Abstract
Purpose
A prior study showed that exogenous galectin-3 (Gal-3) stimulates re-epithelialization of corneal wounds in wild-type (Gal-3+/+) mice but, surprisingly, not in galectin-3–deficient (Gal-3−/−) mice. In an effort to understand why the injured corneas of Gal-3−/− mice are unresponsive to exogenous Gal-3, the present study was designed to determine whether genes encoding the enzymes that regulate the synthesis of glycan ligands of Gal-3 are differentially expressed in Gal-3−/− corneas compared with the Gal-3+/+ corneas.
Methods
Glycogene microarray technology was used to identify differentially expressed glycosyltransferases in healing Gal-3+/+ and Gal-3−/− corneas.
Results
Of ~2000 glycogenes on the array, the expression of 8 was upregulated and that of 14 was downregulated more than 1.3-fold in healing Gal-3−/− corneas. A galactosyltransferase, β3GalT5, which has the ability to synthesize Gal-3 ligands was markedly downregulated in healing Gal-3−/− corneas. The genes for polypeptide galactosaminyltransferases (ppGalNAcT-3 and -7) that are known to initiate O-linked glycosylation and N-aspartyl-β-glucosaminidase, which participates in the removal of N-glycans, were found to be upregulated in healing Gal-3−/− corneas. Microarray data were validated by qRT-PCR.
Conclusions
Based on the known functions of the differentially expressed glycogenes, it appears that the glycan structures on glycoproteins and glycolipids, synthesized as a result of the differential glycogene expression pattern in healing Gal-3−/− corneas may lead to the downregulation of specific counterreceptors for Gal-3. This may explain, at least in part, why, unlike healing Gal-3+/+ corneas, the healing Gal-3−/− corneas are unresponsive to the stimulatory effect of exogenous Gal-3 on re-epithelialization of corneal wounds.
Re-epithelialization is the first step in the wound-repair process. Impaired or delayed re-epithelialization and associated nonhealing epithelial defects and ulceration constitute a serious medical problem in many organs, including cornea, skin and the gastrointestinal tract.1-4 In most cases, defects in cell migration over the wound bed, rather than cell proliferation, contribute to the failure of re-epithelialization. 5,6 Cell migration is a complex cyclical process in which dynamic changes in cell–matrix interactions play a crucial role.7 More recent studies have suggested that the members of the galectin class of β-galactoside– binding proteins have the potential to mediate cell–matrix interactions by a novel, carbohydrate-based, recognition system (reviewed in Refs. 8, 9). To date, 15 mammalian galectins have been identified (galectin-1 to -15). Among the members of the galectin family, galectin-3 (Gal-3) is structurally unique and contains a single carbohydrate recognition domain (CRD) connected via a collagen-like domain to a non-lectin N-terminal region that promotes oligomerization of the lectin. It is expressed in a variety of inflammatory and epithelial cells including corneal and skin epithelial cells.8-12 Recent studies in our laboratory have shown that: (1) Gal-3 is expressed in mouse corneal epithelium at sites of cell–matrix and cell– cell adhesion, (2) Gal-3 expression is increased at the leading edge of the migrating epithelium of healing corneas, (3) re-epithelialization of corneal wounds is significantly slower in galectin-3– deficient (Gal-3−/−) mice than in wild-type (Gal-3+/+) mice, and (4) exogenous Gal-3 stimulates re-epithelialization of corneal wounds in the wild-type mouse animal model. Of particular interest is our finding that exogenous Gal-3 accelerates re-epithelialization of wounds in Gal-3+/+ mice but, surprisingly, not in the Gal-3−/− mice.13 The reason that Gal-3−/− mice are unresponsive to Gal-3-induced wound closure is not well understood. In a prior study, we have shown that the CRD of Gal-3 is directly involved in the beneficial effect of Gal-3 on corneal wound closure in Gal-3+/+ mice.13 Also, it is well established that the extracellular functions of Gal-3 are mediated by a carbohydrate-mediated interaction between the lectin and its counterreceptors (i.e., glycoproteins, which bear the saccharide ligands of galectins) on the cell surface or in extracellular matrix (ECM).8,9 Thus, the lack of efficacy of exogenous Gal-3 on re-epithelialization of corneal wounds in Gal-3−/− mice may suggest that the Gal-3−/− mice are deficient in the counterreceptors of the lectin itself. Accordingly, we hypothesize that Gal-3 in fact modulates the expression of glycosyltransferases, which, in turn, regulate glycosylation of the proteins that serve as cell surface or ECM counterreceptors of Gal-3 itself. To test this hypothesis, in the present study, we used glycogene microarrays to compare the expression profiles of glycosyltransferases and glycosidases in Gal-3−/− and Gal-3+/+ healing corneas. We report here for the first time that compared with healing Gal-3+/+ corneas, healing Gal-3−/− corneas show a distinct glycogene expression pattern that potentially leads to the downregulation of Gal-3-specific glycans.
Methods
Animals and Wound-Healing Experiments
All animal experiments conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and to the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Gal-3−/− mice generated by targeted disruption14 were kindly provided by Fu-Tong Liu (University of California Davis School of Medicine, Sacramento, CA). Six- to eight-week-old Gal-3−/− and Gal-3+/+ mice (10 animals/group in triplicate, a total of 30 animals) were used. Before the corneas were wounded, the mice were anesthetized by an intraperitoneal injection of 1.25% Avertin (0.2 mL/10 g body weight; Aldrich Chemical Co., Milwaukee, WI). Proparacaine eye drops (Alcain; Alcon Laboratories, Inc., Fort Worth, TX) were applied to the cornea as a topical anesthetic. We created 2-mm corneal wounds on the right eye of each animal by transepithelial ablation (2-mm optical zone; 42–44-μm ablation depth, phototherapeutic keratectomy mode) with an excimer laser (Apex Plus; Summit Technology, Waltham, MA), whereas the left eye served as the unwounded control. Buprenorphine (intramuscular, 0.2 mL of 0.3 mg/mL Buprenex; Reckitt and Colman Pharmaceuticals, Inc., Richmond, VA) was used to control pain after the surgery. In addition, antibiotic ointment (Vetropolycin; Pharmaderm, Melville, NY) was applied. The corneas were allowed to heal partially in vivo for 18 to 22 hours, and the animals were then killed. The corneas were excised and immediately placed in liquid nitrogen until used.
RNA Isolation
Total RNA isolation was performed per the manufacturer’s recommendations (RNeasy; Qiagen, Chatsworth, CA). In brief, frozen corneas were homogenized, suspended in guanidine thiocyanate buffer (Buffer RLT) and loaded onto a column (QIAshredder; Qiagen). Eluent from the column was subjected to RNA extraction, and RNA that selectively bound to the silica gel–based column was eluted with RNase-free water. The yield and quality of samples was determined on a bioanalyzer (RNA 6000 LabChip kit; model 2100 bioanalyzer; Agilent Technologies, Waldbronn, Germany).
The average yield of total RNA from each group of 10 healing Gal-3+/+ and Gal-3−/− corneas was 8.8 ± 1.3 μg (n = 3) and 9.5 ± 1.3 μg (n = 3), respectively. The quality of RNA was good across samples with an rRNA ratio (28S/18S) of 1.7 (Fig. 1A). The representative electropherograms and gel-like images are shown in Figure 1B, which demonstrates that 18S and 28S gel bands and graph peaks are dominant, and the amount of low-molecular-weight RNA is insignificant, indicating minimal RNA degradation in the samples. Thus, all RNA preparations used in this study were deemed satisfactory for hybridization to the chips.
Figure 1.
RNA quality from healing Gal-3+/+ and Gal-3−/− mouse corneas used in this study was satisfactory. (A) Table showing yield and 28S/18S ratios of all RNA preparations used in the study. (B) Representative electropherograms and gel-like images of RNA preparations of healing Gal-3+/+ (Bi) and Gal-3−/− (Bii) corneas. Note that in all samples, 18S and 28S gel bands and graph peaks are dominant.
Microarray Experiment and Data Analysis
The hybridization probes were prepared according to the protocol described earlier.15 In brief, RNA samples were subjected to two rounds of T7 RNA polymerase amplification, and the resultant cDNA was used as the starting material for in vitro transcription incorporating biotin-labeled ribonucleotides. Labeled cRNA was hybridized to a glycogene microarray (GLYCOv2; Affymetrix, Santa Clara, CA). All arrays were then scanned (model 3000; Affymetrix).
The GLYCOv2 gene chip is an oligonucleotide microarray, custom made by Affymetrix for the Consortium for Functional Glycomics at the Scripps Institute. The array contains probe sets for ~2000 murine and human gene transcripts representing glycosyltransferases, glycosidases, and other glycan-processing enzymes related to nucleotide synthesis and transport, proteoglycans, glycan-binding proteins, cytokines, chemokines, and several housekeeping proteins. A full list of the genes monitored by the array is available at http://www.functionalglycomics.org/static/consortium/resources/resourcecoree.shtml. In this array, three identical probe sets were used to detect the transcripts for each glycogene. Each probe set consisted of eleven 25-bp perfect-match and eleven 25-bp mismatched oligonucleotide probe pairs. The mismatched oligos have a single-base mismatch at the center position.
The RMA algorithm was used to generate expression signal values (http://www.stat.berkeley.edu/~bolstad/RMAExpress/RMAExpress.html/ provided in the public domain by the University of California Berkeley, Berkeley, CA). This algorithm models the performance of the perfect match probe sets with all chips used in this study. Quantile normalization and background subtraction were performed to generate base 2 log-transformed expression values for all probe sets used in the analysis. Then, the gene expression patterns in the three replicates of each group (Gal-3−/− and Gal-3+/+ corneas) were analyzed using hierarchical clustering function in BRB ArrayTools 3.2.2 software (http://linus.nci.nih.gov/BRB-ArrayTools.html/ provided in the public domain by the National Cancer Institute, Frederick, MD).
The differential gene expressions in healing Gal-3−/− and Gal-3+/+ mouse corneas were analyzed as described by Diskin et al.15 In brief, a single-expression value for each probe set was generated by averaging transformed expression values for replicated probe sets. The BRB ArrayTools 3.2.2 software was used to identify statistically significant changes in gene expression. The class comparison test was conducted with a randomized variance model and a multivariate, permutation-based, false-discovery rate calculation. The false-discovery rate calculation was set at a confidence level of 80%, and the predicted proportion of false discoveries was preset at 10%.
The differentially expressed genes were visualized by heat maps generated using Cluster and Tree View software (http://rana.lbl.gov/EisenSoftware.htm/ provided in the public domain by Eisen Lab, University of California Berkeley) and were subjected to further analysis to gain insight into their biological functions. For this, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) software (http://apps1.niaid.nih.gov/david/), GeneCards (http://www.genecards.org/) and Entrez gene (http://www.ncbi.nlm.nih.gov/entrez) were used (provided in the public domain by the National Institutes of Health (DAVID and Entrez) and Xennex, Inc. (Cambridge MA. Provided without charge to academic institutions. Others must obtain a license.).
Quantitative RT-PCR Analysis
qRT-PCR was performed on a real-time PCR system (model Mx3000P; Stratagene, La Jolla, CA). Briefly, cDNA was synthesized from 100 ng total RNA (High Capacity kit; Applied Biosystems [ABI], Foster City, CA) according to the manufacturer’s instructions. Real-time PCR was performed in triplicate with 2.5 μL of cDNA (derived from 2.5 ng total RNA), MGB probes and primer sets (TaqMan; ABI), and PCR master mix (20-μL total reaction volume; TaqMan Universal; ABI). The ABI inventory primer sets used included ribosomal protein L8 (RPL8) (Mm00657299_m1), β1,3-galactosyltransferase (β3GalT5) (Mm00473621_s1), polypeptide N-acetylgalactosaminyltransferase 3 (ppGalNAcT-3) (Mm00489348_m1), polypeptide N–acetylgalactosaminyltransferase 7 (ppGalNAcT-7) (Mm00519998_m1), N-aspartylglucosaminidase (Mm01208044_m1), cytidine monophospho-N-acetylneuraminic acid hydroxylase (cmah) (Mm01133427_m1), interleukin-(IL)1β (Mm004324228_m1), and mannose receptor, C type 2 (Endo180) (Mm00485184_m1). Reactions performed in the absence of template served as the negative control. After an initial denaturation step (95°C for 10 minutes), the reactions were subjected to 50 cycles involving denaturation (95°C for 15 seconds) and annealing plus extension (60°C for 1 minute). Fluorescent signals were recorded with a detector corresponding to FAM, and data analysis was performed (Mx3000P software; Stratagene). The FAM fluorescent signals were measured against the ROX (internal reference dye) signal to normalize the non–PCR-related fluctuations. The amplification plots showing the increase in FAM fluorescence with each cycle of PCR (ΔRn) were generated for all samples, and the threshold cycle values (Ct) were calculated for all samples. Quantification data of each gene were normalized to the expression of a housekeeping gene, RPL8. A value of 1.0 was assigned to the expression level of each gene in the healing Gal-3+/+ mouse corneas, which served as the calibrator. The values for healing Gal-3−/− mouse corneas were expressed as a change in expression levels (x-fold) with respect to Gal-3+/+ mouse corneas.
Results
Gene Expression Profiles of Healing Gal-3−/− and Gal-3+/+ Corneas
To identify glycogenes that are differentially expressed in healing Gal-3−/− compared with healing Gal-3+/+ mouse corneas, biotinylated cRNA was hybridized to the microarray (GLY-COv2; Affymetrix), and expression signal values were obtained by using the RMA algorithm. An unsupervised hierarchical clustering by sample, which provides a step-wise analysis of the similarity in overall gene expression profiles between individual samples, was used to elucidate the relationship between sample data sets. The relationship is presented graphically in a dendrogram (Fig. 2), wherein the samples cluster closer to each other the more their respective gene expression patterns resemble one another. As shown in Figure 2, all healing Gal-3−/− cornea samples clustered together on one side of the dendrogram, whereas all Gal-3+/+ cornea samples clustered on the other side of the dendrogram, suggesting that there are differences in the expression profile between the healing Gal-3−/− and healing Gal-3+/+ corneas. The short branches within each of the three replicates for healing Gal-3+/+ and Gal-3−/− corneal tissues show that there was high correlation among samples.
Figure 2.
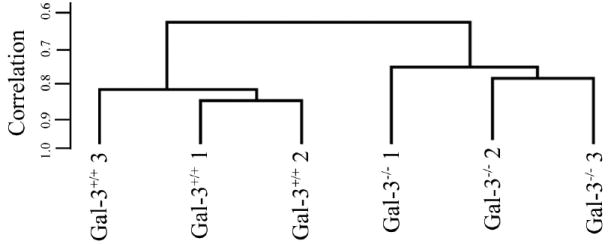
Dendrogram showing hierarchical cluster analysis of gene expression profiles of Gal-3+/+ and Gal-3−/− mouse corneas. The individual samples are clustered in the branches of the dendrogram based on overall similarity in patterns of gene expression. All healing Gal-3−/− corneas clustered together on the right side of the dendrogram, whereas healing Gal-3+/+ corneas clustered on the left.
Differentially Expressed Glycogenes in Gal-3−/− Corneas after Excimer Laser Injury
Of the ~ 2000 genes on the microarray, the expression of 8 was upregulated and that of 14 was downregulated more than 1.3-fold in the healing Gal-3−/− compared with the healing Gal-3+/+ corneas (parametric P < 0.01; Table 1). These differentially expressed genes were visualized using the Cluster and Tree View software for heat map creation (Fig. 3). These analyses also revealed that healing Gal-3−/− corneas show a distinct profile of glycogene expression from that of the healing Gal-3+/+ corneas. All 22 differentially expressed genes are listed in Table 1. For clarity, these genes are grouped according to their involvement in specific cellular processes or functions as determined using GeneCard, Entrez gene, and David software. Among the differentially expressed genes in healing Gal-3−/− corneas, the largest group (41%) were glycosyltransferases and glycosidases that regulate glycosylation of proteins and lipids. Specifically, the expression of a galactosyltransferase, β3-galactosyltransferase 5 (β3GalT5) that synthesizes Galβ1,3GlcNAc (type I chain) to create lactosamine repeats on glycoproteins and glycolipids and thereby has the potential to synthesize ligands for Gal-3,16-18 was downregulated in healing Gal-3−/− compared with Gal-3+/+ corneas. The enzymes that were upregulated include polypeptide N-acetylgalactosaminyltransferases-3 and -7 (ppGalNAcTs-3 and -7) that initiate mucin-type O-glycosylation19 and N-aspartylglucosaminidase that removes N-glycans.20 The upregulation of ppGalNAcTs and N-aspartylglucosaminidase in healing Gal-3−/− corneas may result in glycoproteins that are hyperglycosylated with O-glycans and underglycosylated with N-glycans. Furthermore, the expression of genes that are well known to be upregulated during wound healing were largely downregulated in healing Gal-3−/− compared with Gal-3+/+ corneas. These genes include transforming growth factor-β receptor I (TGF-βRI), interleukin-(IL)1β, erbB3, a member of family of epidermal growth factor receptors, mincle, fibromodulin, sulfatase 1 (SULF1), and Endo180 (Table 1).
Table 1.
Genes Differentially Expressed in Healing Gal-3−/− Compared to Healing Gal-3+/+ Mouse Corneas*
| Difference (x-Fold) | Gene Name | Accession Number |
|---|---|---|
| Cell Adhesion Proteins and Receptors | ||
| 1.7 | Gap junctional protein, gamma 1 (Gjc1) | AF283254 |
| −1.5 | C-type lectin domain family 4, member e (Mincle) | BC003218 |
| −1.5 | Galectin-1 (Lgals1) | NM_008495 |
| Cytokines/Growth Factors and Receptors | ||
| 1.4 | Glial cell line derived neurotrophic factor (GDNF) | NM_010275 |
| −1.7 | Transforming growth factor, beta receptor 1 (TGF-βR1) | NM_009370 |
| −1.5 | Interleukin-1 beta (IL-1β) | M15131 |
| −1.4 | INF-gamma R2 (IFN-γ R2) | NM_008338 |
| −1.4 | ErbB3 | L47240 |
| Regulators of Glycosylation | ||
| 1.5 | N-aspartylglucosaminidase (AGA) | AI853851 |
| 1.6 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (ppGalNAcT3) | NM_015736 |
| 1.4 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (ppGalNAcT7) | AF349573 |
| 1.5 | UDP-N-acetyl-alpha-D-galactosamine:(N-acetylneuraminyl)-galactosylglucosylceramide-beta-1, 4-N-acetylgalactosaminyltransferase (β4GalNAcT2) | NM_008080 |
| 1.5 | Beta-1,3-N-acetylgalactosaminyltransferase 2 (β3GalNAcT2) | AK084275 |
| −1.7 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (β3GalT5) | NM_033149 |
| −1.5 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (ST6GalNAc1) | NM_011371 |
| −1.4 | Cytidine monophospho-N-acetylneuraminic acid hydroxylase (cmah) | NM_007717 |
| −1.4 | Golgi UDP-GlcNAc transporter (SLC35A3) | BC024110 |
| Extracellular Matrix Proteins | ||
| −2.1 | Fibromodulin (Fmod) | X94998 |
| Miscellaneous Transducers, Effectors and Modulators | ||
| 1.4 | Killer cell lectin-like receptor, subfamily A, member 18 (Ly49R) | AF288377 |
| −1.5 | Phosphoglucomutase 1 (PGM1) | BC008527 |
| −1.7 | Sulfatase 1 (SULF1) | NM_172294 |
| −1.4 | Mannose receptor, C type 2 (Endo180) | U56734 |
Genes are considered differentially expressed when there is a difference of >1.3-fold between the geometric mean signal of the healing Gal-3−/− group (n = 3) and the healing Gal-3+/+ group (n = 3). Results of glycogenes marked in bold were confirmed by qRT-PCR.
Figure 3.
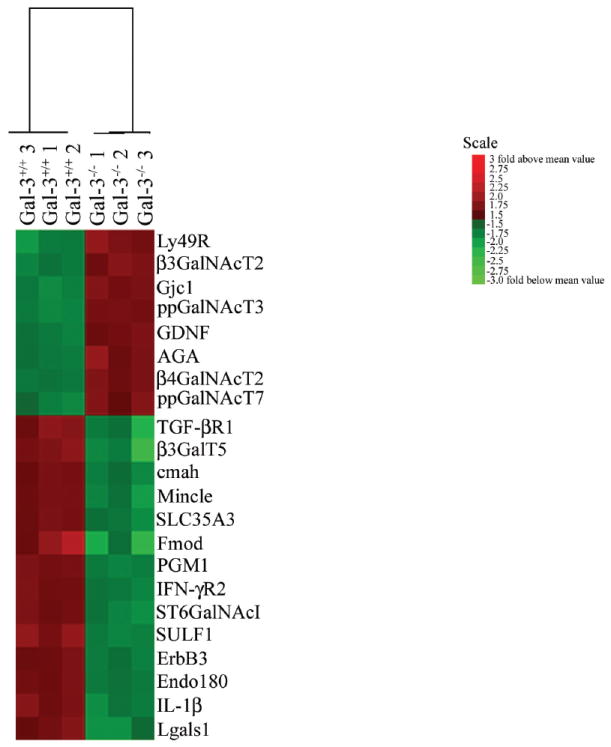
Heat map of 22 differentially expressed genes (change, >1.3-fold; P < 0.01). All signals are compared to a median value, and change (x-fold) from the median is visually represented by color assignment (Scale at right). Healing Gal-3-deficient (Gal-3−/−) and healing wild-type (Gal-3+/+) corneas showed visibly distinct profiles of gene expression.
Validation of Differentially Expressed Genes by qRT-PCR
We used gene-specific qRT-PCR to confirm the differential expression of seven selected genes (Table 2). Of these seven genes, four were upregulated and three were downregulated based on the microarray analysis. We chose these seven genes because they are likely to have an impact on the expression of Gal-3 counterreceptors and are therefore most relevant to the present study. RPL8, a housekeeping gene, was used as a reference gene in the present study, as its expression was similar between Gal-3+/+ and Gal-3−/− healing mouse corneas (change, 1.03-fold). The qRT-PCR showed expression patterns very similar to those obtained by glycogene array hybridization for six of the seven genes tested (86%; Table 2). In agreement with the microarray data, we quantified significant upregulation of ppGalNAcT-3 and -7, and N-aspartylglucosaminidase, and significant downregulation of β3GalT5, IL-1β, and Endo180 in healing Gal-3−/− compared to Gal-3+/+ corneas by qRT-PCR (Table 2). On the other hand, cmah mRNA levels were found to be 1.4-fold upregulated in healing Gal-3−/− corneas by qRT-PCR, which contradicted the microarray analyses (1.4-fold downregulation). No PCR products were amplified in control reactions performed in the absence of reverse transcriptase.
Table 2.
Comparison of Differential Expression Data Obtained by Microarray Hybridization and qRT-PCR
| Gene | qRT-PCR* | Glycogene Microarrays |
|---|---|---|
| β1,3-galactosyltransferase 5 (β3GalT5) | 1.4↓ | 1.7↓ |
| UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 3 (ppGalNAcT3) | 1.7↑ | 1.6↑ |
| UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 (ppGalNAcT7) | 1.3↑ | 1.4↑ |
| N-Aspartylglucosaminidase | 1.5↑ | 1.4↑ |
| Cytidine monophospho-N-acetylneuraminic acid hydroxylase (cmah) | 1.4↑ | 1.4↓ |
| Interleukin-1 β (IL-1 β) | 1.4↓ | 1.5↓ |
| Mannose receptor, C type 2 (Endo180) | 1.4↓ | 1.4↓ |
Data are x-fold change.
Change in expression after normalization to housekeeping gene, RPL8.
Discussion
The goal of the present study was to determine whether glycosyltransferases and glycosidases are differentially expressed in the healing Gal-3−/− compared with the Gal-3+/+ mouse corneas. Although creative strategies have been implemented to select differentially expressed genes, none has gained widespread acceptance in the analysis of microarray data and, to date, the change cutoff (x-fold) used is arbitrary. In the present study, differentially expressed genes were selected using the cutoff value of >1.3-fold. At first sight, it may appear that the changes detected in the present study (1.4- to 2.1-fold), while statistically significant (P < 0.01), are not very large. This is, however, not surprising. Large changes are expected if the groups being compared are dramatically different such as normal versus healing corneas.18 On the other hand, low changes may be expected if the groups being compared are related. In the present study, the two groups (Gal-3+/+ and Gal-3−/−) are both healing corneas, and, therefore, changes are likely to be subtle. Thus, the arbitrary cutoff value of >1.3-fold (P < 0.01) was used. Many published studies have reported statistically and biologically significant microarray data using a low cutoff rate similar to that used in the present study.21-26 Moreover, many differential gene expression studies have shown low changes in glycogenes, specifically glycosyltransferases, to be biologically relevant.27-30 For example, Comelli et al.29 showed that a 1.3- to 1.5-fold decrease in ST3GalI expression between fresh and activated T cells (and B cells) is sufficient to cause hyposialylation of O-glycans.
Comparison of gene expression profiles revealed that healing Gal-3−/− mouse corneas have a unique glycogene expression pattern that defines them as a group distinct from the healing Gal-3+/+ mouse corneas (Figs. 2, 3). A major finding of the present study is that β3GalT5 is downregulated in healing Gal-3−/− corneas. Type 1 chain oligosaccharides found in N- and O-glycans, as well as in glycolipids synthesized by β1,3GalTs, contain the distinctive Galβ1,3GlcNAc disaccharide as their core structure.31 The biological role of β1,3GalT5 includes type I chain elongation of core 2 and core 3 O-glycans, N-glycans,16,17 lactoceramides,31 and globosides32 and create lactosamine residues, which are known to have affinity for Gal-3.8,9 In the wild-type mice, β1,3GalT5 is markedly upregulated (~12-fold) in healing mouse corneas compared with the normal, unwounded corneas.18 The downregulated expression of β1,3GalT5 in healing Gal-3−/− corneas suggests that the glycan ligands on glycoproteins and glycolipids specific for Gal-3 may be reduced in Gal-3−/− corneas, which may be the reason for the absence of effects of exogenous Gal-3 in promoting wound closure in Gal-3−/− mouse corneas.
Another interesting result of the microarray analysis is that the expression of N-acetylgalactosaminyltransferases, ppGalNAcT-3 and -7 is upregulated in Gal-3−/− healing corneas compared with the healing Gal-3+/+ corneas. These enzymes belong to a family of polypeptide N-acetylgalactosaminyltransferases (ppGalNAcTs) that add GalNAc to the Ser/Thr residues of the peptide backbones, and thereby initiate the synthesis of O-glycans and regulate the density and position of O-linked oligosaccharides in glycoproteins. 19 In human, 24 putative ppGalNAcTs have been identified, and each isoform varies in its spatial and temporal regulation.33 Different ppGalNAcTs show preferences for different polypeptide sequences, and some of them—for example, ppGalNAcT-3 and -7, require the presence of O-glycans on the polypeptide before its action to add O-glycan chains at new locations.34,35 The upregulation of these galactosaminyltransferases in Gal-3−/− corneas suggests that some glycoproteins in Gal-3−/− mouse corneas may be hyperglycosylated with O-glycans. However, based on our previous study showing that exogenous Gal-3 does not stimulate re-epithelialization in Gal-3−/− corneas,13 it appears that the hyperglycosylated glycoproteins, because of overexpression of ppGalNAcTs, if present in Gal-3−/− mice during re-epithelialization, are not serving as wound-healing–relevant counterreceptors for Gal-3. Nevertheless, this is the first report to demonstrate upregulation of ppGalNAcTs in Gal-3−/− mice, and its significance in the context of the role of galectins in the regulation of glycosyltransferases is worthy of further investigation.
A lysosomal enzyme that is involved in the catabolism of N-glycosylated proteins, N-aspartyl-β-glucosaminidase,20 was upregulated in the healing Gal-3−/− corneas. We note that N-aspartyl-β-glucosaminidase expression is upregulated in a congenital disorder of glycosylation type 1, wherein the majority of the proteins are underglycosylated.36 Therefore, it is tempting to hypothesize that many glycoproteins may be underglycosylated with N-glycans in the healing Gal-3−/− corneas, potentially explaining the lack of the stimulatory effect of exogenous Gal-3 on wound closure in Gal-3−/− corneas.
In summary, we have demonstrated that the expression pattern of glycogenes encoding enzymes that regulate glycosylation is aberrant in healing Gal-3−/− corneas. Based on the known function of the differentially expressed glycogenes in healing Gal-3−/− corneas compared with healing Gal-3+/+ corneas, it appears that the glycan structures on glycoproteins and glycolipids, synthesized as a result of the differential glycogene expression pattern in healing Gal-3−/− corneas, have a different glycoform (a glycoform is defined as a distinct combination of glycan structures on a glycoprotein) which may lead to the downregulation of specific counterreceptors for Gal-3. In the future, we are planning to use the glycan profiling approach to identify differentially expressed N- and O-linked glycans in healing Gal-3−/− and Gal-3+/+ corneas to establish the extent of correlation between the glycogene and glycan expression patterns.
As regards the reason Gal-3−/− mice are unresponsive to the stimulatory effect of exogenous Gal-3 and why the rate of re-epithelialization of wounds is delayed in Gal-3−/− mice, clearly, the idea that Gal-3 itself modulates the expression of its glycan counterreceptors, which, in turn, regulate the function of the lectin is very attractive. However, other equally appealing possibilities exist. For example, the lectin may influence the expression of key proteins (e.g., integrins)37 that play a role in the re-epithelialization of corneal wounds.38 In this respect, apart from the enzymes regulating glycosylation, the present study demonstrates that healing Gal-3−/− corneas show differential expression of several genes, including those coding for TGF-βRI, IL-1β, erbB3, mincle, fibromodulin, and Endo180.
IL-1β, which is known to be upregulated during corneal wound healing and play a key role in the healing process,39,40 was found to be downregulated in healing Gal-3−/− compared to Gal-3+/+ corneas. Relevant to this, Jawahara et al.41 have also reported that in an animal model of colitis, a marked reduction in the secretion of IL-1β occurs in Gal-3−/− mice compared with the Gal-3+/+ mice.
In the present study, Endo180 expression was also found to be downregulated in healing Gal-3−/− corneas. Endo180 is a membrane glycoprotein, and its expression is upregulated during skin wound healing.42 In endosomes, Endo180 regulates myosin-light chain activation and contractile signals to promote adhesion disassembly.43 In a recent study, we have demonstrated that Gal-3 promotes cell scattering44 and migration of corneal epithelial cells and Endo180 binds to Gal-3 in a β-lactose–inhibitable manner (Saravanan and Panjwani, unpublished data, 2008). These findings in conjunction with our earlier observation that re-epithelialization of corneal wounds is impaired in Gal-3−/− mice13 and the observation reported in the present study that Endo180 is expressed in reduced amounts in healing Gal-3−/− corneas compared with Gal-3+/+ corneas lead us to speculate that during re-epithelialization of wounds, Gal-3 binds to Endo180 to promote cell contraction, which is a critical step in cell migration.7
Regarding how Gal-3 regulates gene expression, it is known that the lectin, as an extracellular protein, regulates the function of transcription factors by modulating the signal transduction pathways mediated by cell surface receptors.45 Also, as a nuclear matrix protein, Gal-3 may directly modulate gene expression through the regulation of transcription and/or mRNA splicing.46-49 Future studies should shed more light on the molecular mechanism involved in Gal-3-mediated regulation of gene expression, specifically glycosyltransferases and glycosidases, to synthesize the glycan ligands of the lectin itself during wound healing.
Acknowledgments
Supported by National Eye Institute Grant EY007088 (NP), a core grant for vision research P30EY13078, New England Corneal Transplant Fund, Mass Lions Eye Research fund, and a challenge grant from Research to Prevent Blindness. The gene microarray was conducted by the Gene Microarray (E) Core of The Consortium for Functional Glycomics funded by the National Institute of General Medical Sciences Grant GM62116.
Footnotes
Disclosure: C. Saravanan, None; Z. Cao, None; S.R. Head, None; N. Panjwani, None
References
- 1.Ma JJ, Dohlman CH. Mechanisms of corneal ulceration. Ophthalmol Clin North Am. 2002;15:27–33. doi: 10.1016/s0896-1549(01)00017-7. [DOI] [PubMed] [Google Scholar]
- 2.Lu L, Reinach PS, Kao WW. Corneal epithelial wound healing. Exp Biol Med (Maywood) 2001;226:653–664. doi: 10.1177/153537020222600711. [DOI] [PubMed] [Google Scholar]
- 3.Raja Sivamani K, Garcia MS, Isseroff RR. Wound re-epithelialization: modulating keratinocyte migration in wound healing. Front Biosci. 2007;12:2849–2868. doi: 10.2741/2277. [DOI] [PubMed] [Google Scholar]
- 4.Dignass AU. Mechanisms and modulation of intestinal epithelial repair. Inflamm Bowel Dis. 2001;7:68–77. doi: 10.1097/00054725-200102000-00014. [DOI] [PubMed] [Google Scholar]
- 5.Hanna C. Proliferation and migration of epithelial cells during corneal wound repair in the rabbit and the rat. Am J Ophthalmol. 1966;61:55–63. [PubMed] [Google Scholar]
- 6.Seiler WO, Stahelin HB, Zolliker R, Kallenberger A, Luscher NJ. Impaired migration of epidermal cells from decubitus ulcers in cell cultures: a cause of protracted wound healing? Am J Clin Pathol. 1989;92:430–434. doi: 10.1093/ajcp/92.4.430. [DOI] [PubMed] [Google Scholar]
- 7.Ridley AJ, Schwartz MA, Burridge K, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 8.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 9.Elola MT, Wolfenstein-Todel C, Troncoso MF, Vasta GR, Rabinovich GA. Galectins: matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell Mol Life Sci. 2007;64:1679–1700. doi: 10.1007/s00018-007-7044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrdlickova-Cela E, Plzak J, Smetana K, Jr, et al. Detection of galectin-3 in tear fluid at disease states and immunohistochemical and lectin histochemical analysis in human corneal and conjunctival epithelium. Br J Ophthalmol. 2001;85:1336–1340. doi: 10.1136/bjo.85.11.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta SK, Masinick S, Garrett M, Hazlett LD. Pseudomonas aeruginosa lipopolysaccharide binds galectin-3 and other human corneal epithelial proteins. Infect Immun. 1997;65:2747–2753. doi: 10.1128/iai.65.7.2747-2753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smetana K, Jr, Dvorankova B, Chovanec M, et al. Nuclear presence of adhesion-/growth-regulatory galectins in normal/malignant cells of squamous epithelial origin. Histochem Cell Biol. 2006;125:171–182. doi: 10.1007/s00418-005-0074-0. [DOI] [PubMed] [Google Scholar]
- 13.Cao Z, Said N, Amin S, et al. Galectins-3 and -7, but not galectin-1, play a role in re-epithelialization of wounds. J Biol Chem. 2002;277:42299–42305. doi: 10.1074/jbc.M200981200. [DOI] [PubMed] [Google Scholar]
- 14.Hsu DK, Yang RY, Pan Z, et al. Targeted disruption of the galectin-3 gene results in attenuated peritoneal inflammatory responses. Am J Pathol. 2000;156:1073–1083. doi: 10.1016/S0002-9440(10)64975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diskin S, Kumar J, Cao Z, Schuman JS, Gilmartin T, Head SR, Panjwani N. Detection of differentially expressed glycogenes in trabecular meshwork of eyes with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47:1491–1499. doi: 10.1167/iovs.05-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holgersson J, Lofling J. Glycosyltransferases involved in type 1 chain and Lewis antigen biosynthesis exhibit glycan and core chain specificity. Glycobiology. 2006;16:584–593. doi: 10.1093/glycob/cwj090. [DOI] [PubMed] [Google Scholar]
- 17.Salvini R, Bardoni A, Valli M, Trinchera M. beta 1,3-Galactosyltransferase beta 3Gal-T5 acts on the GlcNAcbeta 1→3 Galbeta→4GlcNAcbeta 1→R sugar chains of carcinoembryonic antigen and other N-linked glycoproteins and is down-regulated in colon adenocarcinomas. J Biol Chem. 2001;276:3564–3573. doi: 10.1074/jbc.M006662200. [DOI] [PubMed] [Google Scholar]
- 18.Saravanan C, Cao Z, Head S, Panjwani N. Analysis of differential expression of glycosyltransferases in healing corneas by glycogene microarrays. Glycobiology. In press. [Google Scholar]
- 19.Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 20.Makino M, Kojima T, Ohgushi T, Yamashina I. Studies on enzymes acting on glycopeptides. J Biochem. 1968;63:186–192. doi: 10.1093/oxfordjournals.jbchem.a128760. [DOI] [PubMed] [Google Scholar]
- 21.McGlinn AM, Baldwin DA, Tobias JW, Budak MT, Khurana TS, Stone RA. Form-deprivation myopia in chick induces limited changes in retinal gene expression. Invest Ophthalmol Vis Sci. 2007;48:3430–3436. doi: 10.1167/iovs.06-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA. Gene expression profile of aging in human muscle. Physiol Genomics. 2003;14:149–159. doi: 10.1152/physiolgenomics.00049.2003. [DOI] [PubMed] [Google Scholar]
- 23.Hockley SL, Arlt VM, Brewer D, Giddings I, Phillips DH. Time- and concentration-dependent changes in gene expression induced by benzo (a) pyrene in two human cell lines, MCF-7 and HepG2. BMC Genomics. 2006;7:260. doi: 10.1186/1471-2164-7-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handayani R, Rice L, Cui Y, et al. Soy isoflavones alter expression of genes associated with cancer progression, including interleukin-8, in androgen-independent PC-3 human prostate cancer cells. J Nutr. 2006;136:75–82. doi: 10.1093/jn/136.1.75. [DOI] [PubMed] [Google Scholar]
- 25.Tree JA, Elmore MJ, Javed S, Williams A, Marsh PD. Development of a guinea pig immune response-related microarray and its use to define the host response following Mycobacterium bovis BCG vaccination. Infect Immun. 2006;74:1436–1441. doi: 10.1128/IAI.74.2.1436-1441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci U S A Suppl. 2003;100(suppl 1):11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo HB, Nairn A, Harris K, et al. Loss of expression of N-acetylglucosaminyltransferase Va results in altered gene expression of glycosyltransferases and galectins. FEBS Lett. 2008;582:527–535. doi: 10.1016/j.febslet.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbott KL, Nairn AV, Hall EM, et al. Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics. 2008;8:3210–3220. doi: 10.1002/pmic.200800157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comelli EM, Sutton-Smith M, Yan Q, et al. Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. J Immunol. 2006;177:2431–2440. doi: 10.4049/jimmunol.177.4.2431. [DOI] [PubMed] [Google Scholar]
- 30.Smith FI, Qu Q, Hong SJ, Kim KS, Gilmartin TJ, Head SR. Gene expression profiling of mouse postnatal cerebellar development using oligonucleotide microarrays designed to detect differences in glycoconjugate expression. Gene Expr Patterns. 2005;5:740–749. doi: 10.1016/j.modgep.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Amado M, Almeida R, Schwientek T, Clausen H. Identification and characterization of large galactosyltransferase gene families: galactosyltransferases for all functions. Biochim Biophys Acta. 1999;1473:35–53. doi: 10.1016/s0304-4165(99)00168-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhou D, Henion TR, Jungalwala FB, Berger EG, Hennet T. The beta 1,3-galactosyltransferase beta 3GalT-V is a stage-specific embryonic antigen-3 (SSEA-3) synthase. J Biol Chem. 2000;275:22631–22634. doi: 10.1074/jbc.C000263200. [DOI] [PubMed] [Google Scholar]
- 33.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 34.Pratt MR, Hang HC, Ten Hagen KG, et al. Deconvoluting the functions of polypeptide N-alpha-acetylgalactosaminyltransferase family members by glycopeptide substrate profiling. Chem Biol. 2004;11:1009–1016. doi: 10.1016/j.chembiol.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Tenno M, Ohtsubo K, Hagen FK, et al. Initiation of protein O glycosylation by the polypeptide GalNAcT-1 in vascular biology and humoral immunity. Mol Cell Biol. 2007;27:8783–8796. doi: 10.1128/MCB.01204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson M, Clayton P, Grunewald S, et al. Elevation of plasma aspartylglucosaminidase is a useful marker for the congenital disorders of glycosylation type I (CDG I) J Inherit Metab Dis. 2005;28:1197–1198. doi: 10.1007/s10545-005-0157-z. [DOI] [PubMed] [Google Scholar]
- 37.Matarrese P, Fusco O, Tinari N, et al. Galectin-3 overexpression protects from apoptosis by improving cell adhesion properties. Int J Cancer. 2000;85:545–554. [PubMed] [Google Scholar]
- 38.Stepp MA. Corneal integrins and their functions. Exp Eye Res. 2006;83:3–15. doi: 10.1016/j.exer.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Sotozono C, He J, Matsumoto Y, Kita M, Imanishi J, Kinoshita S. Cytokine expression in the alkali-burned cornea. Curr Eye Res. 1997;16:670–676. doi: 10.1076/ceyr.16.7.670.5057. [DOI] [PubMed] [Google Scholar]
- 40.Planck SR, Rich LF, Ansel JC, Huang XN, Rosenbaum JT. Trauma and alkali burns induce distinct patterns of cytokine gene expression in the rat cornea. Ocul Immunol Inflamm. 1997;5:95–100. doi: 10.3109/09273949709085057. [DOI] [PubMed] [Google Scholar]
- 41.Jawhara S, Thuru X, Standaert-Vitse A, et al. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J Infect Dis. 2008;197:972–980. doi: 10.1086/528990. [DOI] [PubMed] [Google Scholar]
- 42.Honardoust HA, Jiang G, Koivisto L, et al. Expression of Endo180 is spatially and temporally regulated during wound healing. Histopathology. 2006;49:634–648. doi: 10.1111/j.1365-2559.2006.02559.x. [DOI] [PubMed] [Google Scholar]
- 43.Sturge J, Wienke D, Isacke CM. Endosomes generate localized Rho-ROCK-MLC2-based contractile signals via Endo180 to promote adhesion disassembly. J Cell Biol. 2006;175:337–347. doi: 10.1083/jcb.200602125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saravanan C, Liu F-T, Gipson IK, Panjwani N. Galectin-3 promotes lamellipodia formation in epithelial cells by interacting with complex N-glycans on alpha3beta1 integrin. J Cell Sci. doi: 10.1242/jcs.045674. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortegano I, Pozo V, Cardaba B, et al. Interaction between galectin-3 and FcgammaRII induces down-regulation of IL-5 gene: implication of the promoter sequence IL-5REIII. Glycobiology. 2000;10:237–242. doi: 10.1093/glycob/10.3.237. [DOI] [PubMed] [Google Scholar]
- 46.Lin HM, Pestell RG, Raz A, Kim HR. Galectin-3 enhances cyclin D(1) promoter activity through SP1 and a cAMP-responsive element in human breast epithelial cells. Oncogene. 2002;21:8001–8010. doi: 10.1038/sj.onc.1205820. [DOI] [PubMed] [Google Scholar]
- 47.Paron I, Scaloni A, Pines A, et al. Nuclear localization of galectin-3 in transformed thyroid cells: a role in transcriptional regulation. Biochem Biophys Res Commun. 2003;302:545–553. doi: 10.1016/s0006-291x(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 48.Song S, Byrd JC, Mazurek N, Liu K, Koo JS, Bresalier RS. Galectin-3 modulates MUC2 mucin expression in human colon cancer cells at the level of transcription via AP-1 activation. Gastroenterology. 2005;129:1581–1591. doi: 10.1053/j.gastro.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Mourad-Zeidan AA, Melnikova VO, Wang H, Raz A, Bar-Eli M. Expression profiling of galectin-3–depleted melanoma cells reveals its major role in melanoma cell plasticity and vasculogenic mimicry. Am J Pathol. 2008;173:1839–1852. doi: 10.2353/ajpath.2008.080380. [DOI] [PMC free article] [PubMed] [Google Scholar]