Summary
The specialized ribonuclease Dicer plays a central role in eukaryotic gene expression by producing small regulatory RNAs – miRNAs and siRNAs – from larger double stranded RNA (dsRNA) substrates. Although Dicer will cleave both imperfectly base-paired hairpin structures (pre-miRNAs) and perfect duplexes (pre-siRNAs) in vitro, it has not been clear whether these are mechanistically equivalent substrates and how dsRNA binding proteins such as TRBP influence substrate selection and RNA processing efficiency. We show here that human Dicer is much faster at processing a pre-miRNA substrate compared to a pre-siRNA substrate under both single and multiple turnover conditions. Maximal cleavage rates (Vmax) calculated by Michaelis-Menten analysis differed by more than 100-fold under multiple turnover conditions. TRBP was found to enhance dicing of both substrates to similar extents, and this stimulation required the two N-terminal dsRNA binding domains of TRBP. These results demonstrate that multiple factors influence dicing kinetics. While TRBP stimulates dicing by enhancing the stability of Dicer-substrate complexes, Dicer itself generates product RNAs at rates determined at least in part by the structural properties of the substrate.
Introduction
MicroRNAs (miRNAs) and short interfering RNAs (siRNAs) play central roles in controlling eukaryotic gene expression and have attracted increasing attention as potential therapeutic agents. In the cytoplasm, many of these RNAs are generated from cleavage of longer double-stranded RNA (dsRNA) substrates by the enzyme Dicer, a large multi-domain, Ribonuclease III-family protein 1; 2. However, the precursors to these RNAs are structurally distinct: pre-miRNAs are predicted to be imperfectly base-paired hairpins with a variable-size loop, whereas pre-siRNAs are usually perfectly base-paired duplexes of variable length with no loop. Some species have functionally distinct Dicers that process different kinds of substrates. For example, in D. melanogaster Dcr-1 and Dcr-2 are responsible for the biogenesis of miRNAs and siRNAs, respectively 3. However, there is only one known Dicer in the human system, which putatively processes both pre-miRNAs and pre-siRNAs. Understanding how processing rates for human Dicer are affected by substrate differences and by Dicer’s protein binding partners is paramount to identifying the molecular mechanisms that underlie regulatory RNA production.
The catalytic core of Dicer, represented by the structure of the enzyme from Giardia intestinalis, comprises a PAZ domain N-terminal to two Ribonuclease III domains 4. This structure, likely to be common to all Dicer enzymes, led to a model for dsRNA recognition in which substrates are positioned for cleavage to produce dsRNA products of a specific 21–27 bp length 1. However, it remains unclear how Dicer accommodates substrate RNAs with the different helical structures and thermodynamic stabilities that characterize pre-miRNAs and pre-siRNAs.
In most eukaryotes, Dicer contains additional domains beyond those found in the Giardia enzyme, including an N-terminal DExH/D-box helicase, a domain of unknown function (DUF 283) and a C-terminal dsRNA binding domain (dsRBD) (Fig. 1A). These domains function at least in part to recruit dsRNA binding proteins that are central players in RNA-mediated gene silencing 5; 6; 7; 8.
Figure 1.
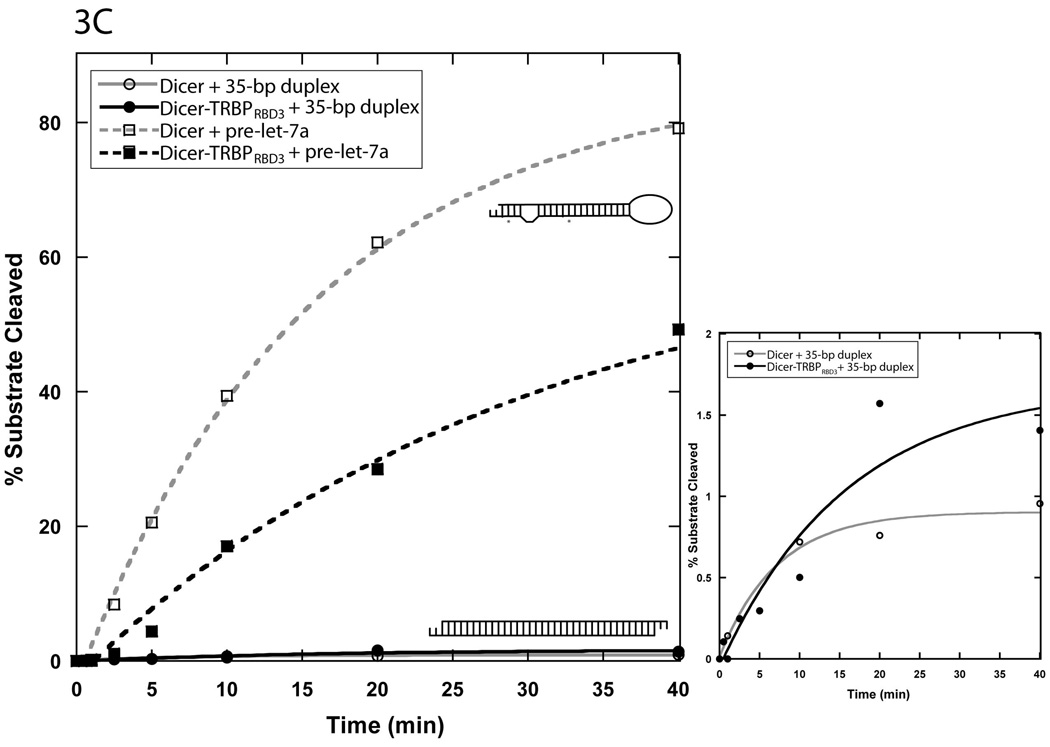
(a) Schematic representation of the domain organization of the proteins involved in this study: full-length human dicer and TRBP. Mutations made to generate the phospho-mimic mutant of TRBP (TRBPpm) are denoted. Also shown are cartoon representations of the two substrates used in this study, pre-let-7a and a 35-bp perfectly complementary duplex with 3’ two-nucleotide overhangs. There are four G-U wobbles in the stem of pre-let-7a, two of which are predicted by Mfold 37 to be base-paired (depicted with asterisks) and two of which are predicted to be mis-matched (depicted with a bulge). Arrows indicate sites of Dicer-mediated cleavage. (b) Single turnover dicing assay with 35-bp duplex and human pre-let-7a using Dicer alone showing products of expected length based on a single pair of cleavage sites for pre-let-7a and two pairs (one on either end) for the 35-bp duplex. Reactions contained <1 nM RNA and 25 nM Dicer, and were incubated at 37 °C. The 16% urea-polyacrylamide gels corresponding to the plots are shown above the graph. The plots for each substrate are marked by substrate icons, as depicted in (a). (c) Multiple turnover dicing assay with 50 nM 35-bp duplex and pre-let-7a using Dicer alone at a concentration of 5 nM. The plots for each substrate are marked by substrate icons, as depicted in (a). The inset shows an expanded y-axis of cleavage data for the 35-bp duplex.
Studies of RNA samples isolated from mammalian cells having or lacking Dicer show that a majority of Dicer products are miRNAs 9; 10. Some examples of endogenous siRNAs have been identified, but their overall occurrence is relatively uncommon in these cell types 11; 12. Nevertheless, siRNA precursors such as short hairpin RNAs (shRNAs) are thought to be processed by human Dicer, and some studies suggest that such processing has a vital role in enhancing siRNA efficiency by facilitating optimal RNA-induced silencing complex (RISC) loading 13; 14.
In addition to Dicer’s potential for differential substrate recognition, double-stranded RNA binding proteins that directly interact with Dicer could influence its activity 15. These proteins, typically comprising 2–3 consecutive dsRBDs connected by variable-length linkers, include RDE-4 in C. elegans 16, R2D2 and Loquacious/R3D1 in D. melanogaster 17; 18 and TRBP and PACT in H. sapiens 5; 6. In C. elegans, RDE-4 is thought to be exclusively involved in the siRNA pathway 19. In D. melanogaster, the substrate preferences detected for R2D2 and Loquacious/R3D1 suggest roles in selecting pre-siRNA and pre-miRNA substrates, respectively, and in specifying guide strand selection during RISC assembly 18; 20; 21; 22. In humans, the HIV trans-activating response (TAR) RNA binding protein (TRBP) ensures efficient recruitment of miRNA onto Argonaute 2 (Ago-2) in RISC and enhances the stability and efficiency of both Dicer and RISC 5; 13; 23; 24; 25. Perhaps as a result, changes in steady-state levels of certain miRNA families correlate with TRBP truncation mutations that characterize some colorectal cancers 26. PACT has also been shown to bind to human Dicer 6; 25, though its effect on dsRNA binding or processing remains unclear.
To investigate the effects of substrate variants and TRBP on dicing kinetics, we compared the processing rates of two distinct dsRNA substrates containing two-nucleotide 3’ overhangs: a 73-nucleotide hairpin with an imperfect stem (human pre-let-7a) and a 35-bp perfect duplex (Fig. 1A). Dicer showed a marked preference for processing pre-let-7a over the 35-bp duplex, an effect most pronounced under multiple turnover conditions in which substrate is in excess over enzyme. TRBP increased the rate of endonucleolytic cleavage of both substrates by ~4 to 5-fold under multiple turnover conditions. This stimulatory effect required the two N-terminal dsRBDs of TRBP. These results reveal substantial variability in Dicer-catalyzed small RNA processing depending on substrate sequence and secondary structure. Although TRBP stimulates dicing activity, substrate-dependent differences in dicing rates are inherent to human Dicer itself. Our findings imply that levels of miRNAs and siRNAs are influenced at least in part by precursor structure and/or sequence. Furthermore, levels of regulatory RNA in the cell could change as a consequence of mutations that alter substrate RNA structure or stability.
Results and Discussion
Dicing rates vary with different substrates
In vitro, human Dicer will cleave various dsRNA substrates including pre-miRNAs and short or long perfect duplexes to generate miRNAs and siRNAs, respectively. These substrates are structurally distinct, leading to potential differences in dicing rates that could contribute to variability in miRNA and/or siRNA levels in vivo. To directly test dicing rates of these two substrate types, we designed a 73-nucleotide hairpin with the sequence of the human pre-miRNA pre-let-7a and a 35-bp perfect duplex representing a pre-siRNA. Both substrates contained 3’ two-nucleotide overhangs for optimal recognition by Dicer (Fig. 1A).
We performed dicing assays under single turnover conditions, in which Dicer is present in large molar excess over the substrate concentration. Under these reaction conditions, the rate of product formation is not limited by product release since each enzyme reacts at most with one substrate molecule. Cleavage of pre-let-7a was considerably faster than that observed for the 35-bp duplex. The t1/2 (time to cleave half of the starting material) for pre-let-7a was <5 minutes, whereas for the 35-bp duplex it was ~80 minutes (Fig. 1B).
Under multiple turnover conditions, in which substrate concentration is in molar excess over that of the enzyme, an even more striking difference in substrate processing rates was observed. With a 10-fold excess of substrate over Dicer, the t1/2 for pre-let-7a was ~15 minutes, whereas <1% of the 35-bp duplex was cleaved in the same amount of time (Fig. 1C). Thus, Dicer catalyzes cleavage of multiple pre-let-7a substrates per enzyme molecule while being unable to process the 35-bp substrate in multiple turnover fashion under identical reaction conditions.
Equilibrium binding affinities of each substrate for Dicer are similar (Table 1), which suggests that the differences in rate and extent of Dicer-mediated cleavage with these substrates are not simply a consequence of substrate binding affinity. Other possible explanations include inefficient product release or sub-optimal substrate loading, resulting in catalytic defects and/or the formation of non-productive enzyme-substrate complexes. Electrophoretic mobility shift assays (EMSA) performed in prior studies, as well as our own filter binding data, showed that Dicer has a very low affinity for duplexes of length that represents dicing products (Table 1) 23. Additionally, the results from our single turnover experiments demonstrated that cleavage rates already differ by >10-fold under conditions in which product release should not influence the accumulation of product. We therefore speculate that the 35-bp duplex is “mis-loaded” onto Dicer, thereby retarding the progress of the reaction. This might not occur with pre-let-7a if its structural features preclude non-productive binding to Dicer. This possibility is consistent with the observation that in vivo, human Dicer is predominantly responsible for the production of miRNAs 9. It is noteworthy that even among pre-miRNAs, there naturally exist many variations in structure such as the size of the loop and the number of mismatches in the stem, which may lead to considerable differences in the way distinct substrates are recognized and processed by Dicer. Recent studies have revealed pre-miRNAs with structural properties that make them unsuitable substrates for Dicer and are instead processed by Ago-2 27; 28 It is therefore conceivable that Dicer has a specific set of substrate structural requirements that lead to considerable variability in catalytic efficacy.
Table 1.
Equilibrium dissociation constants (Kds) for protein:RNA complexes
| Dicing Substrates | Dicing Product | ||
|---|---|---|---|
| pre-let-7a | 35-bp duplex | 19-bp duplex | |
| TRBP | 1.3 ± 0.2 nM | < 90 pMa | 0.29 ± 0.05 nMb |
| TRBPpm | 2.9 ± 0.6 nM | < 100 pMa | 1.04 ± 0.07 nMb |
| Dicer | 1.8 ± 0.4 nM | 3.4 ± 0.5 nM | 700 ± 100 nMc |
| Dicer-TRBP | < 50 pMa | < 51 pMa | 0.24 ± 0.04 nMb |
| Dicer-TRBPpm | < 40 pMa | < 52 pMa | ND |
| Dicer-TRBPRBD3 | 1.3 ± 0.1 nM | 3.0 ± 0.4 nM | ND |
Equilibrium dissociation constants were determined using filter binding as described in the Methods. Data were analyzed using a standard binding isotherm unless otherwise stated, according to the equation: fraction bound = A × [protein] / (Kd + [protein]), where A is the amplitude of the binding curve. ND, not determined. Errors, where included, represent the standard fitting error.
Because of the extremely low equilibrium dissociation constants for these complexes, it was not possible to lower the RNA concentration sufficiently below the Kd. Therefore, binding isotherms were fit with the solution of a quadratic equation describing a bimolecular dissociation reaction, as described previously38. Kds are reported as an upper bound because the RNA concentration in the reactions could not be precisely determined, thereby limiting the accuracy of the linear regression fit.
Binding curves displayed apparent negative cooperativity and were best fit by allowing a variable Hill coefficient (n = 0.4–0.6). Reported Kds are K0.5 values, i.e. the protein concentration at which the fraction RNA bound is equal to 50%.
The low affinity of this complex prevented complete definition of the saturation regime in the binding curve. To avoid underestimating the Kd as a result, the amplitude was fixed (A = 1) when fitting the data. The reported Kd represents an upper bound estimate.
TRBP has a stimulatory effect on dicing kinetics
TRBP binds and stabilizes Dicer, modifies Dicer’s endonucleolytic activity and has been suggested to help load product dsRNAs onto Ago-2 during formation of RISCs 5; 23. In addition, TRBP truncations correlate with changes in the population distributions of certain endogenous miRNAs in vivo 26. Studies using chimeric constructs of RDE-4 and TRBP showed that the two N-terminal dsRBDs contribute largely to the dsRNA binding affinity of TRBP 29. Whether and how TRBP influences Dicer’s selection and speed of dsRNA substrate processing has not been well characterized.
To test the influence of TRBP on dicing kinetics, recombinantly expressed Dicer and TRBP were incubated together to form a heterodimeric complex, which was then purified by size exclusion chromatography to remove any free TRBP 7. Control experiments showed that excess TRBP binds tightly to dsRNA and sequesters it from the Dicer-TRBP complex, inhibiting dicing without necessarily reflecting the behavior of the Dicer-TRBP complex itself (data not shown).
Equilibrium binding analysis of Dicer-dsRNA complexes in the presence and absence of TRBP, using an electrophoretic mobility shift assay, suggested that TRBP enhances complex stability with both pre-let-7a and the 35-bp duplex (Fig. 2A). The Dicer-TRBP complex gave rise to a slowly migrating ternary complex with either dsRNA substrate, whereas Dicer alone did not. To obtain quantitative information about dsRNA binding affinities, we performed filter binding assays to measure equilibrium dissociation constants (Kds) of Dicer alone and the Dicer-TRBP complex bound to both substrates. Although there is little difference in binding affinities between the two substrates, the Dicer-TRBP-RNA complex has a significantly lower Kd compared to the Dicer-RNA complex without TRBP (<50 pM versus ~2 nM for pre-let-7a and <51 pM versus ~3.5 nM for the 35-bp duplex, Table 1).
Figure 2.
(a) Electrophoretic mobility shift assay with both 35-bp duplex and pre-let-7a showing the relative tendencies of Dicer and Dicer-TRBP to form stable protein-RNA complexes. Binding reactions were analyzed with 6% non-denaturing PAGE and stained with ethidium bromide. The gels corresponding to each substrate are marked at the bottom by substrate icons, as depicted in 1a. (b) Single turnover dicing assay with 35-bp duplex and pre-let-7a using a Dicer-TRBP complex under conditions identical to 1b. The plots for Dicer alone are shown in grey for the purpose of comparison. The plots for each substrate are marked by substrate icons, as depicted in 1a. (c) Multiple turnover dicing assays with 25 nM 35-bp duplex and 50 nM pre-let-7a using a Dicer-TRBP complex under conditions identical to 1c. The plots for Dicer alone are shown in grey for the purpose of comparison. The inset shows an expanded y-axis of cleavage data for the 35-bp duplex.
We then performed dicing assays using the Dicer-TRBP complex. Under single turnover conditions, the purified Dicer-TRBP complex showed a slight stimulatory effect on dicing of the 35-bp duplex (Fig. 2B). While the t1/2 for Dicer alone was ~80 minutes, it was ~45 minutes for Dicer-TRBP. In the case of pre-let-7a we found Dicer-TRBP to be identical to Dicer alone (t1/2 of <5 minutes) (Fig. 2B). Under multiple turnover conditions, TRBP caused an increase in the rate and extent of dicing with both substrates. Notably, however, the presence of TRBP did nothing to ameliorate the relatively inefficient processing of the 35-bp duplex compared to pre-let-7a (Fig. 2C). With pre-let-7a, the t1/2 for Dicer alone was ~15 minutes, compared to ~8 minutes for the Dicer-TRBP complex (Fig. 2C).
TRBP-mediated stimulation of dicing could occur at multiple stages of the dicing reaction trajectory. The much higher affinity of the Dicer-TRBP complex for RNA substrates, as compared to Dicer alone (Table 1), strongly suggests that TRBP accelerates dicing in part by assisting in substrate recruitment. It is also possible that substrate binding exerts some amount of conformational strain on Dicer, which TRBP may alleviate via its interaction with Dicer and by properly orienting the substrate in Dicer’s active site. Furthermore, we cannot exclude the possibility that TRBP also plays a role facilitating product release and enzyme turnover. Future experiments will be needed to fully explain the mechanistic basis for TRBP’s stimulatory effect on dicing.
TRBP requires high RNA binding affinity to stimulate dicing
In order to obtain a more detailed picture of the effect of substrate variation and the presence of TRBP on Dicer function, we performed dicing assays under multiple turnover conditions using a series of substrate concentrations. Michaelis-Menten plots obtained from these data revealed that Dicer’s maximum cleavage rate (Vmax) for pre-let-7a is 100-fold higher than that for the 35-bp perfect duplex. We also find that the Dicer-TRBP complex shows a 4–5 fold increase in Vmax over that measured for Dicer alone, irrespective of substrate type (Fig. 3A, B). Thus, the specific sequence and structure of RNA substrates have substantial effects on catalysis rates, resulting in Vmax differences spanning multiple orders of magnitude. Furthermore, TRBP leads to an overall enhancement in cleavage rate, regardless of the specific substrate. Surprisingly, Km values extracted from the Michaelis-Menten curves are in the tens of nanomolar range (Fig. 3A, B), both in the presence and absence of TRBP, whereas the Kds for substrate binding are up to three orders of magnitude lower (Table 1). Because Km does not equal Kd for these complexes, product formation may not be the rate-limiting step. For example, substrate molecules may need to undergo conformational changes after initial binding in order to form productive enzyme-substrate complexes.
Figure 3.
(a) Michaelis-Menten analysis of dicing of the 35-bp duplex. 5 nM enzyme (Dicer, Dicer-TRBP or Dicer-TRBPpm) was incubated with 5’ 32P-labeled substrate at the concentrations shown. (b) Michaelis-Menten analysis of dicing of pre-let-7a. 5 nM enzyme (Dicer, Dicer-TRBP or Dicer-TRBPpm) was incubated with 5’ 32P labeled substrate at the concentrations shown. (c) Multiple turnover assays with the 35-bp duplex and pre-let-7a comparing the activity of Dicer-TRBPRBD3 to that of Dicer alone. Cleavage data for the 35-bp duplex is re-plotted to the right with an expanded y-axis. The plots for each substrate are marked by substrate icons, as depicted in 1a.
The stimulation of dicing activity by TRBP could either be due to its dsRNA binding affinity or a conformational change brought about by protein-protein interactions between TRBP and Dicer, or a combination of both. Previous RNA-binding studies using wild-type or mutated forms of TRBP implicated the first two dsRBDs in RNA binding; while dsRBD2 was clearly shown to have the highest affinity for dsRNA 30, dsRBD1 also enhanced the RNA binding affinity of a TRBP-RDE4 chimera 29. In agreement, studies of truncation mutants showed that the C-terminal dsRBD3 is dispensable for RNA binding 29; 31 and contributes primarily to protein-protein interactions and Dicer binding 5; 32. Furthermore it has also been shown in Drosophila that a truncation mutant of Loquacious containing dsRBD2 and dsRBD3 is sufficient both for binding Dcr-1 (via dsRBD3) and enhancing Dcr-1 mediated miRNA production (via dsRBD2) 33.
In order to investigate the roles of the dsRNA-binding and Dicer-binding dsRBDs of TRBP in stimulating dicing, we designed a truncation construct (TRBPRBD3) that lacked dsRBD1 and dsRBD2 but retained its ability to interact with Dicer. We were able to purify the Dicer-TRBPRBD3 complex, which failed to produce an electrophoretic mobility shift with either the pre-let-7a or the 35-bp dsRNA substrate, similar to the behavior of Dicer alone (data not shown). Additionally, the measured binding affinity of Dicer-TRBPRBD3 for either substrate is within error of the affinity measured for Dicer alone (Table 1). These observations strongly suggest that TRBP confers high dsRNA binding affinity to the Dicer-TRBP complex via its two N-terminal dsRBDs.
The Dicer-TRBPRBD3 complex was tested in dicing assays with both pre-let-7a and the 35-bp duplex under multiple turnover conditions. In both cases, this enzyme complex produced little or no stimulation of dicing relative to the rate observed for Dicer alone (Fig. 3C). However, we have not completely eliminated the possibility that TRBPRBD3 has a lower binding affinity for Dicer compared to that of full-length TRBP, and therefore dissociates appreciably during the reaction. Nevertheless, these data indicate that dsRBD2 – and perhaps dsRBD1 to a lesser extent – are required for TRBP’s stimulatory affect on dicing. In agreement with studies of Loquacious and Dcr-1 from Drosophila 33, dsRBD3 is required for Dicer binding. Future studies of TRBP containing point mutations in dsRBD1 and dsRBD2 that inhibit RNA binding will be needed to investigate whether these domains are also involved in protein-protein interactions that affect dicing kinetics.
A TRBP phosphorylation mimic does not significantly affect dicing kinetics
The post-translational phosphorylation of TRBP in vivo has been shown to increase the apparent half-life of Dicer and affect cellular miRNA levels 24. TRBP expressed in E. coli is devoid of post-translational modifications, whereas TRBP expressed in a baculovirus system exhibits heterogeneous phosphorylation 24Enbo Ma and J.A.D., unpublished data). Therefore, to test whether the TRBP phosphorylation state affects Dicer’s enzymatic activity, we purified a phospho-mimic mutant of TRBP (TRBPpm) containing aspartate residues at each of the four positions shown to be sites of phosphorylation in vivo (Fig. 1A). This protein was shown to recapitulate the effects of native phosphorylated TRBP in cultured human cells 24. We performed dicing assays using a Dicer-TRBPpm complex under multiple turnover conditions at substrate concentrations that were previously used for Michaelis-Menten analysis. We found that for both substrates used in this study, Dicer-TRBPpm – mediated cleavage did not differ significantly from that measured for Dicer-TRBP. While we see a 1.5 fold increase in the Vmax for dicing the 35-bp duplex, the Vmax for dicing pre-let-7a remained virtually identical (Fig. 3A, B).
Paroo et al. showed that the relative abundance of certain miRNAs varied between cells expressing the phospho-mimic TRBP mutant and those expressing a non-phosphorylatable TRBP mutant. Notably, in this study, let-7a was among the miRNAs whose abundance decreased in the TRBPpm-containing cells. The absence of an effect of TRBPpm on dicing rates in this study suggests that instead of altering dicing kinetics, TRBP phosphorylation may influence miRNA levels by an indirect mechanism. A more detailed analysis with pre-miRNAs representative of both the up-regulated and down-regulated populations would nevertheless be desirable to evaluate the possibility that TRBP phosphorylation influences dicing kinetics in other contexts. We note that the dissociation constants for TRBPpm:RNA complexes are comparable to those measured for wild-type TRBP (Table 1), consistent with the idea that TRBP stimulates dicing by a mechanism that generally depends on its RNA binding affinity.
Conclusions
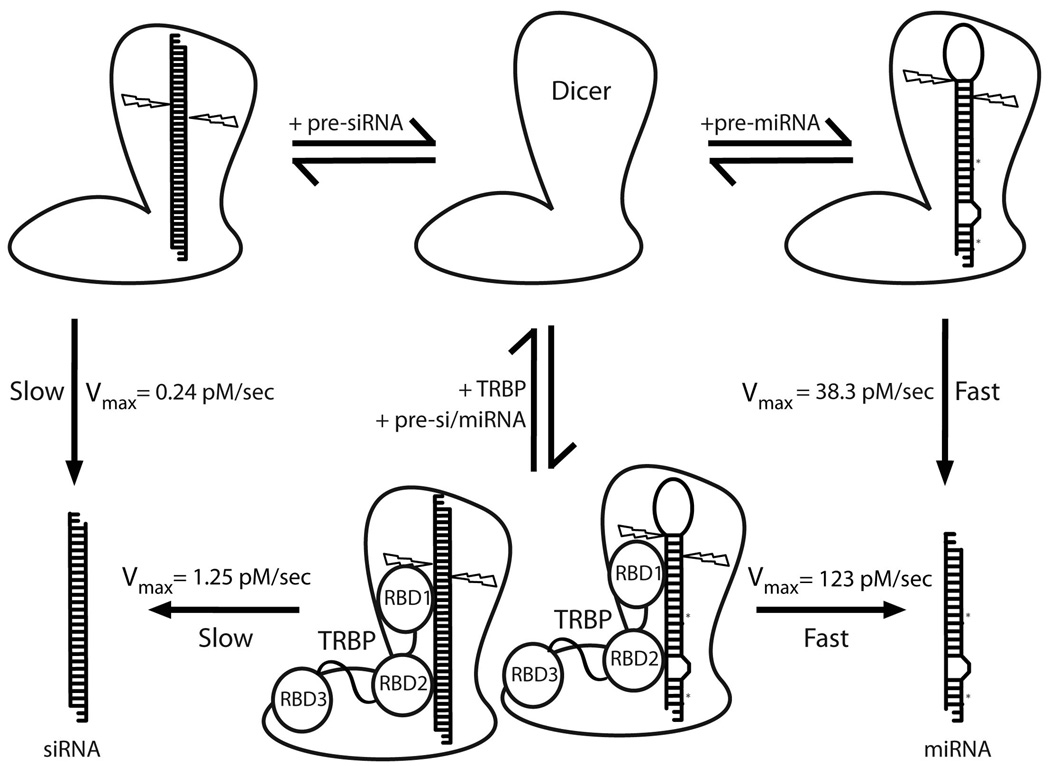
Among Dicer-dependent small RNAs in mammalian cells, miRNAs are by far the most prevalent and well characterized 9; 10. The occurrence of endogenous siRNAs in mammalian cells is a recently discovered and relatively rare phenomenon 11; 12. It is therefore plausible that pre-miRNAs are the natural substrates for Dicer in human cells and that other kinds of small RNAs, if processed in vivo, are generated by Dicer-independent mechanisms. We propose that specific structural features of pre-miRNAs facilitate optimal loading and therefore efficient Dicer-mediated processing, which is precluded in the case of other substrates such as the pre-siRNA analyzed in this study (Fig. 4). The TRBP-dependent increase in dicing rates observed for both substrates, an effect that requires high-affinity RNA binding, suggests that TRBP assists in recruiting substrates to Dicer, as previously suggested in the C. elegans and D.melanogaster systems 29; 33; 34, and/or stabilizes the interaction between Dicer and its RNA substrates (Fig. 4). However, Dicer itself possesses an intrinsic ability to distinguish between substrates with different sequences and structural characteristics. Together, these results show that the specific nature of the dicing substrate heavily influences the catalytic production of regulatory RNAs and could therefore be an important aspect of small RNA biogenesis in vivo and for the development of RNAi-based therapeutics.
Figure 4.
Model depicting Dicer’s substrate selectivity and the effect of TRBP on dicing kinetics. Cartoon representatives of Dicer and Dicer-TRBP are based on a low resolution structure determined by electron microscopy 8. The ~100 fold difference in Vmax for pre-miRNA and pre-siRNA dicing could possibly reflect a difference in the loading efficacy, which in turn may be a function of the nature of the substrate. The N-terminal helicase domain of Dicer is known to bind the C-terminal dsRBD3 of TRBP, which in turn may mediate protein-nucleic acid interactions between the substrate RNA and one or both of the other dsRBDs of TRBP, thus stabilizing the enzyme-substrate complex. This could explain the 4–5 fold increase in activity of Dicer upon binding TRBP.
Materials and Methods
Sample preparation
Dicer, TRBP and Dicer-TRBP complexes were purified using published protocols 35. We used site-directed mutagenesis to incorporate the TRBP point mutants S142D, S152D, S283D and S286D, and purified TRBPpm 24 using the same procedure as for wild-type TRBP. Human pre-let-7a was synthesized by in vitro transcription using T7 RNA polymerase from a DNA template containing a double ribozyme system to ensure homogeneous 5′ and 3′ ends 36. The two strands of the 35-bp duplex (a and b) and those of the 19-bp duplex (a and b) were synthesized by IDT (Integrated DNA technologies, Coralville, IA). All substrates were purified using 16% urea-PAGE, and purified pre-let-7a, 35-bp duplex b and 19-bp duplex a were then 5’ end-labeled using T4 polynucleotide kinase (New England Biolabs Inc., Beverly, MA) and (γ-32P)-ATP. The substrates were heated at 65°C for 10 minutes in annealing buffer containing 3mM MgCl2, 30mM NaCl, and 100mM Tris.HCl (pH 7.5), followed by flash cooling for refolding pre-let-7a and slow cooling to anneal the 35-bp and 19-bp duplexes. The sequences of the substrates are as follows:
pre-let-7a: 5′-UGAGGUAGUAGGUUGUAUAGUUUUAGGGUCACACCCACCACUGGGAGAUAACUAUACAAUCUACUGUCUUACC-3′
35-bp duplex a: 5’-UGAGGUAGUAGGUUGUAUAGUUUGAAAGUUCACGAUU-3’
35-bp duplex b: 5’-UCGUGAACUUUCAAACUAUACAACCUACUACCUCAAA-3’
19-bp duplex a: 5’-GUCACAUUGCCCAAGUCUCTT-3’
19-bp duplex b: 5’-GAGACUUGGGCAAUGUGACTT-3’
Kinetic assays
Single turnover assays were performed in 45 µL mixtures, which consisted of 25 nM enzyme and <1 nM 5’ 32P-labeled substrate. Upon incubation at 37 °C, 5 µL aliquots were removed and mixed with 6 µL of loading buffer at time points of 0.5, 1, 2.5, 5, 10, 20, 40 and 80 minutes. These samples were heated at 70 °C for 10 minutes prior to being run on a 16% urea-polyacrylamide gel and quantified with a phosphorimager (GE Healthcare). The data were plotted using Kaleidagraph (Synergy software). Multiple turnover dicing assays were also performed in 45 µL mixtures containing 5 nM enzyme and 5’ 32P-labeled substrate at concentrations of 25, 50, 75, 100, 150, 225 and 375 nM. Reactions were incubated at 37 °C and 5 µL aliquots were taken after 0.5, 1, 2.5, 5, 10, 20, 40 and 80 minutes. Variations in the time points were required in order to adapt to varying levels of dicing activity with different substrates and different proteins, as indicated in the figure axes. After quantification, initial velocities (vo) at each substrate concentration (S) were determined by linear regression, and Vmax along with standard errors were calculated by fitting to the Michaelis-Menten equation, vo = (Vmax × S)/(Km + S), using Kaleidagraph.
Electrophoretic Mobility Shift Assay
35-bp duplex and pre-let-7a were annealed in 100 mM Tris-HCl (pH 7.5), 3 mM MgCl2 and 30 mM NaCl by heating at 65°C for 10 minutes and either slow cooled (35-bp duplex) or flash cooled (pre-let-7a). ~10.5 picomoles of each of these RNAs were then incubated with equimolar Dicer, Dicer-TRBP or Dicer-TRBPRBD3 for an hour at 4 °C in 20 mM Tris–HCl (pH 7.5), 25 mM NaCl, 5 mM EDTA, 1 mM DTT and 1% glycerol. Reactions were analyzed on a 6% non-denaturing polyacrylamide gel by staining with ethidium bromide.
Filter Binding Assays
Serial dilutions of Dicer, TRBP or Dicer-TRBP constructs were incubated with <1 nM of 5’ 32P-radiolabeled 35-bp duplex, pre-let-7a or 19-bp duplex in binding buffer containing 20 mM Tris-HCl (pH 7.5), 25 mM NaCl, 1 mM DTT, 1% glycerol and 0.01% Igepal CA-630. Reactions with Dicer were supplemented with 5 mM EDTA to prevent RNA cleavage by Mg2+ chelation. After incubating reactions (25 or 50 µL volume) on ice for one hour, samples were applied by vacuum to a dot-blot apparatus containing two pieces of Whatman filter paper below three membranes: 0.2 µm pore size Tuffryn (Pall Co.), 0.1 µm pore size Protran (Whatman) and Hybond-N (Amersham). The membranes were equilibrated in binding buffer for at least 20 minutes prior to use. After applying reactions and washing with 50 µL of binding buffer, membranes were dried and exposed overnight on a phosphor screen. The amounts of free RNA (retained on Hybond-N membrane) and protein-bound RNA (retained on Protran membrane) were quantified using a phosphorimager (GE Healthcare). The extent of complex aggregation, as detected by radioactivity on the Tuffryn membrane, was negligible. The fraction of RNA bound, calculated as the ratio of radioactivity on the Protran membrane to the sum of radioactivity on the Protran and Hybond-N membranes, was plotted as a function of protein concentration. Kds were determined by fitting the data to binding isotherms using KaleidaGraph (see Table 1 for details).
Research Highlights
Human Dicer shows significant variability in dicing efficiency in a substrate sequence and/or structure dependent manner.
We report a clear preference for pre-miRNAs over pre-siRNAs.
The Dicer associated dsRNA binding protein TRBP engenders a ~ 5 – fold increase in endonucleolytic activity with both kinds of substrates.
This stimulatory activity of TRBP is most likely dependent on the high RNA binding affinity attributable primarily to dsRBD2 and perhaps, to a smaller extent, also to dsRBD1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 2.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 5.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 12.Meiri E, Levy A, Benjamin H, Ben-David M, Cohen L, Dov A, Dromi N, Elyakim E, Yerushalmi N, Zion O, Lithwick-Yanai G, Sitbon E. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq376. doi:10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paroo Z, Liu Q, Wang X. Biochemical mechanisms of the RNA-induced silencing complex. Cell Res. 2007;17:187–194. doi: 10.1038/sj.cr.7310148. [DOI] [PubMed] [Google Scholar]
- 16.Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 17.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 19.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 20.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 23.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 26.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, Ferreira B, Liu CG, Villanueva A, Capella G, Schwartz S, Jr, Shiekhattar R, Esteller M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008;384:967–979. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daviet L, Erard M, Dorin D, Duarte M, Vaquero C, Gatignol A. Analysis of a binding difference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur J Biochem. 2000;267:2419–2431. doi: 10.1046/j.1432-1327.2000.01256.x. [DOI] [PubMed] [Google Scholar]
- 31.Gatignol A, Buckler C, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniels SM, Melendez-Pena CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Laine S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- 34.Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferre-D'Amare AR, Doudna JA. Use of cis- and transribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]