Abstract
In recent years, new anticancer therapies have accompanied the classical approaches of surgery and radio- and chemotherapy. These new forms of treatment aim to inhibit specific molecular targets namely altered or deregulated proteins, which offer the possibility of individualized therapies.
The specificity and efficiency of these new approaches, however, bring about a number of challenges. First of all, it is essential to specifically identify and quantify protein targets in tumor tissues for the reasonable use of such targeted therapies. Additionally, it has become even more obvious in recent years that the presence of a target protein is not always sufficient to predict the outcome of targeted therapies. The deregulation of downstream signaling molecules might also play an important role in the success of such therapeutic approaches. For these reasons, the analysis of tumor-specific protein expression profiles prior to therapy has been suggested as the most effective way to predict possible therapeutic results. To further elucidate signaling networks underlying cancer development and to identify new targets, it is necessary to implement tools that allow the rapid, precise, inexpensive and simultaneous analysis of many network components while requiring only a small amount of clinical material.
Reverse phase protein microarray (RPPA) is a promising technology that meets these requirements while enabling the quantitative measurement of proteins. Together with recently developed protocols for the extraction of proteins from formalin-fixed, paraffin-embedded (FFPE) tissues, RPPA may provide the means to quantify therapeutic targets and diagnostic markers in the near future and reliably screen for new protein targets.
With the possibility to quantitatively analyze DNA, RNA and protein from a single FFPE tissue sample, the methods are available for integrated patient profiling at all levels of gene expression, thus allowing optimal patient stratification for individualized therapies.
Keywords: Formalin-fixed, paraffin-embedded (FFPE), human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor 1 (EGFR), urokinase-type plasminogen activator (uPA), plasminogen activator inhibitor 1 (PAI-1), personalized cancer therapy, mitogen-activated protein kinase (MAPK)
Introduction
The deregulation of signaling pathways in tumors can lead to enhanced cancer cell growth, proliferation, survival, invasion, and metastasis or reduced apoptosis 1, 2. Such pathways became the focus of the development of targeted cancer therapies during the last decades 3-5. Kinases are of special interest within these systems, either as receptor molecules or downstream regulators of signaling cascades (see Table 1 for an overview of targeted therapies). Examples of receptor kinases to be further discussed in this review are the human epidermal growth factor receptors 1 (EGFR) and 2 (HER2). Both kinases are targets for anticancer drugs and can be analyzed for their expression by clinically approved tests, such as immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). EGFR overexpression due to gene amplification is often found in human cancers; in gliomas, this deregulation is often associated with structural rearrangements leading to in-frame deletions in the extracellular domain of the receptor 6. HER2 overexpression as found in 25 to 30% of human breast cancers can be mediated either by transcriptional activation or gene amplification 7-10. The HER2 status of breast cancer patients does not only have a predictive value, but the receptor itself is also a target for the monoclonal anti-HER2 antibody Trastuzumab 8. Recent evidence has demonstrated that besides being an important therapeutic target in breast cancer, HER2 is a target for the treatment of metastasized gastric cancers 11, 12, and thus, the anticancer drug Herceptin (Trastuzumab) was approved for the treatment of advanced gastric carcinomas. HER2 is currently detected by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH), and protocols for the detection of HER2 in gastric cancers by IHC were recently suggested by Rüschoff et al. 13. However, although HER2 is well-established as a therapeutic target, recent evidence suggests that down-stream signaling molecules may be better predictors for a response to a HER2 directed therapy than the receptor itself. This follows from the fact that the membrane-bound molecule alone does not necessarily lead to an activation of the signaling cascade. For that reason, identifying the activation status of cancer-related signaling cascades might provide a better insight into the mechanisms underlying the success and failure of targeted therapies, thus providing a useful approach to stratify patients for optimal personalized treatment regimens.
Table 1.
Drugs currently used for targeted therapy
| Drug | Tumor type | Target | Detection method |
|---|---|---|---|
| Trastuzumab 3 | Metastatic breast cancer, gastric cancer | HER2 | IHC, FISH, ISH |
| Cetuximab 3 | Metastatic colorectal cancer | EGFR | IHC, FISH |
| Imatinib Mesylate (Gleevec) 3 | CML, GIST with activated c-kit receptor tyrosine kinase, other sarcomas | Bcr/abl, c-kit, PDGFR, | IHC |
| Bevacizumab 3 | Colorectal cancer | VEGF | IHC |
| Gefitinib (Iressa) | Non-small-cell lung cancer | mutant EGFR | Mutational analysis, immunoblotting |
| G3139 (Genta, Berkley) 3 | Hematologic malignancies and malignant melanoma | Antiapoptotic gene bcl-2 | Immunophenotyping by IHC |
| Erlotinib (Tarveca) 3 | Non-small-cell lung cancer | mutant EGFR | Mutational analysis, immunoblotting |
| Rapamycin RAD001 3 | Breast, prostate, renal cancer | mTOR | ELISA |
| BAY43-9006 3 | Melanoma | RAF kinase | Mutational analysis |
| BMS354825 3 | GIST | Kit | IHC |
| Lapatinib 5 | Breast cancer | EGFR, HER2 | IHC, FISH, ISH |
| Sunitinib 5 | Renal cell cancer | VEGFR, PDGFR, cKit, Flt-3 | IHC |
| Pertuzumab 4 | Breast cancer | HER2 | IHC, FISH, ISH |
| Dasatinib 4 | Breast cancer | Bcr/abl | RT-PCR |
Abbreviations: HER2 (human epidermal growth factor receptor 2); EGFR (epidermal growth factor receptor); c-abl (Abelson Murine Leukemia Viral Oncogene Homolog 1); bcr (breakpoint cluster region); Bcr/abl (fusion gene of bcr and abl in Philadelphia Chromosomes); c-kit (stem cell factor-receptor); PDGFR (platelet-derived growth factor receptor); VEGF (vascular endothelial growth factor receptor); bcl-2 (B-cell lymphoma 2); TOR (target of rapamycin); RAF (rapidly growing fibrosarcoma); Flt-3 (fms-like tyrosine kinase receptor-3); IHC (immunohistochemistry); FISH (fluorescence in situ hybridization); ISH (in situ hybridization)
To individualize cancer, biomarker identification has become even more important for the stratification of patients for special treatment regimens. Urokinase-type plasminogen activator (uPA) and its inhibitor plasminogen activator inhibitor‑1 (PAI-1) are prominent examples of such biomarkers. Both are used to provide a more detailed prognosis for nodal-negative breast cancer patients and have reached the highest level of evidence (LOE I) for this purpose 14.
However the level of these two markers can just be determined in fresh-frozen tissue. This is problematic as formalin-fixed, paraffin-embedded (FFPE) tissue is the main source of patient material world wide. Therefore for clinical determination of diagnostic and therapeutic targets new methods are crucially needed to maximize the data that can be gleaned from this kind of tissue. The recently developed method to extract full-length, immunoreactive proteins from FFPE tissues (for review see 15) is a major progress for such approaches and provides the means to quantitate clinically relevant proteins like HER2 and analyze cancer-related pathways by reverse phase protein microarray (RPPA)16.
Because the field of targeted therapy is growing rapidly, it is important to emphasize the necessity of standardization for pre-analytical as well as analytical settings.
In this review, we shed light on new developments for the detection of therapeutic protein targets and diagnostic biomarkers in clinical tissues and comment on suggestions for the standardization of tissue handling and analysis.
Proteomics of formalin-fixed, paraffin-embedded tissues
Formalin is the standard fixative in clinical tissue collections worldwide and the method of formalin fixation was described in detail in a previous article of the authors 17. The protein and nucleic acid cross-linking induced by formalin maintains the tissue in an excellent condition for the pathological examination of the morphological characteristics of diseased tissues 18. With the new interest in macromolecules (DNA, RNA, proteins) as therapeutic targets or biomarkers, scientists began to analyze these entities first in frozen material because FFPE fixation required to preserve the tissue morphology had adverse effects on macromolecules, especially proteins. By adoption of antigen retrieval first described by Ikeda et al. 19 it was possible to develop an assembly of protocols enabling extraction of proteins from FFPE tissues 18-26. An overview of these methods is provided in a recent review by Berg et al., 2010 15. Compared to other recent studies 27-30, using these protocols non-degraded, full-length, and immunoreactive proteins are obtained and may then be used for protein profiling for enhanced diagnosis. However one should keep in mind that all protocols require high SDS concentrations, temperature and pH, which is problematic for some down-stream applications such as ELISA assays. To assure the integrity of isolated proteins we always do a quantification of the total protein obtained by extraction (Bradford-Assay) and additionally check for correct protein masses by control western blots. Other authors suggest controlling for protein integrity by MS 26, 31-33. In the scope of tumor marker research and personalized medicine further aspects of investigation became of major interest: pre-analytical workflow and tissue quality. For molecular diagnostics protein biomarkers need to be precisely measured in high-quality tissue samples. It is known that specimens undergo numerous processing steps from the collection of patient samples to the final diagnostic analysis. This aspect of the sample history is very important with regard to the identification of disease-related biomarkers. But so far there were no detailed investigations concerning for example the influence of temperature and time during transportation, fixation method and storage on the molecular integrity of tissue samples. In order of reproducibility of subsequent diagnosis the impact of pre-analytical parameters on biomolecular integrity and expression needs to be analyzed in detail. Furthermore we need guidelines for the standardized collection, handling, stabilization and storage of biosamples and quality assurance indicators for artificial, post collection changes of biological samples. Two large international initiatives are working toward this goal, one funded by the EU (www.spidia.eu) and one by the US (http://biospecimens.cancer.gov). In Table 2 we highlight the most important problems occurring during the pre-analytical phase of sample preparation.
Table 2.
Pre-analytical parameters influencing the quality of biological samples and hindering the quantitative analysis of potential biomarkers and drug targets
| Pre-analytical parameters | Source of errors | Proposed solutions |
|---|---|---|
| Medication before surgery and type of anaesthesia | Influence on gene or protein expression | Detailed documentation and account for it during analysis |
| Time of vessel ligation | Time is unknown | Detailed documentation |
| Time of specimen removal | Time is unknown | Detailed documentation |
| Location of specimen removal (e.g. centre of a tumour or periphery) | Tumour area not homogeneous | Comparison only between homogeneous pieces, detailed documentation |
| Time and temperature of transport e.g. to pathology | Time and conditions are unknown | As fast as possible, detailed documentation |
| Time until fixation | Time is unknown | As fast as possible, detailed documentation |
| Type and duration of fixation | Cross-linking by formalin | Novel fixation methods |
| Temperature and duration of fixation | Not standardized | Standardized conditions, detailed documentation |
These difficulties are also the main focus of the projects mentioned above. The issue of the evaluation of adverse and variable effects of sample preparation is discussed in more detail in a recent review by Becker and Taylor 34.
Reverse Phase Protein Microarrays
Although morphological parameters, namely tumor size, grade and staging, are the most important parameters for diagnosis, they do not address the complexity and heterogeneity of individual tumors at the molecular level 35, 36. However, since individualized cancer therapies are on gaining ground more detailed patient classifications are necessary to be able to identify the right therapy for each patient. One widely accepted approach is the gene microarray. Anyway, it is important to realize that transcript profiling does not accurately reflect the complexity of cellular protein networks (e.g. protein-protein interactions, protein localization or posttranslational modifications) 37. Additionally, several groups demonstrated that there is no constrained correlation between gene transcript levels and protein expression or the functional state of a protein. 38-41.
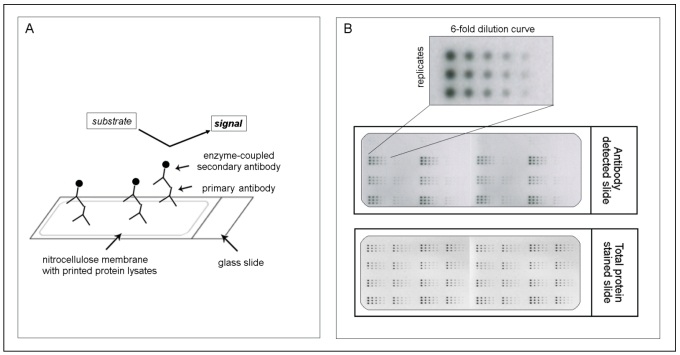
However especially the communication in such a protein network, which is responsible for activation and deactivation of involved proteins, is often altered in cancer and may then lead to aberrant cellular functions that result in proliferation, apoptosis, differentiation, survival, invasion and metastasis. That's why it is of great importance not only to look at the expression of genes or at the activation state of a single protein but to analyze whole protein signaling networks that then can provide fundamental information about the functional state of signaling pathways 42, 43. One method which meets all demands for such a network monitoring is the reverse phase protein microarray (RPPA). This array format allows the simultaneous analysis of multiple samples for the expression of several proteins under the same experimental conditions even from small numbers of tumor cells or small specimens such as biopsies 44, 45. Like this changes in protein expression levels or phosphorylation states, before and after treatment, between disease and non-disease states and between responders and non-responders can be monitored 35. The procedure to generate such an array is simple. After protein extraction from any kind of material (e.g. cells, FFPE tissue or fresh-frozen material), each sample is arrayed in triplicate on nitrocellulose-coated slides using several dilutions to ensure that each analyte/antibody combination can be analyzed in the linear dynamic range. After the spotting each slide may be detected with an antibody against the desired protein (for an overview of the RPPA methodology, see Figure 1). However one should keep in mind that for a successful implementation of RPPA highly specific antibodies are needed as one can't distinguish between different molecular weights (as e.g. in western blot) but only one signal is obtained. Especially for phosphorylated proteins and other posttranslationally modified proteins this seems to be one of the major limiting factors 35, 46.
Figure 1.
The principle of reverse phase protein microarrays (RPPA): A: Protein lysates are spotted onto nitrocellulose-coated glass slides. Single proteins (e.g., PAI-1) can be detected by an antibody assay similar to a western blot analysis; a specific primary antibody binds to the spotted protein. After binding of an enzyme-coupled secondary antibody, protein expression can be measured by light- or fluorescence-based as well as colorimetric methods. B: Each sample is spotted in triplicate and in a six-step dilution series to ensure the quantitative measurement of the target protein in the linear detection range. The normalization of every antibody detected on the slide is performed using a total protein-stained slide (e.g., Sypro-Ruby protein stain).
Another advantage of the RPPA is the possibility to quantify the total amount of protein in the sample via the application of purified recombinant proteins one the same slide 15, 16. This method enables to measure protein expression from FFPE tissues more precisely than it is possible by IHC (e.g., for HER2) and implement the analysis of clinical markers, which until now were only measurable from fresh or frozen tissues (e.g. for uPA/PAI-1). This highly quantitative approach is advantageous for patient selection; however, the correlation between protein abundance and histology, as provided by IHC, is lost.
Nevertheless, we successfully established a set of about 50 antibodies for use in RPPA analysis by stringently selecting for specificity and sensitivity by western blot 15, 16 thus providing a starting point for the analysis of major cancer-related signaling pathways from FFPE tissues of cancer patients. Based on this expertise we recently developed an algorithm to validate antibodies for their simultaneous application to extract based and classical morphology based methods (Schuster C, Malinowsky K et al., in preparation). With this proposal we plan to combine the advantages of both approaches to get broad insight into all aspects of cancer formation and development.
For the application of RPPA generated data for such a purpose in clinical routine it is necessary to be able to compare the results of arrays generated in different hospitals and experimental settings. This can be achieved by reference samples on each slide, to which the signal may be normalized to. Such a control has to be renewable, reproducible in large-scale, successful over a broad range of end points, stable over a long period of time and as closely related to the test sample as possible 47.
To validate the reproducibility of the method, we performed experiments analyzing the variability between sample preparations, array slides and experimental setups (inter-sample as well as inter-assay comparisons) and found the method to be highly reliable 15.
In comparison to the widely used ELISA approaches RPPAs are more sensitive and a two-site antibody sandwich method is not used, hence there won't be any experimental variability introduced due to labeling yield (or) epitope masking.
Deciphering signaling pathways in clinical tissues for personalized medicine
RPPAs can be implemented in nearly all areas regarding the field of personalized medicine. They are used for the rapid and comprehensive analysis of new drug candidates found by in silico approaches or by binding screens for their biological functions 48-50, as well as for biomarker screenings 51.
Many recent studies have demonstrated that RPPA technology is a very promising tool for signaling pathway profiling of human tissues and cell lines to produce valuable information for the development of new therapeutics or patient selection. Feinberg et al. was the first to utilize a microspot technique to detect antigens in serum 52. In 2001, another study used RPPAs to show the activation of pro-survival proteins and pathways during prostate cancer progression 45. In our group, we provided insight into the role of uPA/PAI-1 in cancer related signaling 16. Other prominent examples of cancer-specific signaling deregulation namely the activation of the phosphatidylinositol 3-kinase (PI3K) pathway in a significant number of ovarian and colon tumors 53, 54.and, alterations in the mitogen-activated protein kinase (MAPK) pathway or the overexpression of HER2,8, 53-57 were summarized in an earlier review by the authors 17. These examples demonstrate the diverse signaling-based mechanisms underlying cancer progression and indicate that cancer types (e.g., breast cancer) can be subdivided e.g., into small, well-defined subgroups that express a typical protein profile. Some authors go even further, suggesting that the use of a targeted therapy should not be based on the identity of the cancer but rather on the deregulation of a certain pathway 58. Such an approach would challenge the design of future clinical studies and the approval of new drugs for targeted therapy but could be beneficial for patients by providing highly specific therapies that are only based on the availability of specific targets regardless of the classical characterization of the cancer type. Such a specific treatment could greatly increase the quality of life for patients by minimizing adverse side effects of cancer treatment.
In addition to classical biomarker screenings RPPA is also an adequate tool to compare different types of samples regarding their protein expression profiles. One prominent example is the expression analysis of HER2 from resected tumors versus core needle biopsies. Currently, core biopsies are routinely used for diagnosis of breast cancer and they are often the only sample for providing prognostic and predictive markers before treatment. However, biopsies may not accurately reflect protein expression profiles from the whole tumor. After extraction of full-length proteins from FFPE tissues, Berg et al. used RPPAs to compare HER2, estrogen receptor (ERalpha), and progesterone receptor (PGR) expression levels in a series of 35 FFPE breast cancer surgical specimens and their corresponding core biopsies. We found a high concordance between protein expression in core biopsies and surgical specimens. In this study, the authors could show that HER2, ERalpha, and PGR expression can be assessed reliably on core biopsies of FFPE breast cancer tissues using RRPA. These results might further strengthen the implementation of RPPA technology in routine clinical settings (Berg et al. Protein microarray-based comparison of HER2, estrogen receptor and progesterone receptor status in core biopsies and surgical specimens from FFPE breast cancer tissues. Appl Immunohistochem Mol Morphol., in press 2011).
Our vision: Combined protein and RNA profiling of cancer tissue from the same sample
Gene expression profiles (e.g., genes regulating cell cycle, invasion or metastasis) have been proposed as potential biomarkers for the prognosis, prediction of treatment response and the identification of potentially new drugs 59. In addition to the well-established mRNA profiling, the analysis of small non-coding RNAs called miRNAs has become a focus in biomarker research. In recent years, evidence has suggested that whole groups or families of miRNAs are deregulated in different diseases, especially cancer 60, 61.
The quantitative analysis of protein expression described in this review may enable the parallel identification of protein and RNA profiles (mRNA and miRNA) using only one FFPE tissue sample. With this method, it is possible to simultaneously determine mutations in critical genes and relate them to expression profiles. Gene mutations frequently lead to a loss of gene function, although the mutated gene is still expressed at the RNA and protein levels. A classical example for such a mutation is the cell adhesion molecule E-cadherin, in which a loss of E-cadherin function is found in many diffuse-type gastric cancers, although the protein is expressed at high levels in these tumors 62. Another useful application of the combined DNA and protein analysis is the utilization of EGFR as a therapeutic target; although EGFR expression is detectable on a protein level, many patients with colorectal cancers do not respond to Cetuximab therapy 63-65. The downstream target of EGFR is mutated in many of the non-responders, therefore blocking the EGFR does not inhibit proliferative signaling. Therefore, the parallel assessment of the level of EGFR protein and the mutational status of the EGFR gene is one important prerequisite for successful Cetuximab treatment 63-65. The remaining EGFR-positive patients express the wild-type form of the protein and provide a good example for the need of additional biomarkers for the prediction of therapeutic response. Many of these patients do not respond to Cetuximab therapy, probably due to further alterations in pathways downstream of EGFR. In recent years, several additional candidate biomarkers of EGFR resistance were suggested. Molecular aberrations occur in BRAF, PIK3CA and PTEN, which are known downstream effectors of EGFR 66-69. Somatic mutations in these EGFR effectors correlate with the reduced efficacy of Cetuximab in patients with metastatic colorectal cancers 70, 71. An activating mutation of BRAF, which lies downstream of the EGFR signaling cascade, was found in a proportion of patients with wild-type EGFR that do not respond to Cetuximab treatment 66. For that reason, BRAF mutational analysis is currently recommended by the National Comprehensive Cancer Network (NCCN) clinical guidelines for patients with “wild type” metastatic or recurrent colorectal cancers that are receiving Cetuximab 72.
Recently, we succeeded in the parallel extraction of proteins and RNA, including miRNA, from the same FFPE tissue sample (Malinowsky et al.; unpublished data), demonstrating the proof of principle of our vision. While FFPE samples are a large challenge of the clinical application of the combined analysis of protein and RNA, new formalin-free fixatives, e.g. PAXgene tissue fixation and stabilization reagents 73, HOPE 74-76, RCL2 77-79, Finefix 80, metacarn 81 or Umfix 82 show great potential to serve as novel multimodal fixatives for modern pathology, enabling extensive protein biomarker studies on clinical tissue samples. Regardless of the nature of the fixative, our approach allows the integration of analysis at all three levels of gene expression (DNA, RNA and protein), providing insight into the whole spectrum of potential biomarkers (Figure 2).
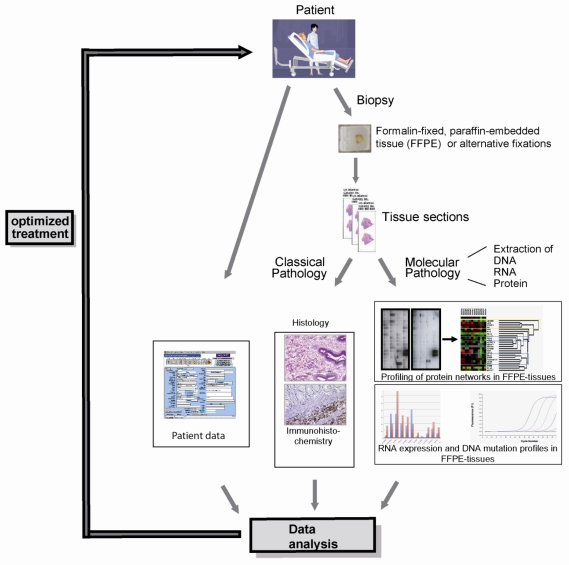
Figure 2.
The integration of DNA, RNA and protein profiling into a routine clinical workflow: The parallel analysis of the morphology as well as DNA, RNA and protein expression profiles can provide a very conclusive description of any given tumor entity, thereby selecting patients for targeted, individualized therapy.
Conclusions
In the scope of new targeted cancer therapy approaches new technologies which meet the issues of fast and precise target detection and quantification are desperately needed. RPPA is a very promising tool in this regard. It has already been implemented in several biomarker studies and becomes even more powerful when combined with the huge sample reservoir provided by FFPE tissues. It is clear now that proteins can be extracted from FFPE tissues and reliably analyzed. We believe that applying these new techniques - probably in conjunction with novel tissue fixatives - will greatly facilitate the search for new biomarkers and therapeutic targets in the near future. More importantly, the great translational potential of the new approaches are apparent as the methodologies discussed can easily combine classic histopathology and modern multiplex protein profiling. However, in depth evaluation of antibodies and the use and design of proper controls for tissue quality are essential.
Acknowledgments
This study is supported by the German Federal Ministry of Education and Research (BMBF) Grant No. 01GR0805 to K.F.B. The authors thank Kerstin Schragner, Kai Tran and Christina Schott for excellent technical support.
References
- 1.Hanahan D, Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Solimini N.L, Elledge S.J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietel M, Sers C. Personalized medicine and development of targeted therapies: The upcoming challenge for diagnostic molecular pathology. A review. Virchows Arch. 2006;448(6):744–55. doi: 10.1007/s00428-006-0189-2. [DOI] [PubMed] [Google Scholar]
- 4.Widakowich C. et al. Molecular targeted therapies in breast cancer: where are we now? Int J Biochem Cell Biol. 2007;39(7-8):1375–87. doi: 10.1016/j.biocel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Widakowich C. et al. Review: side effects of approved molecular targeted therapies in solid cancers. Oncologist. 2007;12(12):1443–55. doi: 10.1634/theoncologist.12-12-1443. [DOI] [PubMed] [Google Scholar]
- 6.Hynes N.E, Lane H.A. receptors ERBB and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 7.Emens L.A. Trastuzumab: targeted therapy for the management of HER-2/neu-overexpressing metastatic breast cancer. Am J Ther. 2005;12(3):243–53. [PubMed] [Google Scholar]
- 8.Piccart M. et al. The predictive value of HER2 in breast cancer. Oncology. 2001;61(Suppl 2):73–82. doi: 10.1159/000055405. [DOI] [PubMed] [Google Scholar]
- 9.Slamon D.J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 10.Slamon D.J. et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 11.Wang J. et al. Pathological complete response after neoadjuvant chemotherapy with trastuzumab-containing regimen in gastric cancer: a case report. J Hematol Oncol. 2010;3(1):31. doi: 10.1186/1756-8722-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan B. et al. A study of HER2 gene amplification and protein expression in gastric cancer. J Clin Pathol. 2010;63(9):839–42. doi: 10.1136/jcp.2010.076570. [DOI] [PubMed] [Google Scholar]
- 13.Ruschoff J. et al. HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch. 2010;457(3):299–307. doi: 10.1007/s00428-010-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janicke F. et al. Randomized adjuvant chemotherapy trial in high-risk, lymph node-negative breast cancer patients identified by urokinase-type plasminogen activator and plasminogen activator inhibitor type 1. J Natl Cancer Inst. 2001;93(12):913–20. doi: 10.1093/jnci/93.12.913. [DOI] [PubMed] [Google Scholar]
- 15.Berg D. et al. Molecular profiling of signalling pathways in formalin-fixed and paraffin-embedded cancer tissues. Eur J Cancer. 2010;46(1):47–55. doi: 10.1016/j.ejca.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Wolff C. et al. Signalling networks associated with urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in breast cancer tissues: new insights from protein microarray analysis. J Pathol. 2010;223(1):54–63. doi: 10.1002/path.2791. [DOI] [PubMed] [Google Scholar]
- 17.Malinowsky K. et al. Deciphering signaling pathways in clinical tissues for personalized medicine using protein microarrays. J Cell Physiol. 2010;225(2):364–70. doi: 10.1002/jcp.22307. [DOI] [PubMed] [Google Scholar]
- 18.Becker K.F. et al. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J Pathol. 2007;211(3):370–8. doi: 10.1002/path.2107. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda K. et al. Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections. J Histochem Cytochem. 1998;46(3):397–403. doi: 10.1177/002215549804600314. [DOI] [PubMed] [Google Scholar]
- 20.Addis M.F. et al. Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. Proteomics. 2009;9(15):3815–3823. doi: 10.1002/pmic.200800971. [DOI] [PubMed] [Google Scholar]
- 21.Becker KF, Mack H, Schott C, Hipp S, Rappl A, Piontek G, Höfler H. Extraction of phosphorylated proteins from formalin-fixed cancer cells and tissues. TOPATJ. 2008;2:44–52. [Google Scholar]
- 22.Becker KF, Schott C, Becker I, Höfler H. Guided protein extraction from formalin-fixed tissues for quantitative multiplex analysis avoids detrimental effects of histological stains. Proteomics Clin Appl. 2008;2:737–743. doi: 10.1002/prca.200780106. [DOI] [PubMed] [Google Scholar]
- 23.Chu W.S. et al. A nondestructive molecule extraction method allowing morphological and molecular analyses using a single tissue section. Lab Invest. 2005;85(11):1416–28. doi: 10.1038/labinvest.3700337. [DOI] [PubMed] [Google Scholar]
- 24.Chung J, Lee S J, Kris Y, Braunschweig T, Traicoff J L, Hewitt SM. A well-based reverse-phase protein array applicable to extracts from formalin-fixed paraffin-embedded tissue. Proteomics Clin Appl. 2008;2(10-11):1539–1547. doi: 10.1002/prca.200800005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nirmalan N.J. et al. Development and validation of a novel protein extraction methodology for quantitation of protein expression in formalin-fixed paraffin-embedded tissues using western blotting. J Pathol. 2009;217(4):497–506. doi: 10.1002/path.2504. [DOI] [PubMed] [Google Scholar]
- 26.Shi S.R. et al. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem. 2006;54(6):739–43. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- 27.Crockett D.K. et al. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 2005;85(11):1405–15. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 28.Hood B.L. et al. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4(11):1741–53. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Palmer-Toy D.E. et al. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4(6):2404–11. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 30.Sage L. Proteomics gets out of a fix. J Proteome Res. 2005;4(6):1903–4. doi: 10.1021/pr0505300. [DOI] [PubMed] [Google Scholar]
- 31.Ostasiewicz P. et al. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 2010;9(7):3688–700. doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 32.Scicchitano M.S. et al. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J Histochem Cytochem. 2009;57(9):849–60. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprung RWJr. et al. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol Cell Proteomics. 2009;8(8):1988–98. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker K.F, Taylor C.R. "Liquid Morphology": Immunochemical Analysis of Proteins Extracted From Formalin-fixed Paraffin-embedded Tissues: Combining Proteomics With Immunohistochemistry. Appl Immunohistochem Mol Morphol. 2010 doi: 10.1097/PAI.0b013e3181f50883. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Wulfkuhle J.D. et al. Technology insight: pharmacoproteomics for cancer--promises of patient-tailored medicine using protein microarrays. Nat Clin Pract Oncol. 2006;3(5):256–68. doi: 10.1038/ncponc0485. [DOI] [PubMed] [Google Scholar]
- 36.Baak J.P. et al. Genomics and proteomics in cancer. Eur J Cancer. 2003;39(9):1199–215. doi: 10.1016/s0959-8049(03)00265-x. [DOI] [PubMed] [Google Scholar]
- 37.Espina V. et al. Protein microarray detection strategies: focus on direct detection technologies. J Immunol Methods. 2004;290(1-2):121–33. doi: 10.1016/j.jim.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Petricoin EF3rd. et al. Medical applications of microarray technologies: a regulatory science perspective. Nat Genet. 2002;32(Suppl):474–9. doi: 10.1038/ng1029. [DOI] [PubMed] [Google Scholar]
- 39.Nishizuka S. et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100(24):14229–34. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calvo K.R, Liotta L.A, Petricoin E.F. Clinical proteomics: from biomarker discovery and cell signaling profiles to individualized personal therapy. Biosci Rep. 2005;25(1-2):107–25. doi: 10.1007/s10540-005-2851-3. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell P. A perspective on protein microarrays. Nat Biotechnol. 2002;20(3):225–9. doi: 10.1038/nbt0302-225. [DOI] [PubMed] [Google Scholar]
- 42.Espina V. et al. Protein microarrays: molecular profiling technologies for clinical specimens. Proteomics. 2003;3(11):2091–100. doi: 10.1002/pmic.200300592. [DOI] [PubMed] [Google Scholar]
- 43.Liotta L.A. et al. Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell. 2003;3(4):317–25. doi: 10.1016/s1535-6108(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 44.Grubb R.L. et al. Signal pathway profiling of prostate cancer using reverse phase protein arrays. Proteomics. 2003;3(11):2142–6. doi: 10.1002/pmic.200300598. [DOI] [PubMed] [Google Scholar]
- 45.Paweletz C.P. et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20(16):1981–9. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 46.Wulfkuhle J. et al. Genomic and proteomic technologies for individualisation and improvement of cancer treatment. Eur J Cancer. 2004;40(17):2623–32. doi: 10.1016/j.ejca.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Sheehan K.M. et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4(4):346–55. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Bartholomeusz C. et al. PEA-15 inhibits tumorigenesis in an MDA-MB-468 triple-negative breast cancer xenograft model through increased cytoplasmic localization of activated extracellular signal-regulated kinase. Clin Cancer Res. 2010;16(6):1802–11. doi: 10.1158/1078-0432.CCR-09-1456. [DOI] [PubMed] [Google Scholar]
- 49.Hong B. et al. Reverse phase protein array identifies novel anti-invasion mechanisms of YC-1. Biochem Pharmacol. 2010;79(6):842–52. doi: 10.1016/j.bcp.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 50.Sikora A.G. et al. Targeted inhibition of inducible nitric oxide synthase inhibits growth of human melanoma in vivo and synergizes with chemotherapy. Clin Cancer Res. 2010;16(6):1834–44. doi: 10.1158/1078-0432.CCR-09-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu M. et al. Ultrasensitive, multiplexed detection of cancer biomarkers directly in serum by using a quantum dot-based microfluidic protein chip. Nano ACS. 2010;4(1):488–94. doi: 10.1021/nn901404h. [DOI] [PubMed] [Google Scholar]
- 52.Feinberg J.G. A 'microspot' test for antigens and antibodies. Nature. 1961;192:985–6. doi: 10.1038/192985a0. [DOI] [PubMed] [Google Scholar]
- 53.Philp A.J. et al. The phosphatidylinositol 3'-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61(20):7426–9. [PubMed] [Google Scholar]
- 54.Shayesteh L. et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21(1):99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 55.Fang J.Y, Richardson B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–7. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 56.Gulmann C. et al. Quantitative cell signalling analysis reveals down-regulation of MAPK pathway activation in colorectal cancer. J Pathol. 2009;218(4):514–9. doi: 10.1002/path.2561. [DOI] [PubMed] [Google Scholar]
- 57.Lobke C. et al. Contact spotting of protein microarrays coupled with spike-in of normalizer protein permits time-resolved analysis of ERBB receptor signaling. Proteomics. 2008;8(8):1586–94. doi: 10.1002/pmic.200700733. [DOI] [PubMed] [Google Scholar]
- 58.Proffitt B. Profiles in Personalized Cancer Therapy. Bio-IT World. 2010.
- 59.van 't Veer L.J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 60.De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for cancer CNS and other disorders. Brain Res. 2010;1338:100–11. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 61.Sioud M, Cekaite L. Profiling of miRNA expression and prediction of target genes. Methods Mol Biol. 2010;629:257–71. doi: 10.1007/978-1-60761-657-3_16. [DOI] [PubMed] [Google Scholar]
- 62.Handschuh G. et al. Tumour-associated E-cadherin mutations alter cellular morphology, decrease cellular adhesion and increase cellular motility. Oncogene. 1999;18(30):4301–12. doi: 10.1038/sj.onc.1202790. [DOI] [PubMed] [Google Scholar]
- 63.Amado R.G. et al. Wild-Type KRAS Is Required for Panitumumab Efficacy in Patients With Metastatic Colorectal Cancer. Journal of Clinical Oncology. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 64.Karapetis C.S. et al. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. New England Journal of Medicine. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 65.Liavre A. et al. KRAS Mutation Status Is Predictive of Response to Cetuximab Therapy in Colorectal Cancer. Cancer Research. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 66.Di Nicolantonio F. et al. Wild-Type BRAF Is Required for Response to Panitumumab or Cetuximab in Metastatic Colorectal Cancer. Journal of Clinical Oncology. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 67.Laurent-Puig P. et al. Analysis of PTEN, BRAF, and EGFR Status in Determining Benefit From Cetuximab Therapy in Wild-Type KRAS Metastatic Colon Cancer. Journal of Clinical Oncology. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 68.Razis E. et al. Potential value of PTEN in predicting cetuximab response in colorectal cancer: An exploratory study. BMC Cancer. 2008;8(1):234. doi: 10.1186/1471-2407-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sartore-Bianchi A. et al. PIK3CA Mutations in Colorectal Cancer Are Associated with Clinical Resistance to EGFR-Targeted Monoclonal Antibodies. Cancer Research. 2009;69(5):1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 70.Loupakis F. et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101(4):715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perrone F. et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Annals of Oncology. 2009;20(1):84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 72.Engstrom P.F, Arnoletti J.P, Benson A.BIII, Chen Y.-J. et al. National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: colon cancer. J Natl Compr Canc Netw. 2009;7(8):778–831. doi: 10.6004/jnccn.2009.0056. [DOI] [PubMed] [Google Scholar]
- 73.Ergin B. et al. Proteomic analysis of PAXgene-fixed tissues. J Proteome Res. 2010;9(10):5188–96. doi: 10.1021/pr100664e. [DOI] [PubMed] [Google Scholar]
- 74.Kahler D. et al. Proteomics out of the archive: Two-dimensional electrophoresis and mass spectrometry using HOPE-fixed, paraffin-embedded tissues. J Histochem Cytochem. 2010;58(3):221–8. doi: 10.1369/jhc.2009.954065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vollmer E. et al. The HOPE technique opens up a multitude of new possibilities in pathology. Rom J Morphol Embryol. 2006;47(1):15–9. [PubMed] [Google Scholar]
- 76.Witchell J. et al. isolation RNA and quantitative from HOPE- PCR and formalin-fixed bovine lymph node tissues. Pathol Res Pract. 2008;204(2):105–11. doi: 10.1016/j.prp.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 77.Belief V. et al. Proteomic analysis of RCL2 paraffin-embedded tissues. J Cell Mol Med. 2008;12(5B):2027–36. doi: 10.1111/j.1582-4934.2008.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bellet V. et al. Proteomic analysis of RCL2 paraffin-embedded tissues. J Cell Mol Med. 2008 Oct;12(5B):2027–36. doi: 10.1111/j.1582-4934.2008.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mange A. et al. Liquid chromatography-tandem and MALDI imaging mass spectrometry analyses of RCL2/CS100-fixed, paraffin-embedded tissues: proteomics evaluation of an alternate fixative for biomarker discovery. J Proteome Res. 2009;8(12):5619–28. doi: 10.1021/pr9007128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stanta G. et al. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive's tissues. Diagn Mol Pathol. 2006;15(2):115–23. doi: 10.1097/00019606-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 81.Dotti I. et al. Effects of formalin, methacarn, and fineFIX fixatives on RNA preservation. Diagn Mol Pathol. 2010;19(2):112–22. doi: 10.1097/PDM.0b013e3181b520f8. [DOI] [PubMed] [Google Scholar]
- 82.Vincek V. et al. A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab Invest. 2003;83(10):1427–35. doi: 10.1097/01.lab.0000090154.55436.d1. [DOI] [PubMed] [Google Scholar]