Abstract
The TonB system energizes transport of nutrients across the outer membrane of Escherichia coli using cytoplasmic membrane proton motive force (PMF) for energy. Integral cytoplasmic membrane proteins ExbB and ExbD appear to harvest PMF and transduce it to TonB. The carboxy terminus of TonB then physically interacts with outer membrane transporters to allow translocation of ligands into the periplasmic space. The structure of the TonB carboxy terminus (residues ~150 to 239) has been solved several times with similar results. Our previous results hinted that in vitro structures might not mimic the dimeric conformations that characterize TonB in vivo. To test structural predictions and to identify irreplaceable residues, the entire carboxy terminus of TonB was scanned with Cys substitutions. TonB I232C and N233C, predicted to efficiently form disulfide-linked dimers in the crystal structures, did not do so. In contrast, Cys substitutions positioned at large distances from one another in the crystal structures efficiently formed dimers. Cys scanning identified seven functionally important residues. However, no single residue was irreplaceable. The phenotypes conferred by changes of the seven residues depended on both the specific assay used and the residue substituted. All seven residues were synergistic with one another. The buried nature of the residues in the structures was also inconsistent with these properties. Taken together, these results indicate that the solved dimeric crystal structures of TonB do not exist. The most likely explanation for the aberrant structures is that they were obtained in the absence of the TonB transmembrane domain, ExbB, ExbD, and/or the PMF.
IMPORTANCE
The TonB system of Gram-negative bacteria is an attractive target for development of novel antibiotics because of its importance in iron acquisition and virulence. Logically, therefore, the structure of TonB must be accurately understood. TonB functions as a dimer in vivo, and two different but similar crystal structures of the dimeric carboxy-terminal ~90 amino acids gave rise to mechanistic models. Here we demonstrate that the crystal structures, and therefore the models based on them, are not biologically relevant. The idiosyncratic phenotypes conferred by substitutions at the only seven functionally important residues in the carboxy terminus suggest that similar to interaction of cytochromes P450 with numerous substrates, these residues allow TonB to differentially interact with different outer membrane transporters. Taken together, data suggest that TonB is maintained poised between order and disorder by ExbB, ExbD, and the proton motive force (PMF) before energy transduction to the outer membrane transporters.
INTRODUCTION
The outer membrane (OM) of Gram-negative Escherichia coli is a diffusion barrier to the transport of nutrients greater than 600 Da through porin proteins. This common limitation is circumvented by the TonB system, which is present in virtually all Gram-negative bacteria (1). A TonB system includes a characteristic OM active transporter specific for one transport substrate or a few transport substrates (22-stranded β-barrel with N-terminal cork that fills the lumen, known as a TonB-gated transporter) and three integral cytoplasmic proteins, ExbB, ExbD, and TonB. ExbB and ExbD appear to harvest the proton motive force (PMF) of the cytoplasmic membrane, which is then transmitted to TonB. In turn, the energized TonB physically interacts with TonB-gated transporters. TonB is capable of interaction with multiple OM transporters to enable transport of diverse substrates across the OM. For example, Xanthomonas campestris has over 70 predicted TonB-gated transporters and six sets of tonB-exbB-exbD genes. In addition, TonB-dependent transport of maltodextrins, nickel, sucrose, and potentially sulfate has been characterized (2–6). Thus, iron and vitamin B12 acquisition may represent only the “tip of the iceberg” (7, 8). Clues about the role of a given TonB system can arise from understanding the regulation of each tonB gene. In E. coli, tonB expression is regulated by iron availability (9).
In E. coli, the 239-residue cytoplasmic membrane protein TonB consists of an amino-terminal signal anchor with a single important residue, His20. The bulk of TonB occupies the periplasmic space (10). The topology of the 141-residue ExbD protein is similar to that of TonB, and ExbD is proposed to be a chaperone for TonB conformational changes in vivo (11). The three transmembrane domains of ExbB share homology with MotA, a protein that translocates protons to energize flagellar rotation (12). Since the TonB system requires the PMF, it is likely that ExbB also translocates protons.
Full-length TonB fractionates with both the cytoplasmic membrane and the OM and is a stable protein. Since it is much less abundant than the various OM transporters it serves, TonB must undergo cyclic contact and disengagement from the TonB-gated transporters during energy transduction (9, 13, 14). During the energy transduction cycle, TonB undergoes conformational changes both before and after an energy transduction event; however, if transport ligands are not present, energy is not transduced (15, 16). It is clear that TonB directly interacts with a conserved region at the extreme amino terminus of the transporters known as the TonB box (17, 18).
The carboxy-terminal ~90 amino acids of TonB have been cocrystallized with the BtuB and FhuA transporters (19, 20). The structure of monomeric TonB in the complexes is similar to the crystal structures of dimeric TonB and to a monomeric configuration from nuclear magnetic resonance (NMR) studies (21–24). In the TonB-BtuB structure, the BtuB TonB box interacts with TonB residues 225 to 232 and residues 158 to 171, suggesting that the residues in those domains would prove to be important for TonB function. In the TonB-FhuA structure, the TonB box was not strictly visible.
Using a genetic approach, we had previously identified 5 out of 20 residues assayed in the TonB carboxy terminus as being functionally important. In that case, “functionally important” meant that a substitution mutation resulted in a nearly complete loss of function in at least one of the several available assays for TonB activity (26). To identify additional important residues and to test whether the dimeric crystal structures reflect actual conformations of TonB, the remaining 70 amino acids were replaced with cysteinyl residues and the resulting phenotypes in a panel of TonB-dependent assays and their ability to form disulfide-linked dimers were analyzed. For all identified functionally important residues, their ability to synergize with one another was also tested as discussed below.
Our resulting data support two conclusions: first, and most importantly, neither of the two dimeric crystal structures represents the conformation of the TonB carboxy terminus, and second, while there are functionally important residues in TonB, no single residue is irreplaceable.
RESULTS AND DISCUSSION
TonB conformations represented by dimeric crystal structures do not exist in vivo.
The dimeric crystal structures, which represent stable forms by their very nature, are the most relevant, since TonB is known to function as a dimer in vivo (15, 25). Previous work with a limited set of 20 Cys substitutions in the TonB carboxy terminus suggested the possibility that the dimeric crystal structures might not represent in vivo conformations. Four Cys substitutions, each distantly located from the cognate partner in the dimeric crystal structures, spontaneously formed a set of three disulfide-linked dimers in vivo. These so-called triplet dimers did not form if TonB was not energized, indicating that they were biologically relevant (15, 26). Since the crystal and NMR structures represent only a snapshot of each protein’s conformation, it is possible that the stable conformation of TonB depicted in the crystal structures was only one of at least three conformations that TonB is known to inhabit (16).
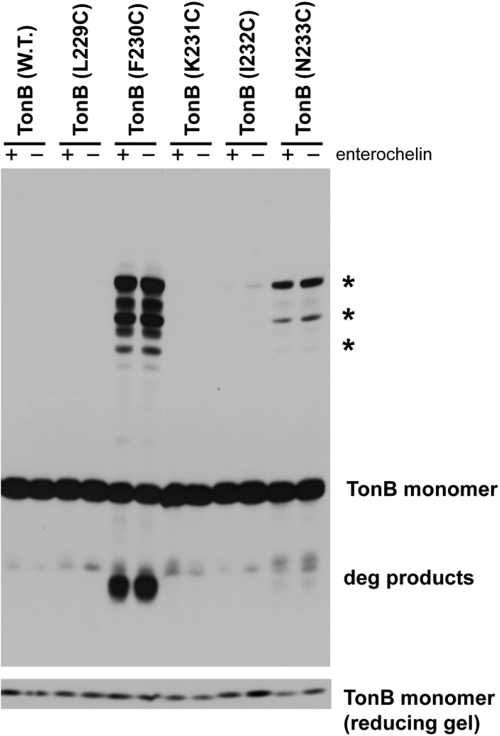
To further examine the relevance of the crystal structures, predicted residues at the dimeric interface, close enough to form disulfide bonds, were identified. In both of the existing crystal structures, Cys substitutions at I232 and N233 would be diagnostic. These substitutions were individually engineered in TonB, along with nearby L229C, F230C (known to cross-link strongly), and K231C and examined for their ability to spontaneously form disulfide-linked dimers. The maximum distance between α-carbons of Cys residues that can form disulfide-linked dimers is ~7 Å (27). TonB L229C and K231C did not form cross-linked dimers and would not be predicted to form them on the basis of the distances between their α-carbons (Fig. 1 and Table 1). However, the distances between I232 α-carbons in structures 1IHR and 1U07 are 4.4 and 6.5 Å, respectively, and the distances between sulfur atoms of optimized rotamers of TonB I232C are 3.7 and 2.7 Å, respectively. If the conformations represented by the crystal structures existed, TonB I232C would dimerize efficiently. In contrast, virtually no disulfide-linked dimers of TonB I232C were detected except on extended overnight exposure of the immunoblots, circumstances under which detectable levels of the largest of the triplet dimers can be observed in a majority of the 90 TonB proteins with Cys substitutions (data not shown). As seen previously, the absence of the sole TonB-dependent ligand produced by E. coli K-12, enterochelin, did not affect the extent of cross-linking in the aroB strain, KP1406 (Fig. 1). Similarly, TonB N233C residues, while slightly more distant in 1IHR (8.1 Å) than in 1U07 (5.7 Å) would be predicted to form dimers strongly and formed them only weakly.
FIG 1 .
In vivo, TonB does not exhibit the crystal structure-predicted disulfide cross-link at residue I232C.
TonB proteins were expressed at chromosomal levels from plasmids carrying genes encoding wild-type (W.T.) TonB or TonB Cys substitutions in pKP632 (L229C), pKP570 (F230C), pKP628 (K231C), pKP629 (I232C), and pKP630 (N233C) and processed in nonreducing sample buffer containing iodoacetamide as described in Materials and Methods. E. coli strain KP1344 (W3110 ∆tonB::blaM) produced enterochelin (+), while strain KP1406 (W3110 aroB ∆tonB::blaM) did not (−). Samples were resolved on an 11% nonreducing SDS-polyacrylamide gel and immunoblotted with TonB-specific monoclonal antibodies and are labeled according to the Cys substitution being examined. The positions of disulfide-linked dimers are indicated by asterisks to the right of the gel. The positions of degradation products (deg products) are also indicated to the right of the gel. The levels of total TonB protein in the nonreducing samples were determined on a reducing gel shown at the bottom of the figure.
TABLE 1 .
Lack of correlation between α-carbon distances in the crystal structures and ability of TonB Cys substitutions to form disulfide-linked dimers in vivo
| Residue | Distance (Å)a |
Dimer formationb | |
|---|---|---|---|
| 1IHR | 1U07 | ||
| G186 | 51.4 | 59.2 | Strong |
| F202 | 6.5 | 31.4 | Strong |
| W213 | 35.6 | 45.1 | Strong |
| Y215 | 47.0 | 51.4 | Strong |
| L229 | 20.7 | 25.1 | - |
| F230 | 12.6 (8.0) | 19.4 (18.4) | Strong |
| K231 | 8.6 (7.2) | 13.6 (12.4) | - |
| I232 | 4.4 (3.7) | 6.5 (2.7) | - |
| N233 | 8.1 (5.2) | 5.7 (3.7) | Weak |
Predicted distance between α-carbon atoms was calculated using the Swiss-PDBViewer, DeepView v4.0 (http://spdbv.vital-it.ch/).The distance between sulfur groups of optimized Cys rotamers is shown in parentheses. 1IHR and 1U07 are Protein Data Bank (PDB) identifiers for solved structures.
Relative levels of triplet dimer formation are shown in Fig. 1 and 2. A hyphen indicates that the dimers were relatively undetectable. The maximum distance between α-carbons of Cys residues that can form disulfide-linked dimers is ~7 Å (27). For a summary of similarities between the 1IHR and1U07 structures, see reference 24.
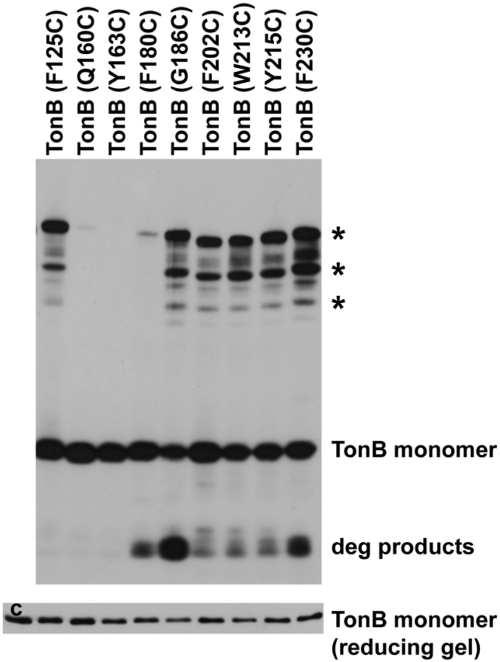
As shown previously, several substituted cysteines spontaneously cross-link efficiently but are distantly located from their cognate partners in the crystal structures and would not be predicted to do so: F202C, W213C, Y215C, and F230C. To this list, TonB G186C (characterized below) can be added (Fig. 2). It cross-linked efficiently, yet according to the crystal structures, these residues are >50 Å apart (Table 1). Thus, we conclude that none of the conformations TonB assumes is represented by the dimeric crystal structures.
FIG 2 .
Six of the 91 Cys substitutions can efficiently and spontaneously form disulfide-linked dimers in vivo. E. coli strain KP1344 (W3110 ∆tonB) carrying plasmids (see Table S2 in the supplemental material) expressing TonB Cys substitutions at chromosomal levels were processed in nonreducing sample buffer containing iodoacetamide as described in Materials and Methods. Samples of TonB proteins encoded by genes carried on plasmids pKP1070 (F125C), pKP588 (Q160C), pKP586 (Y163C), pKP569 (F180C), pKP612 (G186C), pKP415 (F202C), pKP472 (W213C), pKP474 (Y215C), and pKPK570 (F230C) were resolved on an 11% nonreducing SDS-polyacrylamide gel and immunoblotted with TonB-specific monoclonal antibodies and are labeled according to the Cys substitution being examined. TonB Q160C is included as an example of a Cys substitution at a nonaromatic amino acid that does not cross-link efficiently. The positions of disulfide-linked dimers are indicated by asterisks to the right of the gel. The positions of degradation products (deg products) are also indicated to the right of the gel. The identities of the intermediate bands between asterisks are not known. The levels of total protein in the nonreducing samples were determined on a reducing gel (bottom gel) and compared to chromosomally encoded TonB (c).
There are 7 functionally important, but not irreplaceable, residues in the TonB carboxy terminus.
We had previously identified 5 functionally important aromatic residues (F180, F202, W213, Y215, and F230) in the region of the TonB carboxy terminus for which a crystal structure existed (21). Those aromatic amino acids appeared to play a role in discriminating among the many ligand/transporter combinations that TonB contacts (15, 26). Substitution of each residue with either Ala or Cys residues resulted in idiosyncratic profiles of sensitivities to various colicins and phage and ability to transport ferrichrome. Thus, each TonB protein with a specific substitution was often fully active with respect to one agent but virtually inactive with respect to another agent. Sensitivity to different agents varied with each different substitution. Any pair-wise combination of the 5 Ala substitutions in a double mutant cycle analysis resulted in a single synergistically inactive phenotype with respect to all assays (28). It was proposed that the aromatic amino acids all worked together in some fashion, most likely in receptor/colicin recognition (15, 26). This suggests that these aromatic residues are relatively close to each other. Cys substitutions supported this hypothesis. In 4 of the 5 Cys substitutions at these aromatic residues, disulfide-linked triplet dimers were demonstrated. The biological relevance of the disulfide-linked dimers was demonstrated by the fact that dimers did not form if the transmembrane domain carried the inactivating tonB∆V17 mutation or if the ExbB/D and TolQ/R functions were absent (15). These studies did not, however, preclude the existence of more mechanistically important, irreplaceable residues that could overturn that hypothesis.
Indeed, many residues in the TonB carboxy terminus are highly conserved, suggesting that they could represent irreplaceable residues (1). To identify such residues, 70 previously untested residues from residues 150 to 239, and the sole remaining aromatic residue in the entire carboxy terminus, F125, were substituted with Cys residues in a plasmid expressing TonB C18G, which lacks the sole naturally occurring Cys residue. The 71 individual substitutions were expressed at chromosomally encoded levels in a ∆tonB strain, and each Cys substitution was assayed for colicin B, Ia, and M sensitivity and for the ability to energize [55Fe]ferrichrome transport. This combination of assays reflected interactions with three different transporters, each with their different TonB boxes (FepA, Cir, and FhuA, respectively), and three colicins, each of which also has a different TonB box. Sensitivity to colicins can register as little as 1 molecule of TonB per cell, while ferrichrome transport can discriminate in the range between 100% TonB activity (340 molecules per cell in media containing high levels of iron [9]) and 10% TonB activity (29). Because these comprehensive assays represent a spectrum of sensitivities and TonB recognition of different transporters and their colicin ligands, they ensured that few, if any, important mutant phenotypes were overlooked.
The results indicated that no single residue in the TonB carboxy terminus was essential for activity. Of the 71 Cys substitutions, 69 supported essentially full activity with respect to the colicin assays and from 60 to 100% [55Fe]ferrichrome transport, including F125C (see Fig. S1 in the supplemental material). Because Cys side chains could functionally substitute for Ser or Thr side chains, the TonB S151, S157, T183, S195, S222, T235, and T236 residues were individually replaced with Ala residues and assayed. Like the Cys substitutions, each of the Ala substitutions retained wild-type activity, confirming that the Ser and Thr residues were not important for TonB activity (data not shown).
The only two previously undiscovered Cys substitutions that did not support nearly full activity in all assays were Y163C and G186C. Interestingly, these also exhibited idiosyncratic phenotypic profiles (Table 2). TonB Y163C exhibited full sensitivity to colicin Ia and the ability to energize [55Fe]ferrichrome transport at ~60% of the rate of normal wild-type levels. However, it exhibited tolerance (insensitivity) to both colicins B and M. TonB G186C exhibited a different profile of activities, being only slightly impaired in colicin B and Ia sensitivity, completely tolerant to colicin M, and capable of energizing <10% of the initial rate of normal wild-type [55Fe]ferrichrome transport.
TABLE 2 .
Idiosyncratic phenotypic profiles of the 7 functionally important TonB Cys substitutionsa
| Mutant | Activityb |
% Fe transport (mean ± SEM)c | ||
|---|---|---|---|---|
| Col B | Col Ia | Col M | ||
| ΔtonB | T,T,T | T,T,T | T,T,T | 0 |
| pKP568d (wt) | 9,9,9 | 8,8,8 | 6,6,6 | 100 ± 4 |
| Y163C | T,T,T | 8,8,8 | T,T,T | 58 ± 2 |
| F180C | 7,7,7 | 4,4,4 | 4,4,4 | 48 ± 2 |
| G186C | 7,7,7 | 5,5,5 | T,T,T | 10 ± 1 |
| F202C | 4,4,4 | 8,8,8 | T,T,T | 38 ± 1e |
| W213C | 6,6,6 | 5,5,5 | 3,3,3 | 20 ± 1 |
| Y215C | 8,8,8 | 3,3,3 | 3,3,3 | 35 ± 2 |
| F230C | 7,7,7 | 8,8,8 | 2,2,2 | 38 ± 1 |
Spot titer assays were conducted in strain KP1406 (tonB aroB).
Numbers indicate the last 5-fold dilution at which sensitivity to the agent (colicin [Col] B, Ia, or M) was apparent in triplicate assays. T indicates tolerance (no sensitivity) to the particular agent tested. The results for three assays are shown.
Initial rates of [55Fe]ferrichrome transport were determined in strain KP1344 (tonB).
pKP568 carries a gene encoding TonBC18G, which lacks the sole Cys residue in native TonB. wt, wild type.
TonB F202A exhibits 95% transport (26).
The full set of 91 Cys substitutions from this and previous work was also assayed for spontaneous dimer formation. Other than the 4 cross-linkable Cys substitutions previously identified, only two residues, F125 and G186, formed triplet dimers slightly less efficiently than F230C, the most efficient dimer former (Fig. 2 and data not shown). Weak formation of the dimers of highest mass in the triplet was observed for several residues from amino acids (aa) 150 to 170. This region immediately follows an unstructured region in the NMR structure (aa 103 to 149) (23). There were essentially no cross-linked complexes observed from aa 171 to 239 with the exception of weak formation of the dimers of highest mass by P184C, D185C, P198C, and N233C (data not shown). These results indicate that the only 7 residues important for TonB activity share the characteristic of idiosyncratic phenotypic profiles and a subset of these participate in triplet dimer formation.
(i) Conservation does not correlate with functional importance in the TonB carboxy terminus.
Interestingly, there was only partial correlation between the ~30 highly conserved residues in the TonB carboxy terminus and the functionally important residues. As an example, Chu et al. identified TonB Y163 and P164 as constituting a highly conserved motif (1). Consistent with that, TonB Y163 showed 82% conservation and was one of the newly identified functionally important residues in this study. In contrast, P164 exhibited comparable conservation at 86% conservation, but TonB P164C showed only minor decreases in activity (see Fig. S1 in the supplemental material). TonB G186 is highly conserved at 96% and was also functionally important. In contrast, the remaining 5 previously identified functionally important residues are much less well conserved: F180 (45%), F202 (53%), W213 (26%), Y215 (23%), and F230 (22%). These residues are, however, highly conserved among enteric bacteria (26). (The overall percentages of conservation of residues in this discussion were generously provided by B. Chu and H. Vogel [personal communication].)
(ii) The 7 functionally important amino acids play roles in recognition.
On the basis of their individual idiosyncratic phenotypic profiles and the lack of any irreplaceable residues in the carboxy terminus, it seems clear that TonB uses the 7 residues identified as functionally relevant to discriminate among the various TonB-gated transporters and their colicin ligands. Aromatic residues often constitute part of a protein-protein binding interface (30). Precedence for idiosyncratic phenotypic profiles indicating differential binding interactions can be found in the cytochrome P450s, which adopt different conformations to interact with different substrates (31). These results are also consistent with an earlier hypothesis by Sauter et al. (25) based on different phenotypes conferred by Y163C and F230V substitutions in various assays. The fact that the side chains of these 7 functionally important residues are buried in the crystal structures was consistent with the finding that those structures did not exist.
Perhaps, based on their high levels of sequence conservation, Y163 and G186 reflect residues with more intrinsic mechanistic function in transporter recognition. The relatively low overall level of sequence conservation of TonB F202C, W213C, Y215C, or F230C, coupled with a high level of conservation among closely related bacteria, suggests that the residues directly involved in recognition have evolved as the transporter sequences evolved.
The relationship of these 7 residues to the TonB box remains to be determined. TonB residues 158 to 162, virtually all of the region through which TonB interactions with TonB boxes occur, can be deleted and still retain 60% ferrichrome transport activity, indicating that additional amino acids must be involved in interaction with some transporters (32). Furthermore, it has been demonstrated that TonB interacts in vivo with sites other than the transporter TonB boxes before interacting with the TonB boxes themselves (33). In addition, phage display experiments have identified regions of in vitro interactions between the TonB carboxy terminus and outer membrane (OM) transporters outside the TonB box (34).
Each of the functionally important residues exhibits synergy in a double mutant cycle analysis.
Like the previously identified functionally important residues, a double mutant cycle analysis expanded to include Y163A and G186A indicated that they also exhibited synergy, showing complete insensitivity to colicins B, D, M, and Ia (see Table S1 in the supplemental material). Trivial explanations for the inactivity of the double mutants were ruled out. The double Ala substitutions were efficiently exported as determined by their proteinase K accessibility in spheroplasts (see Fig. S2 in the supplemental material). They displayed the ability to form abundant and apparently random formaldehyde-mediated cross-links as described previously for a subset of the double mutants (26) (see Fig. S3, far left, middle, in the supplemental material). Two of the TonB double Ala substitutions showed reduced cross-linking to ExbD, while the remainder were too unstable to survive 15 min at room temperature during the formaldehyde cross-linking assay but did not otherwise affect the cross-linking profile of ExbD (see Fig. S3, far right, in the supplemental material).
The concept of synergy has been classically applied to enzymatic reaction mechanisms where measurements of ∆∆G are feasible. The interpretation in those cases is that the two wild-type residues in question either interact directly or they facilitate the same non-rate-limiting step, such that when each is mutated, a new slower rate-limiting step is imposed (28, 35). While all possible combinations of the mutations (912) were not assayed to develop a synergy matrix, selected combinations of residues whose mutation had no effect on function were examined, and no synergy was detected. When TonB F125A was combined with each of the other Ala substitutions in this study, the phenotypes exhibited additivity but not synergy: for example, TonB F125A F180A exhibited the phenotype that is characteristic of TonB F180A (see Table S1 in the supplemental material). Likewise, the pairing of M201C and E216C, each at a residue next to a Cys substitution that exhibited synergy, exhibited only additivity (data not shown). While mechanistic interpretations of these data were not yet possible, they did indicate that the set of 7 amino acids all have special properties when considered within the context of carboxy-terminal residues and, most importantly, that they all functioned as a group. Their interaction and functional importance would not be predicted to occur if current crystal structures represented the structure of TonB.
There are no irreplaceable residues in TonB.
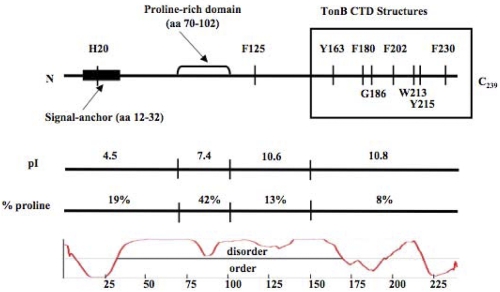
The TonB protein has unusual properties. Approximately 17% of its amino acid residues are prolines compared to the average E. coli TonB protein that contains ~4% proline (36). While many of the proline residues are located in the proline-rich region from residues 70 to 102 (42% proline), there are 7 prolines in the carboxy-terminal 90 amino acids (~8%) and still higher proportions in the rest of the protein (Fig. 3). In addition, the TonB carboxy terminus from residues 102 to 239 is highly basic, with an overall calculated pI of 10.9.
FIG 3 .
Features of the TonB primary amino acid sequence. The top line displays some of the key features of the TonB primary amino acid sequence that have been studied by mutational analysis. The TonB sequence has been divided into 4 regions: (i) residues 1 to 69, containing the essential amino-terminal signal anchor and showing the position of the H20 residue within it; (ii) residues 70 to 102, constituting the proline-rich region; (iii) residues 103 to 149; and (iv) residues 150 to 239, the only region for which structures have been determined thus far (CTD, C-terminal domain). The next line displays the calculated pI for each of the 4 regions. The calculated percentage of prolyl residues found in each region is shown on the next line. At the bottom is a prediction of unstructured regions in TonB protein determined by the PONDR program, with the corresponding amino acid numbers on the x axis. Predicted unstructured regions are shown above the line, and structured regions are shown below the line. A slightly different view of the PONDR graph for TonB is shown in reference 10.
Another unusual feature of TonB is that there are no irreplaceable residues in the entire protein. Residues 1 to 12 preceding the uncleaved signal sequence are replaceable with the first 25 residues of the soluble protein TrpC (37). The only important residue in the transmembrane domain (aa 12 to 32) is the highly conserved H20 residue (10). Nonetheless, overexpression of TonB H20A or other inactivating mutations in the transmembrane domain restores low-level activity (16, 38; unpublished data). The periplasmic region proximal to the membrane (aa 33 to 64) can be deleted without inactivating TonB entirely. Moderate sensitivity to colicins B and Ia and ~2% [55Fe]ferrichrome transport are retained (this study). The triple proline motif from residues 63 to 65 can be deleted without significant effect (this study); the proline-rich domain from residues 66 to 100 (or residues 70 to 102 [this study]) can be deleted without effect on TonB activities unless the periplasmic space is temporarily expanded, resulting in ~50% decrease in TonB activity (39, 40). Consistent with the nonessentiality of residue F125, the region from residues 103 to 149 can be deleted with retention of nearly full colicin sensitivity and an initial rate of [55Fe]ferrichrome transport of 14% of the wild-type rate (this study). This study showed that 83 of the 90 residues from residues 150 to 239 are replaceable. The importance of the remaining 7 residues depends entirely on the assay, and no single mutation inactivates TonB completely. By comparison, complete Cys scanning of E. coli lactose permease yielded 6 irreplaceable residues out of 417 (41). We are unaware of any proteins that have proven to be as tolerant of mutation as TonB.
Final thoughts.
It might not be surprising that in vivo TonB conformations are not represented by the dimeric crystal structures. First, the crystal structures were solved in the absence of the TonB transmembrane domain, ExbB, ExbD, and the PMF—components all known to influence its conformation in vivo (11, 15, 16, 42). Furthermore, while interactions could not be detected between the purified periplasmic domains of TonB and ExbD, PMF-dependent interactions were detected in vivo (11, 43). Second, the TonB carboxy terminus appears poised between predicted structured and unstructured tendencies (Fig. 3), bearing many of the hallmarks that characterize other intrinsically unstructured proteins (44), such as the fact that TonB is modular with two independent functional domains, the amino terminus and the carboxy terminus. Like other such proteins, part of the linker region that separates these two functional domains is rich in proline and can assume a rigid extended conformation (45). Such a linker region often serves to allow an efficient search for the correct target, which in this case would be ligand-bound transporters. For intrinsically unstructured proteins, coupled folding and binding often result in high specificity with low affinity to allow cyclic binding and release. TonB, as the limiting protein in the system, must interact with the numerous available transporters with high specificity to ensure uptake of needed nutrients (13). It must, however, also have low affinity to allow rapid cyclic binding and release from target transporters, of which there are 7 in E. coli K-12 (9, 14, 46). A model that accommodates the data is one where TonB identifies and binds to ligand-bound transporters by means of conformational sampling.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains W3110, KP1270 (W3110 aroB), KP1344 (W3110 ∆tonB::blaM), and KP1406 (W3110 aroB ∆tonB::blaM) were used in this study (16). The plasmid strains used in this study are all derivatives of pKP325 (16), where the araBAD promoter regulates TonB expression. Cysteinyl codon substitutions were created in the tonB gene by two different methods as previously described and are listed in Table S2 in the supplemental material (26, 32). For each mutant, the entire tonB open reading frame was sequenced to rule out unintended base changes (Penn State Genomics Core Facility, University Park, PA).
Media and culture conditions.
Luria-Bertani (LB), tryptone (T), and M9 minimal salts were prepared as previously described (47). Liquid cultures, agar plates, and tryptone top agar were supplemented with 34 µg ml−1 chloramphenicol and/or 100 µg ml−1 ampicillin and plasmid-specific levels of l-arabinose as needed for expression of plasmid-encoded TonB variants at chromosomally encoded levels, determined by immunoblot analysis. M9 salts were supplemented with 0.5% glycerol (wt/vol), 0.4 µg ml−1 thiamine, 1 mM MgSO4, 0.5 mM CaCl2, 0.2% Casamino Acids (wt/vol), 40 µg ml−1 tryptophan, and 1.85 µM FeCl3. Cultures were grown with aeration at 37°C. Adapted M9 medium for growth of aroB strains contained additional aromatic amino acids (0.4% each of tyrosine, phenylalanine, and tryptophan) and iron (37 µM FeCl3).
Activity assays.
Spot titer assays were performed in triplicate as previously described (29, 48). The initial rates of [55Fe]ferrichrome uptake were determined as described previously (48).
In vivo disulfide cross-linking assay.
Saturated overnight cultures of strains carrying plasmids encoding TonB substitutions were subcultured 1:100 in T broth containing chloramphenicol and supplemented with l-arabinose. Cultures were harvested in mid-exponential phase and precipitated with trichloroacetic acid (TCA). Cell pellets were suspended in nonreducing Laemmli sample buffer containing 50 mM iodoacetamide, as previously described (15). Samples were resolved on 11% nonreducing SDS-polyacrylamide gels and evaluated by immunoblot analysis using anti-TonB monoclonal antibody 4F1.
In vivo formaldehyde cross-linking.
Saturated overnight cultures were subcultured 1:100 into M9 minimal medium (above) supplemented with arabinose, and when the cultures reached mid-exponential phase, they were treated with formaldehyde as previously described (48). Cross-linked complexes were detected by immunoblotting with monoclonal TonB 4F1 antiserum or ExbD-specific polyclonal antibodies (9).
Proteinase K accessibility assays.
Spheroplasts of strains expressing TonB double Ala mutants at chromosomally encoded levels were prepared and treated with proteinase K as described previously (16). Samples were visualized on immunoblots of 11% SDS-polyacrylamide gels with anti-TonB monoclonal antibody 4F1 (49).
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We thank Amber Dillhof and Lisa Van Gemert for performing spot titer assays on many of the mutants and Gail Deckert for screening and identifying most of the engineered tonB mutants. Helpful discussions with Lisa Gloss are gratefully acknowledged.
This work is supported by grant GM42146 from the National Institute of General Medical Sciences to K.P.
Footnotes
Citation Postle, K., K. A. Kastead, M. G. Gresock, J. Ghosh, and C. D. Swayne. 2010. The TonB dimeric crystal structures do not exist in vivo. mBio 1(5):e00307-10. doi:10.1128/mBio.00307-10.
REFERENCES
- 1. Chu B. C., Peacock R. S., Vogel H. J. 2007. Bioinformatic analysis of the TonB protein family. Biometals 20:467–483 [DOI] [PubMed] [Google Scholar]
- 2. Blanvillain S., Meyer D., Boulanger A., Lautier M., Guynet C., Denance N., Vasse J., Lauber E., Arlat M. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lohmiller S., Hantke K., Patzer S. I., Braun V. 2008. TonB-dependent maltose transport by Caulobacter crescentus. Microbiology 154:1748–1754 [DOI] [PubMed] [Google Scholar]
- 4. Neugebauer H., Herrmann C., Kammer W., Schwarz G., Nordheim A., Braun V. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187:8300–8311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schauer K., Gouget B., Carriere M., Labigne A., de Reuse H. 2007. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63:1054–1068 [DOI] [PubMed] [Google Scholar]
- 6. Tralau T., Vuilleumier S., Thibault C., Campbell B. J., Hart C. A., Kertesz M. A. 2007. Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J. Bacteriol. 189:6743–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Postle K., Larsen R. 2004. The TonB, ExbB, and ExbD proteins, p. 96–112 In Crosa J. H., Mey A. R., Payne S. M., Iron transport in bacteria. ASM Press, Washington, DC [Google Scholar]
- 8. Schauer K., Rodionov D. A., de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the “tip of the iceberg?” Trends Biochem. Sci. 33:330–338 [DOI] [PubMed] [Google Scholar]
- 9. Higgs P. I., Larsen R. A., Postle K. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271–281 [DOI] [PubMed] [Google Scholar]
- 10. Larsen R. A., Deckert G. E., Kastead K. A., Devanathan S., Keller K. L., Postle K. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 189:2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ollis A. A., Manning M., Held K. G., Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou J., Sharp L. L., Tang H. L., Lloyd S. A., Billings S., Braun T. F., Blair D. F. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kadner R. J., Heller K. J. 1995. Mutual inhibition of cobalamin and siderophore uptake systems suggests their competition for TonB function. J. Bacteriol. 177:4829–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letain T. E., Postle K. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Gram-negative bacteria. Mol. Microbiol. 24:271–283 [DOI] [PubMed] [Google Scholar]
- 15. Ghosh J., Postle K. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 55:276–288 [DOI] [PubMed] [Google Scholar]
- 16. Larsen R. A., Thomas M. G., Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809–1824 [DOI] [PubMed] [Google Scholar]
- 17. Braun V., Endriss F. 2007. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals 20:219–231 [DOI] [PubMed] [Google Scholar]
- 18. Cadieux N., Kadner R. J. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. U. S. A. 96:10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pawelek P. D., Croteau N., Ng-Thow-Hing C., Khursigara C. M., Moiseeva N., Allaire M., Coulton J. W. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399–1402 [DOI] [PubMed] [Google Scholar]
- 20. Shultis D. D., Purdy M. D., Banchs C. N., Wiener M. C. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396–1399 [DOI] [PubMed] [Google Scholar]
- 21. Chang C., Mooser A., Pluckthun A., Wlodawer A. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535–27540 [DOI] [PubMed] [Google Scholar]
- 22. Kodding J., Killig F., Polzer P., Howard S. P., Diederichs K., Welte W. 2005. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem. 280:3022–3028 [DOI] [PubMed] [Google Scholar]
- 23. Peacock S. R., Weljie A. M., Howard S. P., Price F. D., Vogel H. J. 2005. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol. 345:1185–1197 [DOI] [PubMed] [Google Scholar]
- 24. Wiener M. C. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15:394–400 [DOI] [PubMed] [Google Scholar]
- 25. Sauter A., Howard S. P., Braun V. 2003. In vivo evidence for TonB dimerization. J. Bacteriol. 185:5747–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh J., Postle K. 2004. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol. Microbiol. 51:203–213 [DOI] [PubMed] [Google Scholar]
- 27. Lynch B. A., Koshland D. E. 1991. Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:10402–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mildvan A. S., Weber D. J., Kuliopulos A. 1992. Quantitative interpretations of double mutations of enzymes. Arch. Biochem. Biophys. 294:327–340 [DOI] [PubMed] [Google Scholar]
- 29. Larsen R. A., Chen G. J., Postle K. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J. Bacteriol. 185:4699–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janin J., Bahadur R. P., Chakrabarti P. 2008. Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 41:133–180 [DOI] [PubMed] [Google Scholar]
- 31. Zhao Y., Halpert J. R. 2007. Structure-function analysis of cytochromes P450 2B. Biochim. Biophys. Acta 1770:402–412 [DOI] [PubMed] [Google Scholar]
- 32. Vakharia-Rao H., Kastead K. A., Savenkova M. I., Bulathsinghala C. M., Postle K. 2007. Deletion and substitution analysis of the Escherichia coli TonB Q160 region. J. Bacteriol. 189:4662–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Devanathan S., Postle K. 2007. Studies on colicin B translocation: FepA is gated by TonB. Mol. Microbiol. 65:441–453 [DOI] [PubMed] [Google Scholar]
- 34. Carter D. M., Gagnon J. N., Damlaj M., Mandava S., Makowski L., Rodi D. J., Pawelek P. D., Coulton J. W. 2006. Phage display reveals multiple contact sites between FhuA, an outer membrane receptor of Escherichia coli, and TonB. J. Mol. Biol. 357:236–251 [DOI] [PubMed] [Google Scholar]
- 35. Mildvan A. S. 2004. Inverse thinking about double mutants of enzymes. Biochemistry 43:14517–14520 [DOI] [PubMed] [Google Scholar]
- 36. Neidhardt F. C., Umbarger H. E. 1996. Chemical composition of Escherichia coli, p. 13–16 In Neidhardt F. C., et al., Escherichia coli and Salmonella: cellular and molecular biology, vol. 1 ASM Press, Washington, DC [Google Scholar]
- 37. Skare J. T., Roof S. K., Postle K. 1989. A mutation in the amino terminus of a hybrid TrpC-TonB protein relieves overproduction lethality and results in cytoplasmic accumulation. J. Bacteriol. 171:4442–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fischer E., Günter K., Braun V. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsen R. A., Wood G. E., Postle K. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10:943–953 [DOI] [PubMed] [Google Scholar]
- 40. Seliger S. S., Mey A. R., Valle A. M., Payne S. M. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801–812 [DOI] [PubMed] [Google Scholar]
- 41. Guan L., Kaback H. R. 2006. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35:67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Larsen R. A., Thomas M. T., Wood G. E., Postle K. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (∆V17) by a missense mutation in ExbB. Mol. Microbiol. 13:627–640 [DOI] [PubMed] [Google Scholar]
- 43. Garcia-Herrero A., Peacock R. S., Howard S. P., Vogel H. J. 2007. The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol. Microbiol. 66:872–889 [DOI] [PubMed] [Google Scholar]
- 44. Dyson H. J., Wright P. E. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197–208 [DOI] [PubMed] [Google Scholar]
- 45. Kohler S. D., Weber A., Howard S. P., Welte W., Drescher M. 2010. The proline-rich domain of TonB possesses an extended polyproline II-like conformation of sufficient length to span the periplasm of Gram-negative bacteria. Protein Sci. 19:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Braun V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:s1409–s1421 [DOI] [PubMed] [Google Scholar]
- 47. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 48. Postle K. 2007. TonB system, in vivo assays and characterization. Methods Enzymol. 422:245–269 [DOI] [PubMed] [Google Scholar]
- 49. Larsen R. A., Myers P. S., Skare J. T., Seachord C. L., Darveau R. P., Postle K. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.