | - | - | - |
| Rectum | 119 | 18.1% | 0 | 0.0% | - | - | - | - | |
| Mixed | 0 | 0.0% | 0 | 0.0% | - | - | - | - | |
| Not specified | 3 | 0.5% | 0 | 0.0% | - | - | - | - | |
| Adenoma | Colon | - | - | - | - | 343 | 66.3% | 848 | 67.5% |
| Rectum | - | - | - | - | 67 | 13.0% | 244 | 19.4% | |
| Mixed | - | - | - | - | 103 | 19.9% | 148 | 11.8% | |
| Not specified | - | - | - | - | 4 | 0.8% | 17 | 1.4% | |
In the WHI study, rs719725 was statistically significantly associated with risk of colorectal cancer, with an OR of 1.19 per A allele (95% CI 1.01–1.40; p-trend 0.04; Table 3). Similarly, the PLCO data provided a marginally statistically significant finding in the same direction for risk of advanced adenoma (OR 1.11 per A allele; 95% CI 0.99–1.25; p-trend 0.07). In both MinnCCS and DALS, rs719725 was not associated with risk of colorectal tumors. The OR estimates from the unrestricted models were generally consistent in their direction and trend with the results from the additive models.
Table 3.
Association between 9p24 risk locus rs719725 and colorectal tumor.†
| Cases | Controls | Adjusted OR | 95% CI | P trend | P diff | |||
|---|---|---|---|---|---|---|---|---|
| WHI (Cancer) | Colorectal | CC | 75 | 92 | 1.00 | (ref) | 0.04 | |
| AC | 281 | 310 | 1.11 | 0.79 – 1.57 | ||||
| AA | 263 | 235 | 1.37 | 0.96 – 1.95 | ||||
| per A allele | 1.19 | 1.01 – 1.40 | ||||||
| Colon Only | CC | 60 | 92 | 1.00 | (ref) | 0.11 | 0.07 | |
| AC | 241 | 310 | 1.18 | 0.82 – 1.70 | ||||
| AA | 207 | 235 | 1.34 | 0.92 – 1.95 | ||||
| per A allele | 1.15 | 0.97 – 1.37 | ||||||
| Rectum Only | CC | 15 | 92 | 1.00 | (ref) | 0.04 | ||
| AC | 39 | 310 | 0.80 | 0.42 – 1.52 | ||||
| AA | 55 | 235 | 1.47 | 0.79 – 2.74 | ||||
| per A allele | 1.38 | 1.01 – 1.88 | ||||||
| DALS (Cancer) | Colon Only | CC | 207 | 258 | 1.00 | (ref) | 0.96 | |
| AC | 659 | 808 | 1.01 | 0.82 – 1.25 | ||||
| AA | 587 | 724 | 1.01 | 0.81 – 1.25 | ||||
| per A allele | 1.00 | 0.91 – 1.11 | ||||||
| MinnCCS (Adenoma) | Colorectal | CC | 82 | 81 | 1.00 | (ref) | 0.82 | |
| AC | 209 | 277 | 0.72 | 0.51 – 1.08 | ||||
| AA | 215 | 246 | 0.86 | 0.59 – 1.26 | ||||
| per A allele | 0.98 | 0.82 – 1.17 | ||||||
| Colon Only | CC | 54 | 81 | 1.00 | (ref) | 0.83 | 0.66 | |
| AC | 132 | 277 | 0.72 | 0.47 – 1.10 | ||||
| AA | 149 | 246 | 0.91 | 0.60 – 1.39 | ||||
| per A allele | 1.02 | 0.84 – 1.25 | ||||||
| Rectum Only | CC | 11 | 81 | 1.00 | (ref) | 0.41 | ||
| AC | 31 | 277 | 0.83 | 0.40 – 1.74 | ||||
| AA | 24 | 246 | 0.72 | 0.34 – 1.55 | ||||
| per A allele | 0.86 | 0.60 – 1.23 | ||||||
| PLCO (Adenoma) | Colorectal | CC | 163 | 196 | 1.00 | (ref) | 0.07 | |
| AC | 575 | 674 | 1.03 | 0.81 – 1.30 | ||||
| AA | 506 | 509 | 1.20 | 0.94 – 1.53 | ||||
| per A allele | 1.11 | 0.99 – 1.25 | ||||||
| Colon Only | CC | 117 | 196 | 1.00 | (ref) | 0.05 | 0.15 | |
| AC | 367 | 674 | 0.93 | 0.71 – 1.21 | ||||
| AA | 355 | 509 | 1.18 | 0.91 – 1.55 | ||||
| per A allele | 1.14 | 1.00 – 1.29 | ||||||
| Rectum Only | CC | 26 | 196 | 1.00 | (ref) | 0.58 | ||
| AC | 129 | 674 | 1.43 | 0.91 – 2.25 | ||||
| AA | 88 | 509 | 1.29 | 0.81 – 2.07 | ||||
| per A allele | 1.06 | 0.87 – 1.30 | ||||||
Models are adjusted for age, sex, and study center, as appropriate. For colon- and rectum-specific analyses, excluded cases where tumor location was unknown (WHI 3, MinnCCS 4, PLCO 17) or in both the colon and the rectum (MinnCCS 103, PLCO 148).
To further evaluate the association between rs719725 and colorectal neoplasia risk, we performed logistic regression analyses stratified by tumor location (Table 3). These stratified analyses did not show any statistically significant differences between colon and rectal tumors in any study (p for difference > 0.07). Stratifying these analyses by sex did not produce any significantly different results (Table 4).
Table 4.
Association between 9p24 risk locus rs719725 and colorectal tumor, stratified by sex.‡
| Study | Sex | Colorectal | Colon | Rectum | |||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | OR | 95% CI | ||
| WHI (Cancer) | Female | 1.19 | 1.01 – 1.40 | 1.15 | 0.97 – 1.37 | 1.38 | 1.01 – 1.88 |
| DALS (Cancer) | Overall | - | - | 1.00 | 0.91 – 1.11 | - | - |
| Male | - | - | 1.02 | 0.89 – 1.17 | - | - | |
| Female | - | - | 0.98 | 0.84 – 1.13 | - | - | |
| MinnCCS (Adenoma) | Overall | 0.98 | 0.82 – 1.17 | 1.02 | 0.84 – 1.25 | 0.86 | 0.60 – 1.23 |
| Male | 0.90 | 0.71 – 1.15 | 0.90 | 0.69 – 1.17 | 1.00 | 0.61 – 1.63 | |
| Female | 1.08 | 0.83 – 1.41 | 1.24 | 0.91 – 1.70 | 0.68 | 0.40 – 1.17 | |
| PLCO (Adenoma) | Overall | 1.11 | 0.99 – 1.25 | 1.14 | 1.00 – 1.29 | 1.06 | 0.87 – 1.30 |
| Male | 1.11 | 0.96 – 1.28 | 1.13 | 0.96 – 1.32 | 1.15 | 0.87 – 1.51 | |
| Female | 1.09 | 0.90 – 1.32 | 1.13 | 0.91 – 1.41 | 0.95 | 0.70 – 1.29 | |
Models are adjusted for age and study center, as appropriate. For colon- and rectum-specific analyses, excluded cases where tumor location was unknown (WHI 3, MinnCCS 4, PLCO 17) or in both the colon and the rectum (MinnCCS 103, PLCO 148).
Only three interactions were found to be statistically significant, and all were in MinnCCS (Table 5). These interactions were between rs719725 and each of folate intake (p = 0.02); HRT use (p = 0.001); and duration of HRT use (p = 0.01).
Table 5.
Interaction between environmental risk factors and 9p24 risk locus rs719725 for colorectal tumor.§
| Risk Factor | Variable definition | P for interaction | ||||
|---|---|---|---|---|---|---|
| WHI | DALS | MinnCCS | PLCO | Overall | ||
| Age | 0.39 | 0.71 | 0.25 | 0.68 | 0.73 | |
| Sex | - | 0.65 | 0.32 | 0.87 | 0.54 | |
| Relative with Colorectal cancer | First degree relative | 0.75 | 0.09 | 0.45 | 0.34 | 0.84 |
| Cigarette Smoking | Never/former/current | 0.82 | 0.57 | 0.96 | 0.96 | 0.82 |
| Pack years | 0.90 | 0.57 | 0.79 | 0.64 | 0.72 | |
| NSAIDs | Use yes/no | 0.26 | 0.37 | 0.80 | 0.71 | 0.97 |
| Aspirin | Use yes/no | 0.13 | 0.16 | 0.14 | 0.99 | 0.61 |
| Alcohol consumption | g/day | 0.06 | 0.59 | 0.98 | 1.00 | 0.45 |
| Physical activity | hours modest/vigorous | 0.39 | 0.90 | 0.40 | 0.66 | 0.41 |
| BMI | kg/m^2 | 0.49 | 0.09 | 0.38 | 0.31 | 0.81 |
| Folate intake | Total folate mcg/day | 0.96 | 0.99 | 0.02 | 0.77 | 0.49 |
| Calcium intake | mg/day | 0.47 | 0.82 | 0.63 | 0.94 | 0.80 |
| Red meat intake | g/day | 0.78 | 0.40 | 0.19 | 0.76 | 0.54 |
| HRT use | Ever/never | 0.36 | 0.69 | 0.001 | 0.36 | 0.42 |
| Duration | 0.55 | - | 0.01 | 0.26 | 0.21 | |
The study specific p-value is for the likelihood ratio test adjusted for age, sex and study center, as appropriate. Overall p-value is from meta-analysis of the interaction. WHI is female only, and duration of HRT use was not reported in DALS.
A meta-analysis of the four studies presented here and 13 previous studies (total n = 17, Figure 1) resulted in an overall OR of 1.07 (95% CI 1.03 – 1.12) per A allele (p = 0.001), showing a statistically significant association between rs719725 and colorectal neoplasia across the study populations. There was little evidence for between study heterogeneity (I2 = 24.4%, heterogeneity p = 0.17). Information (where available) on the number of cases and controls, minor allele frequency (MAF), HWE and sex of the other studies included in the meta-analysis are shown in Table 6.
Figure 1.
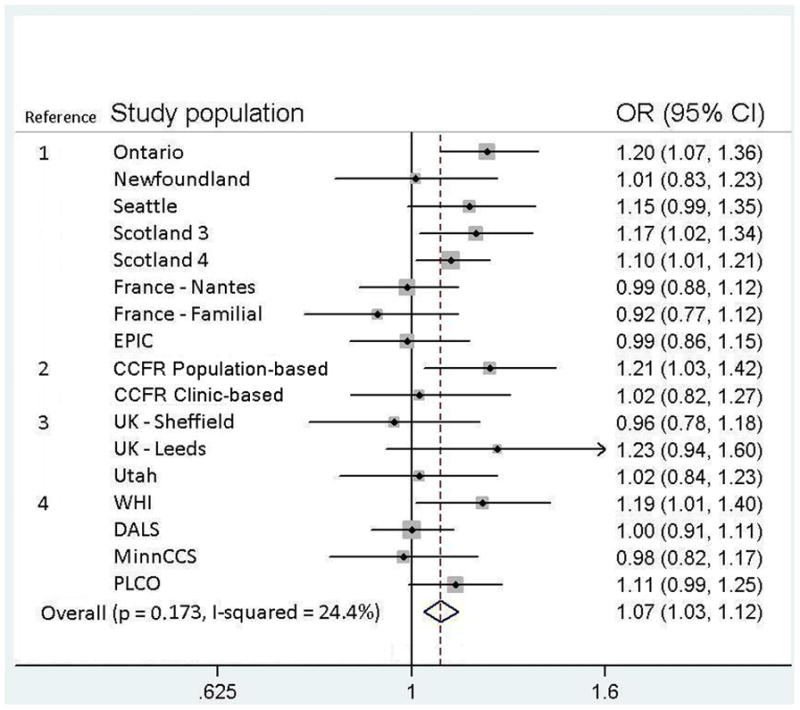
Forest plot for meta-analysis of the association between 9p24 risk locus rs719725 and colorectal cancer and adenoma including the current and previous studies: Reference 1 is Zanke et al. (Nature Genetics 2007); 2 is Poynter et al. (Cancer Research 2007); 3 is Curtin et al. (Cancer Epidemiology, Biomarkers & Prevention 2009); and 4 is this study. The symbol size indicates the weight for each study, and lines indicate the confidence intervals. Weights are from random effects analysis. All studies were of cancer except for MinnCCS and PLCO, which were of adenoma.
Table 6.
Characteristics of study populations used in the meta-analysis of rs719725 and colorectal tumor risk.**
| Study | Population | Location | Cases | Controls | MAF | HWE | % female |
|---|---|---|---|---|---|---|---|
| Zanke et al. | Ontario | Canada | 1226 | 1239 | - | - | - |
| Newfoundland | Canada | 452 | 367 | - | - | - | |
| Seattle | WA | 687 | 688 | - | - | - | |
| Scotland-3 | Scotland | 975 | 1002 | - | - | - | |
| Scotland-4 | Scotland | 1910 | 1985 | - | - | - | |
| France-Nantes | France | 1068 | 1113 | - | - | - | |
| France-Familial | France | 370 | 539 | - | - | - | |
| EPIC | Europe | 761 | 749 | - | - | - | |
| Poynter et al. | Clinic-based | USA, Canada, Australia | 288 | 505 | - | Ok | 51.6 |
| Population-based | USA, Canada, Australia | 1344 | 2196 | - | Ok | 54.0 | |
| Curtin et al. | Utah | Utah | 438 | 439 | ~37 | 0.13 | ~50 |
| Sheffield | United Kingdom | 405 | 403 | ~37 | 0.14 | ~50 | |
| Leeds | United Kingdom | 249 | 218 | ~37 | 0.78 | ~50 | |
| Present study | WHI | 40 US centers | 656 | 664 | 34.8 | 0.53 | 100 |
| DALS | CA, UT, MN | 1461 | 1813 | 37.0 | 0.18 | 45.5 | |
| MinnCCS | Minnesota | 517 | 628 | 36.3 | 0.83 | 50.7 | |
| PLCO | 10 US centers | 1257 | 1385 | 38.7 | 0.26 | 35.8 |
A dash (-) indicates that data was not reported; Ok: Poynter et al. report that all SNPs were in HWE, but do not describe their cutoff value. All studies were of cancer except for MinnCCS and PLCO, which were of adenoma.
Because the interpretation of the results in MinnCCS may be impacted by the large fraction of controls with a positive family history, given that controls were selected among those that chose to be screened for CRC, we also obtained results excluding this study. Taking the MinnCCS study out of the meta-analysis, however, did not produce substantially different results (OR 1.08 per A allele; 95% CI 1.03 – 1.13; p = 0.001). Due to potential differences between adenoma and cancer we also obtained results among studies of cancer only. Taking the adenoma studies PLCO and MinnCCS out of the meta-analysis, however, also did not produce substantially different results (OR 1.08 per A allele; 95% CI 1.03 – 1.13; p = 0.002).
Discussion
Among four well characterized epidemiologic studies we found a statistically significant and marginally significant association in two nested case-control studies of colorectal cancer and adenoma whereas two other case-control studies of colorectal adenoma and cancer did not show evidence for association. We were not able to explain the differences in our results by tumor site or by interactions with environmental risk factors for colorectal cancer. A meta-analysis of 17 studies (including these 4) showed a statistically significant association for this SNP with colorectal tumor. It is important to note that, although the primary distinction among the four studies presented here is the finding of an association in the cohort, but not the case-control studies, that is not what characterizes the rest of the data. Indeed, the first study to report the association was a case-control study.
Our results are similar to other publications that showed inconsistent findings for rs719725 and risk of colorectal cancer. Summarizing findings in a meta-analysis of all available results provided evidence for a positive association between the rs719725 A allele and colorectal tumor (OR 1.07, p = 0.001). However results do not reach genome-wide significance levels of 10−7 to 10−8 (26, 27). It is probably important that only 5 of the 17 reported risk estimates showed no positive association and only 2 of these showed an inverse association (Figure 1). We observed no statistical evidence for heterogeneity, though we may not have had the necessary power to detect it (24, 28). A funnel plot of the estimates of the association used in the meta-analysis did not demonstrate any evidence of publication bias (Egger’s test p = 0.68), though our power to detect such bias may be limited (Figure 2).
Figure 2.
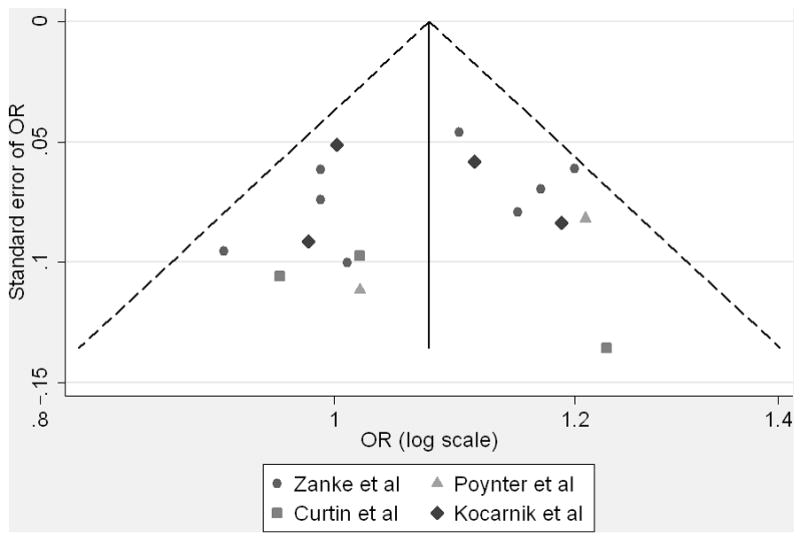
Funnel plot of estimates of association between 9p24 risk locus rs719725 and colorectal cancer from current and previous studies used in the meta-analysis. Please note that zero on the y-axis is at the top, with larger standard errors descending down from the origin.
The weak OR estimates found in this study are not surprising, given the generally weak risk estimates commonly found in GWAS. Although it seems likely that this particular SNP carries only a slight increase in risk of colorectal tumor, its identification, if true, would nevertheless be important for identifying potentially new pathways relevant to preventive and treatment strategies. Furthermore, in concert with many other susceptibility loci, rs719725 could potentially be used to measure an overall risk profile for colorectal tumor. A recent review estimated that ~172 SNPs account for all of the genetic variance for colorectal cancer (7). In this way, rs719725 could potentially resemble a single small piece of the large colorectal cancer genetic risk profile puzzle; the problem of what is sufficient and what is necessary is likely to remain unresolved for a long time.
Among the four study populations, we did not find any consistent evidence for any gene-environment interaction. We observed only three statistically significantly interactions in one study (MinnCCS), for rs719725 and folate intake and HRT use and duration, but these were not replicated in the other three study populations. These three findings do not remain statistically significant after adjustment for multiple comparisons. Further, the lack of a clear pattern across all four studies, along with observing these interactions in the smallest of the four studies, suggests that these findings are likely to be spurious. Our hypothesis that these environmental factors could explain some of the heterogeneity between studies is not supported by our data, though we may have been underpowered to explore such effects or perhaps did not evaluate the correct environmental factor. We included essentially all of the currently known CRC environmental factors. The biological function of this SNP is unknown, and therefore we did not have an a priori hypothesis to examine interactions with specific environmental factors. We chose the approach of evaluating all established environmental risk factors as potential effect modifiers, with the goal of determining if these factors are potentially responsible for the inconsistent results reported in previous papers.
A limitation of this study is the inability to investigate gene-gene interactions, which may be important for this locus. It is possible that such interactions may play a role in explaining the different results across studies, and future research will be necessary to determine how risk is impacted by considering multiple genetic variants simultaneously. We examined only the previously reported SNP in this region and it is possible that other variants are more relevant. While we restricted study participants to only include Caucasians, population stratification is still a potential reason for the inconsistent findings reported across studies. The definition of an adenoma case was also more broadly interpreted in MinnCCS than in PLCO, and this difference in definition could have contributed to the inconsistent association found in these two studies.
Strengths of this study include the use of multiple studies with large sample sizes and the availability of detailed information on environmental risk factors for colorectal cancer, allowing us to evaluate if environment factors modify the association between this SNP and colorectal tumor.
Although we can neither prove nor disprove the existence of a risk locus on chromosome 9p24 that is tagged by SNP rs719725, our analysis, as well as the meta-analysis including previously published studies, provide additional support for an association between variation at 9p24 and colorectal neoplasia. Further replication in larger studies seems to be warranted. If the association continues to be replicated, this could be followed by studies to identify the disease-related polymorphism being tagged and its possible function. It also may be pertinent to evaluate this association among different races and ethnicities, and to further explore potential interactions with other genetic or environmental factors.
Acknowledgments
The project described was supported by Award Numbers R01 CA059045 and R01 CA120582 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. This research was also supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. The authors would like to thank Dr. Roberd Bostick and Ms. Lisa Fosdick for their contributions to the study design and data collection in the MinnCCS. The authors thank Drs Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc., Mr. Tim Sheehy and staff, DNA Extraction and Staging Laboratory, SAIC-Frederick, Inc., and Ms. Jackie King and staff, BioReliance, Inc. Most importantly, we acknowledge the study participants for their contributions to making this study possible.
References
- 1.Colon and Rectal Cancer [Internet] Bethesda (MD): National Cancer Institute, U.S. National Institutes of Health; (cited 2010 Sep 16). Available from: http://www.cancer.gov/cancertopics/types/colon-and-rectal. [Google Scholar]
- 2.American Cancer Society. Colorectal Cancer Facts & Figures 2008–2010. Atlanta: American Cancer Society; 2008. [Google Scholar]
- 3.What are the key statistics for colorectal cancer? [Internet] Atlanta (GA): American Cancer Society; (cited 2009 Aug 10). Available from: http://www.cancer.org/docroot/CRI/content/CRI_2_4_1X_What_are_the_key_statistics_for_colon_and_rectum_cancer.asp. [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 6.Kemp Z, Thirlwell C, Sieber O, Silver A, Tomlinson I. An update on the genetics of colorectal cancer. Hum Mol Genet. 2004;13(2):R177–85. doi: 10.1093/hmg/ddh247. [DOI] [PubMed] [Google Scholar]
- 7.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10(6):353–8. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 8.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 9.Poynter JN, Figueiredo JC, Conti DV, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the colon cancer family registry. Cancer Res. 2007;67(23):11128–32. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 10.Curtin K, Lin WY, George R, et al. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer Epidemiol Biomarkers Prev. 2009;18(2):616–21. doi: 10.1158/1055-9965.EPI-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 13.Lenz H, Lurje CA, Haiman D, et al. Colorectal cancer susceptibility variants and clinical outcome in adjuvant and metastatic colorectal cancer. Am J Clin Oncol; ASCO Annual Meeting Proceedings (Post-Meeting Edition); 2009. supplement; abstract 4051. [Google Scholar]
- 14.Tanaka T. Colorectal carcinogenesis: review of human and experimental animal studies. J Carcinog. 2009;8(5):1–19. doi: 10.4103/1477-3163.49014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langer R, White E, Lewis C, Kotchen J, Hendrix S, Trevisan M. The WHI OS: baseline characteristics of participants and reliability of baseline measures. Ann Epidemiol. 2003;13:S107–21. doi: 10.1016/s1047-2797(03)00047-4. [DOI] [PubMed] [Google Scholar]
- 16.Hutter C, Slattery M, Duggan D, et al. Genetic variation at chromosome 8q24 and risk of colon cancer. BMC Cancer. 2010 in press. [Google Scholar]
- 17.Bergstralh EJ, Kosanke JL. Technical Report #56. Department of Health Sciences Research; Mayo Clinic, Rochester, MN: Apr, 1995. Computerized matching of cases to controls. [Google Scholar]
- 18.Slattery ML, Potter J, Caan B, et al. Energy balance and colon cancer – beyond physical activity. Cancer Res. 1997;57(1):75–80. [PubMed] [Google Scholar]
- 19.Potter JD, Bostick RM, Grandits GA, et al. Hormone replacement therapy is associated with lower risk of adenomatous polyps of the large bowel: the Minnesota cancer prevention research unit case-control study. Cancer Epidemiol Biomarkers Prev. 1996;5:779–84. [PubMed] [Google Scholar]
- 20.Berndt SI, Potter JD, Hazra A, et al. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum Mol Genet. 2008;17(17):2665–72. doi: 10.1093/hmg/ddn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prorok PC, Adnriole GL, Bresalier RS, et al. Design of the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 22.Stukel TA, Demidenko E, Dykes J, Karagas MR. Two-stage methods for the analysis of pooled data. Stat Med. 2001;20(14):2115–30. doi: 10.1002/sim.852. [DOI] [PubMed] [Google Scholar]
- 23.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10(1):101–29. [Google Scholar]
- 24.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;450(7171):887–92. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioannidis JPA, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2(9):e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]


