Abstract
Dishevelled-associated activator of morphogenesis 1 (Daam1), a member of the formin protein family, plays an important role in regulating the actin cytoskeleton via mediation of linear actin assembly. Previous functional studies of Daam1 in lower species suggest its essential role in Drosophila trachea formation and Xenopus gastrulation. However, its in vivo physiological function in mammalian systems is largely unknown. We have generated Daam1-deficient mice via gene-trap technology and found that Daam1 is highly expressed in developing murine organs, including the heart. Daam1-deficient mice exhibit embryonic and neonatal lethality and suffer multiple cardiac defects, including ventricular noncompaction, double outlet right ventricles and ventricular septal defects. In vivo genetic rescue experiments further confirm that the lethality of Daam1-deficient mice results from the inherent cardiac abnormalities. In-depth analyses have revealed that Daam1 is important for regulating filamentous actin assembly and organization, and consequently for cytoskeletal function in cardiomyocytes, which contributes to proper heart morphogenesis. Daam1 is also found to be important for proper cytoskeletal architecture and functionalities in embryonic fibroblasts. Biochemical analyses indicate that Daam1 does not regulate cytoskeletal organization through RhoA, Rac1 or Cdc42. Our study highlights a crucial role for Daam1 in regulating the actin cytoskeleton and tissue morphogenesis.
Keywords: Congenital heart defects, Daam1, Formin protein, Heart development, Cytoskeletal architecture, Sarcomere organization, Actin, Planar cell polarity (PCP), Mouse
INTRODUCTION
Actin is one of the most highly conserved and abundant proteins in all eukaryotes (Pollard and Cooper, 2009). It is essential for cell survival and for many fundamental cellular processes, such as cytokinesis, vesicle trafficking and cell locomotion (Campellone and Welch, 2010). Additionally, actin filaments are major determinants of cell morphology and adhesion, which contribute to proper tissue morphogenesis (Li and Gundersen, 2008). There are over 100 accessory proteins engaged in actin regulation in eukaryotes, e.g. nucleation, polymerization and turnover (Pollard and Cooper, 2009). Interestingly, many of these proteins are specialized in unique aspects of actin regulation, rather than being functionally redundant. For example, Arp2/3 complex is committed to the initiation and elongation of branched actin filaments, whereas formins are required for the linear addition of actin monomers to the barbed ends of growing filaments (Sarmiento et al., 2008).
Characteristic features of the formin protein family include two shared domains: forming homology 1 (FH1) and forming homology 2 (FH2), which are essential for mediating linear actin polymerization (Goode and Eck, 2007). Eukaryotic species have multiple formin proteins, including Dia, Fmn, Fmnl and Daam, the activities of which are regulated by diverse signals (Schonichen and Geyer, 2010). However, our current understanding of their physiological functions is quite limited, particularly in mammals. Through their actin regulating capacity, formins probably impact tissue morphogenesis, a biological/developmental process involving complex signaling pathways mediated by cytoskeletal dynamics (Zallen, 2007). Indeed, Fmn1 was found to be crucial for limb development in mouse (Zhou et al., 2009). Surprisingly, Dia1 (Diap1 – Mouse Genome Informatics) knockout mice were developmentally/morphologically normal, albeit with an age-dependant myeloproliferative defect (Peng et al., 2007). Despite that, Dia1 displays significantly stronger activity in promoting actin nucleation than do other formin family members (Higashi et al., 2008). These findings strongly suggest that each formin plays a unique and specific physiological function, largely depending on its individual expression profile, upstream activators, localization and inherent activity.
Dishevelled-associated activator of morphogenesis 1 (Daam1) was initially identified as a protein interacting with Dishevelled (Dvl), which mediates the non-canonical Wnt/planar cell polarity (PCP) signaling pathway (Habas et al., 2001). Daam1 localizes to the plasma membrane and cytoplasmic vesicles, and this pattern is tightly regulated by Wnt and Dvl (Habas et al., 2001; Kida et al., 2007; Kim and Han, 2007; Matusek et al., 2008). Daam1 is inactive when it forms a closed loop between the N-terminal DID (diaphanous inhibitory domain) and the C-terminal DAD (diaphanous auto-regulatory domain). Upon binding of Dvl to DAD, Daam1 is robustly activated. It is activated to a lesser extent upon binding to Rho-GTP via the GBD (GTPase-binding domain) (Liu et al., 2008). A previous study in Xenopus suggested xDaam is crucial for gastrulation (Habas et al., 2001), whereas Drosophila Daam mutants exhibit trachea defects (Matusek et al., 2006). These early findings suggest that Daam1 impacts tissue morphogenesis. The physiological function of mammalian Daam1 has not heretofore been fully characterized.
Here, we have found that Daam1 is highly expressed in developing murine organs, including heart. Studies using Daam1-deficient mice reveal that Daam1 is essential for cardiac morphogenesis. At the subcellular level, Daam1 is crucial for filamentous actin (F-actin) assembly and organization, sarcomeric organization, and cell adhesion and alignment. The abnormal heart morphogenesis seen in Daam1 mutants is probably caused by abnormal Daam1-mediated cytoskeletal regulation. Biochemical studies indicate that Daam1 does not mediate its effect on the cytoskeleton through regulating activities of RhoA, Rac1 or Cdc42. Our study highlights a crucial role for Daam1 in regulating the actin cytoskeleton and tissue morphogenesis.
MATERIALS AND METHODS
Generation of Daam1 gene trap mouse model
A promoter trapped embryonic stem (ES) cell line (cell No.: RRT390) containing an insertional mutation in the mouse Daam1 gene was from BayGenomics. The gene trap vector contains a splice-acceptor sequence flanked by loxP sites subcloned 5′ of the β-geo reporter cassette, which encodes a fusion protein of the bacterial lacZ and neomycin phosphotransferase II. RRT390 embryonic stem cells were injected into blastocysts from C57BL/6J mice. The generated chimeric males were then intercrossed with C57BL/6J females to generate F1 offspring. PCR primers for genotyping were: D1 forward (on intron 3 of Daam1), 5′-AGCATTCTGAAAGTCATCGTCTTT-3′; D1 reverse (on intron 3 of Daam1), 5′-CCAAATTTAGAACACAGTATAGCACA-3′; D2 reverse (on the inserted gene trap vector), 5′-TATGCAGTGCTGCCATAACC-3′.
Primary mouse embryonic cardiomyocyte and fibroblast culture
E12.5 ventricular cardiomyocytes were isolated from dissected embryonic hearts and cultured as previously described (Chen et al., 2004). Mouse embryonic fibroblast (MEF) cells (isolated from E12.5-E13.5 embryos) were harvested and cultured as previously described (Weaver et al., 1997).
RT-PCR and RNA in situ hybridization
Total RNA was isolated from various tissues (heart, skeletal muscle, liver and brain) using Trizol (Invitrogen). The primer sets used for Daam1 and Daam1-Geo fusion transcript are as follows: F (forward, on exon 3 of Daam1), 5′-GATGAACTTGACCTCACAGACAA-3′; R1 (reverse 1, on exon 4 of Daam1), 5′-AGCCATGGAATTGAGCTGAT-3′; and R2 (reverse 2, on lacZ of the β-geo reporter cassette), 5′-GACAGTATCGGCCTCAGGAA-3′. The PCR products were analyzed by separation in agarose gels. qRT-PCR analyses were performed using Roche LightCycler 480. Primers are listed in Table S1 in the supplementary material. In situ hybridization was carried out as previously described (Chen et al., 2004). Digoxigenin-labeled Daam1 riboprobes were synthesized using template plasmids kindly provided by Dr T. Yamaguchi (Nakaya et al., 2004). All results represented at least three embryos per embryonic stage.
Immunoblot analyses
Tissues and MEF cells were lysed with RIPA buffer. Proteins were resolved in 8-12% polyacrylamide SDS gels and subsequently transferred and immunoblotted. Antibodies used were: monoclonal antibody (mAb) against Daam1 (Abcam), which recognizes the N terminus of Daam1 protein; polyclonal antibody (pAb) against Daam1 (Proteintech), which recognizes the C terminus of Daam1 protein; mAb against α-catenin, Dvl2 and RhoA, and pAb against N-cadherin, ROCK1, ROCK2 and Cdc42 (Santa Cruz Biotechnology); pAb against phospho-JNK (Thr183/Tyr185), total-JNK and phospho-LIMK1 (Thr508) (Cell Signaling Technology); pAb against phospho-MYPT1 (Thr696); mAb again Rac1 (Upstate); and mAb against α-actin and α-actinin (Sigma). Densitometry of western blots was measured using ImageJ software.
Immunofluorescence staining
Fluorophore conjugated probes for phalloidin and wheat germ agglutinin (WGA) were from Molecular Probes. Alexa Fluor secondary antibodies were from Molecular Probes. Nuclei were stained with DAPI or PI. Photomicrographs were acquired with either a Leica microscope or an Olympus FV1000 confocal microscope using a single exposure time for the same set of sections for each staining.
RhoA, Rac1 and Cdc42 activity assays
E16.5 embryonic hearts were collected for the assays. GST-PBD (for Rac1 and Cdc42) and GST-RBD (for Rho) pull-down binding assays were performed as previously described (Hallett et al., 2003).
Cell adhesion assay
A 96-well cell culture plate was coated with 10 μg/ml of rat tail collagen type I. Either MEF cells or freshly isolated E12.5 mouse cardiomyocytes were seeded at 1×104 cells/well and incubated in a CO2 incubator, 37°C for 60 minutes. The wells were then washed with PBS and fixed with 4% PFA. MEF cells were stained with 2% Crystal Violet. Cardiomyocytes were stained with a sarcomeric α-actinin antibody. Fields of stained adhering cells were randomly selected and photographed. Cells/well were counted using ImageJ software and their relative adhesion was calculated from the ratio of adhering mutant cells/adhering wild-type or heterozygous control cells.
Wound healing assay
Wound healing assays were performed as previously described (Rodriguez et al., 2005). The relative migration distance was calculated by the (gap distance at 0 hours – gap distance at 7 hours or 12 hours)/gap distances at 0 hours.
Echocardiographic analyses
The mice were lightly anesthetized with inhaled 1.5% isoflurane. Echocardiography was performed and analyzed as previously described (Zhu et al., 2009).
Statistical analyses
Results were expressed as mean ± s.e.m. for parametric data. Student's t-test and ANOVA were performed for comparison between two groups or more than two groups of parametric data accordingly. χ2 test was applied for the analysis of ratios of different genotypes versus theoretical Mendelian ratios. P<0.05 was considered to be significant.
RESULTS
Generation and characterization of Daam1-deficient mice
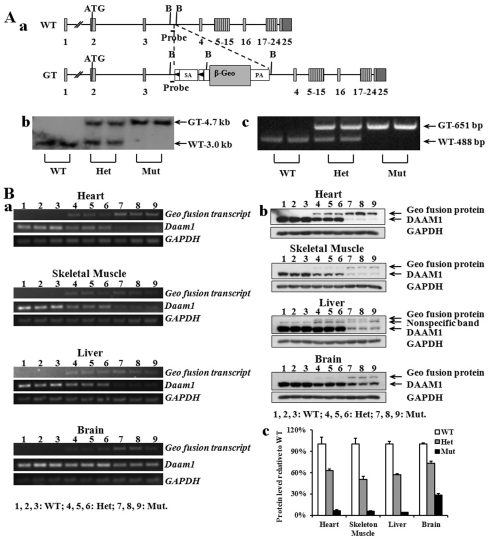
Mice were generated using targeted ES cells containing an insertional gene trap (gt) cassette in intron 3 of the mouse Daam1 gene. The mutant allele was designated Daam1gt. The gene trap vector was designed to terminate the transcription of the endogenous gene through altered splicing. PCR mapping, Southern blot analyses and DNA sequencing were used to characterize the Daam1gt allele (Fig. 1A). Alternative splicing in the Daam1gt allele generates a new transcript in which Daam1 exons 1, 2, 3 and the β-geo reporter form a fusion transcript and subsequently a fusion protein. The fusion protein encodes the first 91 amino acids of Daam1 (which contains about 1/8 of the GBD in the Daam1 N terminus). As Daam1 activity is dependent upon the C-terminal FH1 and FH2 domains (Habas et al., 2001), it is unlikely that the fusion protein retains any actin polymerization activity. Importantly, the gene trap splice acceptor is flanked by 2 loxP sites (Lox71 and LoxP), which are designed to facilitate Cre-mediated cell lineage specific reversion of the mutant trapped allele (Skarnes et al., 1992).
Fig. 1.
Generation and characterization of Daam1 gene trap mice. (A) Generation of Daam1 gene trap mouse line. (a) Schematic representation of wild-type and targeted Daam1 alleles, including the inserted β-galactosidase-neomycin resistance gene-trap cassette. The black solid triangles flanking `SA' represent loxP sites. ATG, translational start site; b, BamHI recognition site; GT, gene trap; PA, poly adenylation site; Probe, Southern probe site; SA, splice acceptor; WT, wild type. (b) Southern blot analysis of DNA prepared from E16.5 embryos. (c) Genotypes of wild type, Daam1gt/+ and Daam1gt/gt by PCR. (B) Characterization of Daam1 gene trap mice. (a) RT-PCR analysis of endogenous Daam1 and the fusion transcript in tissues of heart, skeletal muscle, liver and brain in wild-type (WT), heterozygotes (Het) and homozygous mutant (Mut) E16.5 embryos. Gapdh served as cDNA loading control. (b) Western blot of Daam1 protein and Geo fusion protein in tissues of heart, skeletal muscle, liver and brain in wild-type, Het and Mut E16.5 embryos. GAPDH served as total protein loading control. (c) Quantification of Daam1 protein levels shown in b. Results are mean ± s.e.m.
Loss of function in gene trap models is dependent upon the degree of alternative splicing at the targeted locus. RT-PCR and western blot analyses revealed that, in addition to the expected Daam1-Geo fusion transcript and protein, the wild-type Daam1 transcript (Fig. 1Ba) and protein (Fig. 1Bb) were present in four representative tissues from Daam1 homozygous mutant (Daam1gt/gt) embryos. Western blot quantification revealed the presence of about 5% of the normal levels of wild-type Daam1 protein in Daam1gt/gt embryonic heart, skeletal muscle and liver, and about 25% in brain (Fig. 1Bc), which is consistent with the qRT-PCR evaluation (data not shown). Given that wild-type Daam1 transcripts and protein were present, albeit at significantly lower levels when compared with wild-type controls, the Daam1gt allele is a hypomorphic allele rather than a null mutation. Tissue/cell type-dependent hypomorphic activity commonly occurs in gene trap mouse models and probably reflects alternative splicing events of the endogenous gene in those tissues (Li et al., 2005).
Daam1 expression pattern in mice
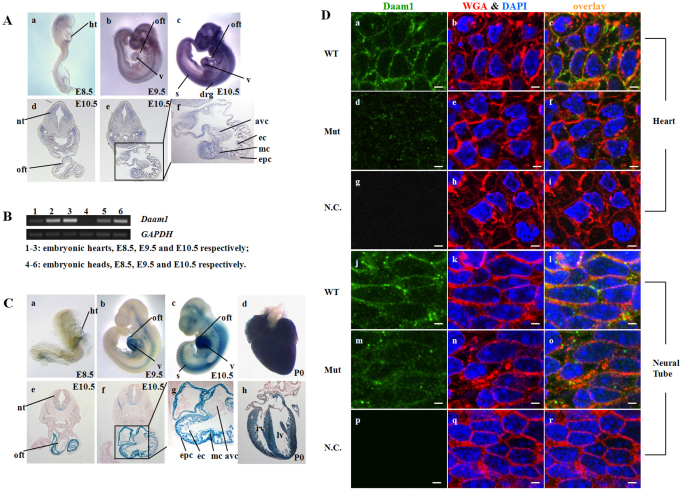
The temporal and spatial expression pattern of Daam1 was evaluated by whole-mount in situ hybridization and by X-Gal staining respectively in wild-type and Daam1 mutant embryos. The expression of Daam1 was weak in the tubular heart and brain at E8.5 (Fig. 2A), but became more prominent at E9.5 and E10.5. An RNA signal was detected in the neural tube at E9.5; at E10.5, Daam1 expression was extended to a broader area, including somites and dorsal root ganglia. In the developing heart, Daam1 expression was broadly distributed in atria, ventricles and outflow tracts. By analyzing sections from whole-mount in situ hybridization, Daam1 expression was found in all three layers of the heart (i.e. the endocardium, myocardium and epicardium, see Fig. 2Af). RT-PCR analyses of isolated hearts and heads from E8.5 to E10.5 further confirmed Daam1 expression, with similar temporal patterns to those observed using whole-mount in situ hybridization analyses (Fig. 2B).
Fig. 2.
The Daam1 expression profile in mice. (A) RNA in situ hybridization analyses of Daam1 in E8.5-10.5 embryos. Lateral views of Daam1 expression in E8.5 (a), E9.5 (b) and E10.5 (c) embryos by whole-mount in situ hybridization. Transverse sections through heart region of a whole-mount in situ hybridization E10.5 embryo are shown in d and e. High magnification of the boxed area in e is shown in f. (B) RT-PCR of Daam1 in isolated hearts and heads of E8.5-E10.5 embryos. The cDNA loading control Gapdh is shown in the lower panel. (C) X-Gal staining in Daam1 gt/gt (a-c) embryos and isolated postnatal day 0 (P0) Daam1gt/+ hearts (d,h). (e,f) Transverse sections of a stained E10.5 embryo. (g) High magnification of the boxed area in f. avc, atrioventricular cushion; drg, dorsal root ganglion; ec, endocardium; epc, epicardium; ht, heart; mc, myocardium; nt, neural tube; oft, outflow tract; s, somites; lv, left ventricle; rv, right ventricle. (D) Daam1 immunofluorescence staining of the developing heart and the neural tube (E12.5) on cryosections using a C-terminus-specific antibody. (g,p) Stained without applying Daam1 antibody as negative controls. Scale bars: 2.5 μm.
X-Gal staining revealed a Daam1 expression pattern spatially and temporally similar to those obtained via whole-mount in situ hybridization (Fig. 2C). A strong X-Gal signal is also observed in hearts from newborn mice, suggesting a potential role for Daam1 in postnatal cardiac development. However, some subtle differences are observed when comparing Daam1 expression patterns determined by whole-mount in situ hybridization with those obtained using X-Gal staining. For example, at E9.5, whole-mount in situ hybridization indicated strong Daam1 expression in the brain, whereas only weak X-Gal signal is observed. This difference can be attributed to the hypomorphic nature of the Daam1gt allele: a specific amount of wild-type Daam1 transcripts are present in the brain of Daam1gt/gt mice (Fig. 1B); consequently, the X-Gal reaction (which only detects the fusion transcript) would underestimate Daam1 expression in this tissue. As hypomorphic activity of the Daam1gt allele is less prevalent in heart, the X-Gal reaction can be used as a faithful surrogate of Daam1 expression in heart.
Last, the subcellular localization of Daam1 in the developing heart and neural tube (E12.5) was examined using a specific antibody against the Daam1 C terminus (see Fig. 2D). Daam1 protein displays a punctate staining pattern in the cytoplasm and cell membrane of cardiomyocytes and neurons, which is consistent with the subcellular localization pattern previously reported in other cell types (Kim and Han, 2007). The Daam1 signal is minimal in Daam1 mutant cardiomyocytes. The staining is present in Daam1 mutant neurons, albeit at a significantly reduced level as compared with wild-type controls. These results are consistent with RT-PCR and western blot results (Fig. 1B) and the hypomorphic feature of the mutant.
Characterization of the phenotype of Daam1gt/gt mice
Daam1 heterozygous (Daam1gt/+) mice are fertile and phenotypically normal. The intercross of Daam1gt/+ mice failed to generate the expected Mendelian ratio of inheritance in the offspring (see Table S2 in the supplementary material). At postnatal day 1 (P1), only five Daam1gt/gt mice were identified out of a total of 153 mice, suggesting that Daam1 deficiency resulted in embryonic and/or neonatal lethality. Some newborn mice were observed to survive for only a few hours, presenting with a cyanotic body color and gasping (Fig. 3). Subsequent PCR analyses identified these animals as Daam1 homozygous mutants, indicating neonatal lethality in homozygous mutants. In order to screen for embryonic lethality, embryos from Daam1gt/+ intercrosses were harvested at E12.5, E14.5 and E16.5 and analyzed. Daam1gt/gt embryos made up 23% of the embryos present at E12.5 (P=0.88 versus the expected Mendelian frequency), 19% at E14.5 (P=0.46) and 16% at E16.5 (P=0.03), suggesting the majority of lethality in Daam1-defficient embryos occurred after E14.5.
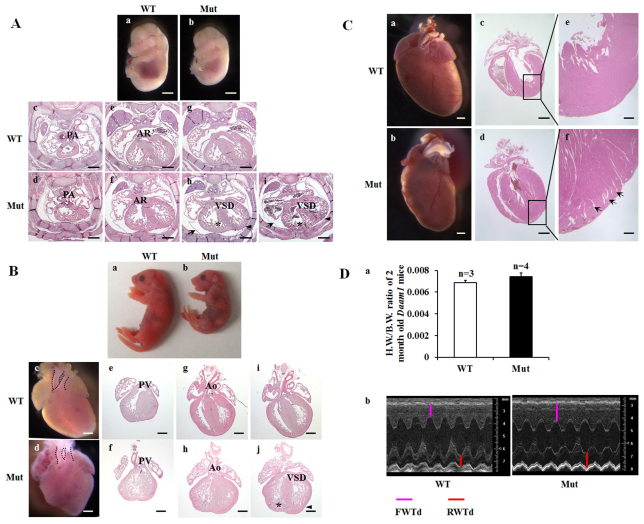
Fig. 3.
Characterization of cardiac phenotypes and function of Daam1gt/gt mice. (A) Histological analyses of Daam1 mice at E14.5. The Daam1 mutant (b) was smaller in body size compared with a wild-type littermate (a). Normal connection of pulmonary artery to right ventricle (c) and normal origin of the aorta from left ventricle in wild type (e). (g) The interventricular septum was completely closed in wild type. (d,f,h) DORV in a Daam1 mutant, showing both pulmonary artery (d) and aorta (f) arising from right ventricle, and an obligate VSD (h). (i) VSD was shown in another mutant. (h,i) Both mutants presented ventricular noncompaction, by exhibiting hypertrophic trabeculae (star) and thin ventricular wall (arrowhead). Daam1 mutants also presented pericardial effusion, indicated by the detachment of pericardial membrane from ventricles (arrows). AR, aortic root; PA, pulmonary artery; VSD, ventricular septal defect. Scale bars: 2 mm in a,b; 0.4 mm in c-i. (B) Histological analyses of neonatal Daam1 mice. A Daam1 mutant pup (b) is smaller than its wild-type littermate (a), and exhibited dark-pink body color (a sign of cyanosis) when compared with red body color in wild type. (c) Normal position and alignment of aorta and pulmonary artery in wild type: the aorta originates more posteriorly and relatively perpendicular to the pulmonary artery compared with (d) the relatively parallel alignment of aorta and pulmonary artery in the mutant. Sections confirmed DORV and revealed ventricular noncompaction in mutant (f,h,j), by exhibiting hypertrabeculation (asterisk) and thin compact layer (arrowhead). Ao, aorta; PV, pulmonary valve. Scale bars: 0.5 mm. (C) Histological analyses of 2-month-old Daam1 mice. A mutant (b,d,f) exhibited deep trabeculae communicating to left ventricle chamber, hypertrabeculation and intraventricular recesses (arrows). Scale bars: 1 mm in a-d; 0.25 mm in e,f. (D) Functional assessment of 2-month-old Daam1 mice. (a) Heart weight/body weight ratio of Daam1 wild-type and mutant mice. Results are mean ± s.e.m. (b) Representative echocardiographic images of Daam1 wild-type and mutant mice, demonstrating thicker left ventricle (LV) wall of mutant heart compared with the LV wall of wild-type heart. FWTd, front wall thickness of LV in diastole; RWTd, rear wall thickness of LV in diastole.
Given the expression of Daam1 in the developing heart, we speculated that the lethality in Daam1gt/gt embryos might be due to cardiac defects. Morphological and histological analyses indicated that at E14.5, most Daam1gt/gt embryos appeared to be significantly smaller in body size compared with their wild-type and Daam1gt/+ littermates. All Daam1gt/gt embryos exhibited cardiac defects, including ventricular noncompaction, double outlet right ventricles (DORV) and ventricular septal defects (VSD, see Table S3 in the supplementary material; Fig. 3). Most Daam1gt/gt embryos also exhibited pericardial effusion, suggesting cardiac insufficiency. Morphological analyses of dying Daam1gt/gt neonates revealed DORV and ventricular noncompaction, which probably account for the symptoms of cyanosis and gasping caused by systemic hypoxemia from the dextroposition of the aorta.
To further characterize Daam1gt/gt survivors, additional Daam1gt/gt intercrosses were performed to generate a larger cadre of homozygous mutant adult animals. Daam1gt/gt survivors (both males and females) were fertile and grossly normal. However, histological analyses of all examined mutant survivors revealed ventricular noncompaction with various degrees of severity (Fig. 3C). There was no significant difference in heart weight/body weight ratios between Daam1gt/gt survivors and wild-type mice (Fig. 3Da). Echocardiographic analyses at 2 and 6 months of age revealed relatively normal systolic cardiac function in Daam1gt/gt mice, despite the increase in diastolic LV wall thickness (Fig. 3Db; see Tables S4 and S5 in the supplementary material), which is consistent with the ventricular noncompaction observed histologically (Fig. 3C).
Genetic rescue of Daam1gt/gt mice
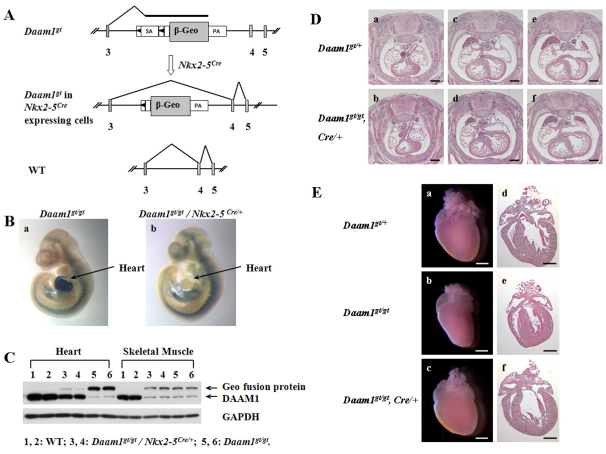
To confirm that the lethality in Daam1gt/gt mice is due to cardiac defects, Daam1gt/+ mice were intercrossed with Nkx2-5Cre/+ mice, in which the Cre activity is under the control of the endogenous Nkx2-5 promoter (Moses et al., 2001). Cre activity can be observed at the cardiac crescent as early as E7.5, becoming stronger in the developing heart by E10.5 (Moses et al., 2001). Cre-mediated recombination of the Daam1gt allele will lead to the excision of the floxed splice acceptor in the gene-trap cassette and consequently restore normal splicing activity (i.e. reversion to wild-type gene expression). Reversion of the allele is readily monitored by the loss of the X-Gal signal (Fig. 4A). Indeed, a marked reduction of the X-Gal signal intensity was observed in the heart and outflow tract of Daam1gt/gt/Nkx2-5Cre/+ mice, whereas the non-cardiac X-gal signal was very similar to that of Daam1gt/gt mice (Fig. 4B). Consistent with this, western blot analysis revealed that Daam1 protein levels in Daam1gt/gt/Nkx2-5Cre/+ hearts are restored to about 50% of wild-type levels (Fig. 4C). By contrast, Daam1 protein in Daam1gt/gt/Nkx2-5Cre/+ skeletal muscle (where the Nkx2-5 Cre activity is absent) remained at low levels, similar to those observed in Daam1gt/gt skeletal muscle. The failure to achieve 100% restoration of Daam1 protein expression in Daam1gt/gt/Nkx2-5Cre/+ hearts is due in part to partial regeneration of wild-type Daam1 transcripts from the reactivated Daam1gt allele and the presence of a significant number of non-Nkx2-5-expressing cells in heart.
Fig. 4.
Rescue of Daam1gt/gt mice. (A) Schematic demonstration of reactivation of Daam1gt allele by crossing with Nkx2-5Cre mouse. (B) X-Gal staining on E10.5 embryos. (C) Western blot of Daam1 protein in tissues from E16.5 embryos. (D) Histological sections of E14.5 embryos. Daam1gt/gt/Nkx2-5Cre/+ embryos (b,d,f) exhibited normal cardiac structure. Scale bars: 0.4 mm. (E) Histological sections of 1-week-old mice. Both Daam1gt/+ (a,d) and Daam1gt/gt/Nkx2-5Cre/+ (c,f) mice showed normal ventricular wall and trabeculation. A Daam1gt/gt mouse (b,e) demonstrated ventricular noncompaction. Scale bars: 0.4 mm in a-c; 1 mm in d-f.
To demonstrate whether partial restoration of cardiac Daam1 would rescue the lethality and cardiac defects of Daam1gt/gt mice, either Daam1gt/gt survivors were intercrossed with Daam1gt/+/Nkx2-5Cre/+ mice or Daam1gt/gt/Nkx2-5Cre/+ mice were intercrossed with Daam1gt/+ mice. In the absence of embryonic or neonatal lethality, 25% of offspring from either cross would be Daam1gt/gt/Nkx2-5Cre/+. At P1, 16 of 67 pups (23.9%) were Daam1gt/gt/Nkx2-5Cre/+ and only four out of 67 (6%) were Daam1gt/gt (see Table S6 in the supplementary material). All Daam1gt/gt/Nkx2-5Cre/+ mice are relatively morphologically normal. However, most Daam1gt/gt/Nkx2-5Cre/+ embryos or pups are smaller than littermates. Histological analyses were performed on E14.5 and P7 Daam1gt/gt/Nkx2-5Cre/+ hearts. At E14.5, Daam1gt/gt/Nkx2-5Cre/+ embryos show normal cardiac structure, with no evidence of DORV, VSD or ventricular noncompaction (Fig. 4D). At P7, Daam1gt/gt/Nkx2-5Cre/+ hearts show normal ventricular compaction, no DORV or VSD, whereas Daam1gt/gt littermate survivors show ventricular noncompaction (Fig. 4E). These data indicate that Daam1 deficiency in heart is the leading cause of lethality in Daam1gt/gt mice. The small body size of rescued Daam1gt/gt mice also suggests that there are additional defects associated with Daam1 deficiency, which are not secondary to the heart defects.
Daam1 deficiency does not affect cardiomyocyte differentiation, proliferation or apoptosis
All Daam1gt/gt mice develop ventricular noncompaction, which may arise from a potential defect in cardiomyocyte differentiation (Chen et al., 2009). To determine whether Daam1 deficiency effected cardiomyocyte differentiation, we investigated whether the expression levels or patterns of cardiogenic transcription factors (e.g. Nkx2-5, Tbx20 and Gata4), morphogenetic protein BMP10 and cardiac chamber specification genes (MLC2A and MLC2V) are altered in Daam1 mutant hearts. We did not find any significant differences in either the expression pattern or expression level of these factors between Daam1gt/gt and Daam1gt/+ mice by either in situ hybridization or qRT-PCR (see Fig. S1 in the supplementary material).
Abnormal cell proliferation and/or apoptosis can result in a variety of heart defects, including DORV, VSD and ventricular noncompaction (Gittenberger-de Groot et al., 2005). Although Formin proteins have been implicated in cell proliferation and apoptosis (Goode and Eck, 2007), Daam1 deficiency does not appear to significantly impact these activities in the developing heart (see Fig. S2 in the supplementary material).
Daam1 is required for proper actin cytoskeleton and sarcomeric organization in cardiomyocytes
Previous in vitro studies have demonstrated that Daam1 regulates filamentous actin (F-actin) assembly (Higashi et al., 2008; Moseley et al., 2006). However, the impact of Daam1 on the actin cytoskeleton has not been examined in an in vivo mammalian system. F-actin is a key cytoskeletal component that is important for cell adhesion, migration and muscle contraction. F-actin can be polymerized by either α-actin or β-actin, the predominance of which depends on cell types. Thus, we first determined the relative abundance of α and β-actin in mouse embryonic cardiomyocytes. Western blot revealed that α-actin is abundant in E12.5 heart lysates, but is not detected in isolated E12.5 mouse embryonic fibroblasts (MEFs), whereas β-actin is robustly expressed in fibroblasts, but expressed at a very low level in E12.5 hearts. α-Actin and β-actin immunostaining of cultured cardiomyocytes and MEF cells confirms the western blot findings (see Fig. S3 in the supplementary material). These findings were expected, as α-actin is one of the fundamental structural components of thin filaments in sarcomeres, which are abundant in cardiomyocytes. Our observations are consistent with previous analyses on actin isoforms in cultured neonatal rat cardiomyocytes (Skwarek-Maruszewska et al., 2009). Thus, F-actin in mouse embryonic cardiomyocytes predominantly comprises α-actin, and alterations in F-actin are primarily associated with changes in α-actin.
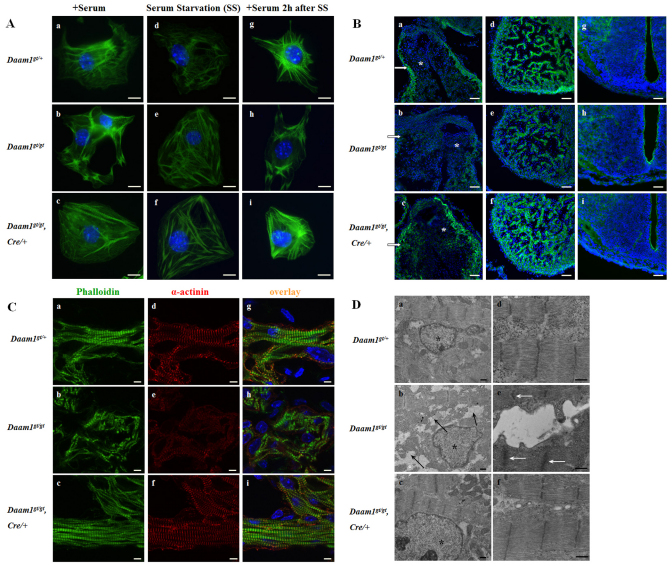
Next, we examined the potential impact of Daam1 deficiency on F-actin formation in cultured embryonic cardiomyocytes in the presence or absence of serum. Lysophosphatidic acid, an important component of serum, regulates the Rho-GDP to Rho-GTP switch (Ridley et al., 1992). Rho-GTP activates Daam1, which in turn regulates actin polymerization (Liu et al., 2008). Thus, similar to the previous study (Liu et al., 2008), serum depletion/replenishment was currently used as a Daam1 activation switch. Under normal culture condition (10% FBS), both Daam1gt/+ and Daam1gt/gt/Nkx2-5Cre/+ cardiomyocytes exhibited a similar F-actin distribution pattern: well-aligned F-actin with striations in sarcomeric regions. By contrast, Daam1gt/gt cardiomyocytes exhibited randomly orientated and aggregated striated F-actin (Fig. 5A). Intriguingly, serum starvation had a similar impact on the cardiomyocyte actin cytoskeleton in all three genotypes, resulting in a randomized and aggregated F-actin pattern. After serum replenishment, both Daam1gt/+ and Daam1gt/gt/Nkx2-5Cre/+ cardiomyocytes establish normal F-actin distribution patterns. By contrast, Daam1gt/gt cardiomyocytes retain the abnormal actin cytoskeletal architecture. Collectively, these observations suggest that Daam1 is required for normal organization of the cardiomyocyte actin cytoskeleton and sarcomeres in response to serum stimulation.
Fig. 5.
Perturbation of the actin cytoskeleton and sarcomeres in Daam1gt/gt cardiomyocytes. (A) Phalloidin staining on cultured E12.5 cardiomyocytes under different culture conditions. (a-c) Cardiomyocytes were cultured under normal conditions: DMEM with 10% FBS; (d-f) cardiomyocytes were serum starved for 72 hours; (g-i) cardiomyocytes were supplemented with 10% FBS for 2 hours, following a 72-hour serum starvation. Phalloidin staining showed normal stretches of well-bundled F-actin in Daam1gt/+ cardiomyocytes (a,g) and Daam1gt/gt/Nkx2-5Cre/+ cardiomyoctyes (c,i), with striations in the sarcomeric region; whereas sarcomeric F-actin in Daam1gt/gt cardiomyocytes (b,e,h) was randomly aggregated, which was also seen in cardiomyocytes of all three genotypes (d-f) under serum starvation. Scale bars: 10 μm. (B) Actin cytoskeleton analyses of E12.5 hearts. Phalloidin staining demonstrated a decrease of F-actin in the myocardial wall of the outflow tract (white arrows) and ventricular myocardia in Daam1gt/gt (b,e) compared with Daam1gt/+ mouse (a,d), whereas F-actin was restored in Daam1gt/gt/Nkx2-5Cre/+ mouse (c,f). The staining is relatively low in the outflow cushion (asterisks) and no significant difference was observed between Daam1gt/+ and Daam1gt/gt mouse in the outflow cushion region. No overt difference was observed in the neural tube region among all three genotypes (g-i). Scale bars: 50 μm. (C) Compromised sarcomeric organization in E12.5 Daam1gt/gt cardiomyocytes. Both Daam1gt/+ (g) and Daam1gt/gt/Nkx2-5Cre/+ cardiomyocytes (i) demonstrated well-formed and elongated sarcomeres revealed by phalloidin and α-actinin staining. In Daam1gt/gt cardiomyocytes, both stainings were relatively weak and exhibited fewer striations with chaotic sarcomeric organization (h). Scale bars: 5 μm. (D) Altered sarcomeric structures in E12.5 Daam1gt/gt cardiomyocytes assessed by TEM. Daam1gt/gt sarcomeres (b) were sparsely distributed with excessive electron-lucent devoid areas (black arrows), as opposed to densely aligned sarcomeres in wild-type (a) and Daam1gt/gt/Nkx2-5Cre/+ (c) cardiomyocytes. Asterisks indicate nuclei. Daam1gt/gt sarcomeres (e) are also shorter in width with widened z-lines (white arrows). Scale bars: 0.5 μm.
To further assess the impact of Daam1 deficiency on F-actin in vivo, we performed phalloidin staining in embryos at E12.5 (the stage before Daam1 mutants exhibit grossly discernable cardiac phenotypes). We found that F-actin is significantly decreased in ventricular myocardia and myocardia around the outflow tract in Daam1gt/gt embryos (Fig. 5B). By contrast, no overt differences in F-actin content are seen in the outflow cushion of Daam1gt/gt hearts when compared with wild-type controls. As expected, F-actin levels are restored to normal levels in Daam1gt/gt/Nkx2-5Cre/+ hearts. By contrast, there are no significant differences observed in the neural tube among all three genotypes. This is expected as neural tissue in Daam1gt/gt mice produce around 25% of the wild-type Daam1 protein (see Fig. 1B).
Sarcomeres are composed of bundles of polymerized actin crosslinked by α-actinin at the Z-line. To further understand the impact of F-actin reduction on ventricular cardiomyocyte sarcomeric organization in vivo, sections were assessed by phalloidin staining and α-actinin immunostaining. In contrast to the well-formed and elongated sarcomeres in wild-type and Daam1gt/gt/Nkx2-5Cre/+ cardiomyocytes, Daam1gt/gt sarcomeres exhibit weaker and more diffuse staining patterns for both α-actin and α-actinin (Fig. 5C), despite normal expression levels (see Fig. S4 in the supplementary material). Additionally, Daam1gt/gt sarcomeres exhibit fewer striations and randomized/chaotic sarcomeric organization. These observations are consistent with aggregated sarcomeric organization seen in cultured Daam1gt/gt cardiomyocytes (Fig. 5A), strongly suggesting that Daam1 is required for normal sarcomeric maturation and organization. The reduction of F-actin (Fig. 5B) and sarcomeric striated staining (Fig. 5C) seen in Daam1gt/gt cardiomyocytes also suggests that Daam1 might influence myofibrillogenesis. Indeed, Daam1gt/gt cardiomyocytes exhibit altered structure and organization of sarcomeres (Fig. 5D). It was difficult to identify sarcomeres in Daam1gt/gt cardiomyocytes, because they are sparsely distributed and significantly shorter in width (2.03±0.07 μm) when compared with wild-type (2.43±0.11 μm) and Daam1gt/gt/Nkx2-5Cre/+ (2.46±0.17 μm) sarcomeres (P<0.05). In addition, Daam1gt/gt sarcomeres exhibit abnormal Z-line architecture. However, the length of Daam1gt/gt sarcomeres is not significantly different from that of wild-type sarcomeres (data not shown). These TEM findings are consistent with the above immunostaining results, i.e. the reduction of F-actin staining correlates with the reduced number and underdeveloped sarcomeres in Daam1gt/gt cardiomyocytes.
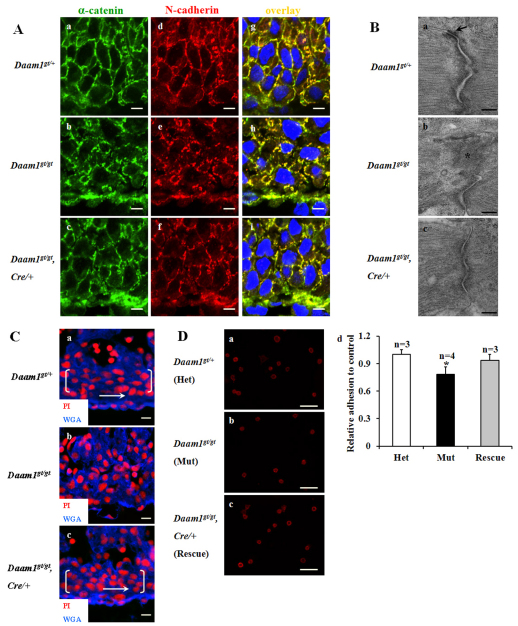
Daam1 is important for cell adhesion in cardiomyocytes
As the actin cytoskeleton is essential for cell-cell and cell-extracellular matrix (ECM) adhesion, we examined whether there are adhesion defects in Daam1gt/gt cardiomyocytes. α-Catenin, an actin-binding protein, links cadherin to the actin cytoskeleton at sites of cell-cell contacts called adherens junctions (Noorman et al., 2009). The expression patterns of α-catenin and N-cadherin in Daam1gt/gt cardiomyocytes are punctate/discontinuous when compared with the homogenous membrane localization observed in Daam1gt/+ and Daam1gt/gt/Nkx2-5Cre/+ hearts (Fig. 6A). Interestingly, the protein expression levels of α-catenin and N-cadherin
Fig. 6.
Perturbation of adhesion junctions in Daam1gt/gt cardiomyocytes. (A) Aberrant staining pattern of α-catenin and N-cadherin in E12.5 Daam1gt/gt cardiomyocytes. The Daam1gt/gt cardiomyocytes of the developing ventricular myocardial wall (b,e,h) have lost their uniform expression of α-catenin and N-cadherin compared with the wild-type (a,d,g) and Daam1gt/gt/Nkx2-5Cre/+ (c,f,i) cardiomyocytes. Scale bars: 5 μm. (B) Aberrant adherens junctions in E12.5 Daam1gt/gt cardiomyocytes. The adherens junction in Daam1gt/gt cardiomyocytes (b) has prominent plaques (asterisk), which are not positioned nicely alongside of the plasma membrane, as seen with adherens junctions in wild-type (a) and Daam1gt/gt/Nkx2-5Cre/+ (c) cardiomyocytes. (a) Arrow indicates a normal desmosome. Scale bars: 0.25 μm. (C) Cell misalignment in Daam1gt/gt hearts. E12.5 heart sections are stained with WGA (for membranes) and PI (for nuclei). Cardiomyocytes in developing ventricular walls (the cells within the bracket) of Daam1gt/+ (a) and Daam1gt/gt/Nkx2-5Cre/+ (c) exhibit fine alignment parallel to the epicardium (arrows), whereas Daam1gt/gt cardiomyocytes (b) manifest a randomized distribution. Scale bars: 10 μm. (D) Abnormal adhesion of cultured E12.5 Daam1gt/gt cardiomyocytes to collagen type I. Cardiomyocytes of three genotypes (a-c) were stained by α-actinin. Scale bars: 50 μm. (d) Quantification of the adhesion assay is shown in D. *Significantly different from heterozygous and rescue, P<0.05. Results are mean ± s.e.m.
are not altered in Daam1gt/gt hearts (see Fig. S4 in the supplementary material). These observations suggest a potential cell-cell junction defect. Indeed, TEM reveals adherens junctions in Daam1gt/gt cardiomyocytes are aberrant, with prominent plaques and misalignment alongside the plasma membrane (Fig. 6B). Interestingly, desmosomes, another type of cell-cell junction structure, which are anchored to intermediate filaments, appeared normal in Daam1gt/gt cardiomyocytes. Normal cell-cell adhesion has been suggested to be a pre-requisite for proper cell alignment in cardiomyocytes (Matsuda et al., 2005). Cell alignment is significantly compromised in the developing ventricular myocardial walls of Daam1gt/gt animals (Fig. 6C). Proper interaction between cardiomyocytes and the ECM is crucial for normal heart structure and function (Weber et al., 1994). Collagen type I is a major ECM component in heart. We found the ability of Daam1gt/gt cardiomyocytes to adhere to collagen type I coated plates is significantly compromised compared with Daam1gt/+ cardiomyocytes (Fig. 6D). By contrast, adhesion is largely restored in Daam1gt/gt/Nkx2-5Cre/+ cells.
Daam1 does not regulate the actin cytoskeleton via small GTPase (RhoA, Rac1 and Cdc42) activities
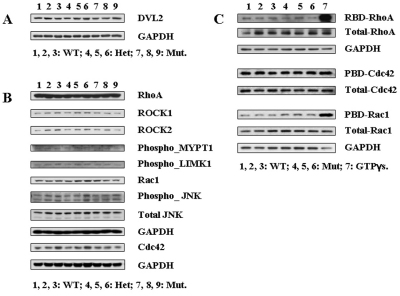
Western blot analyses were performed to understand whether Daam1 controls the actin cytoskeleton by regulating small GTPase activities. First, Daam1 deficiency did not affect total Dvl2 expression, suggesting the absence of a direct feedback loop between Dvl2 and Daam1 (Fig. 7A).
Fig. 7.
Daam1 does not regulate the actin cytoskeleton through modulating small GTPase (RhoA, Rac1 and Cdc42) activities. (A) The expression of Dvl2 in E16.5 hearts. (B) Expression and phosphorylation of substrate proteins of small GTPases in E16.5 hearts. (C) Small GTPase activation assays. GTP-bound RhoA in E16.5 heart lysates was pulled down using GST-RBD and detected with RhoA-specific antibody. GTP-bound Rac1 and Cdc42 were pulled down using GST-PBD and detected with Rac1- and Cdc42-specific antibodies. RhoA, Rac1 and Cdc42 in lysates were detected by the above antibodies. GTPγS treatment served as positive controls for binding assays, and GAPDH served as a total protein loading control.
Small GTPases, including RhoA, Rac1 and Cdc42 all play essential roles in regulating actin cytoskeletal organization (Jenny and Mlodzik, 2006). RhoA has been proposed to be an effector of Daam1 in actin regulation (Habas et al., 2001). Activities of these small GTPases were analyzed in Daam1-deficient hearts where aberrant actin cytoskeletal organization was evident. Interestingly, no overt change in abundance of RhoA, Rac1 and Cdc42 nor in ROCK 1 & 2 (downstream targets of RhoA) was observed in Daam1-deficient hearts when compared with controls (Fig. 7B). Similarly, the phosphorylation status of LIMK1 and MYPT1 (the ROCK 1/2 downstream substrates) (Olson, 2008), as well as JNK (Rac1 and Cdc42 effectors) (Bishop et al., 2000) was not altered in Daam1-deficient hearts. Pull-down assays further revealed that there is no change in the levels of activated RhoA, Rac1 and Cdc42 in Daam1gt/gt hearts when compared with wild-type controls (Fig. 7C). These data collectively suggest that the regulation of actin cytoskeleton assembly and organization by Daam1 is not mediated by regulating these small GTPase activities.
Aberrant cytoskeletal architecture and function in cultured Daam1gt/gt MEFs
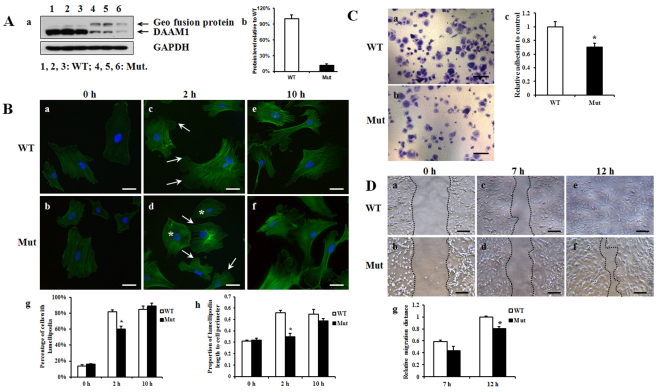
Finally, experiments were performed to determine if Daam1 impacts cytoskeletal architecture and function in cells (i.e. MEFs) in which β-actin predominates. Western blot analysis confirms a marked reduction (about 90%) of Daam1 expression in Daam1gt/gt MEF cells when compared with controls (Fig. 8A), consistent with the hypomorphic nature of the Daam1gt allele. Lamellipodia are projections on the leading edge of motile cells, the formation of which requires actin polymerization (Yang et al., 2007). Therefore, lamellipodia formation reflects the capacity of F-actin formation in cells. During serum starvation, lamellipodia formation is minimal in both wild-type and Daam1gt/gt MEF cells. After 2 hours of serum replenishment, wild-type cells form extensive lamellipodia, whereas the Daam1gt/gt cells develop much smaller lamellipodia and ∼40% of cells do not present any visible lamellipodia formation. After 10 hours, Daam1gt/gt cells eventually develop relatively normal lamellipodia (Fig. 8B). These observations suggest that F-actin assembly is compromised in Daam1gt/gt MEF cells. Cell adhesion and wound healing assays were performed to further assess the impact of Daam1 deficiency on cytoskeleton mediated cellular functions (i.e. adhesion and migration) in MEFs. Both activities are significantly impaired in Daam1gt/gt MEF cells (Fig. 8C,D).
Fig. 8.
Aberrant lamellipodia formation, adhesion and migration in cultured Daam1gt/gt MEF cells. (A) The Daam1 protein level in mutant MEF cells. (b) Densitometric quantification of the western blot in a. Results are mean ± s.e.m. (B) Lamellipodia formation in MEF cells after serum stimulation assessed by phalloidin staining. (a,b) MEF cells were subjected to serum starvation for 48 hours. After 2 hours of culture with 10% serum restoration, wild-type MEF cells (c) demonstrate well formed lamellipodia (arrows); Daam1gt/gt cells (d) develop much smaller lamellipodia (arrows) and some cells (asterisk) do not possess obvious lamellipodia. (e,f) MEF cells after 10 hours culture with 10% FBS media. Scale bars: 50 μm. (g,h) Quantitative data based on three independent experiments. *Significantly different from wild type at 2 hours, P<0.05. Results are mean ± s.e.m. (C) Cell adhesion assay for collagen type I. (a,b) MEF cells were stained with 2% Crystal Violet 1 hour after plating. Scale bars: 0.1 mm. (c) Quantification based on three independent experiments. *Significantly different from wild type, P<0.05. Results are mean ± s.e.m. (D) Wound-healing assay. (a,b) MEF cells (0 hours) were evenly scratched after 24 hours of serum starvation. (c-f) Images of cells 7 hours (7 h) and 12 hours (12 h) after the scratch. Scale bars: 0.2 mm.The cell migration properties were calculated and are shown in g. *Significantly different from wild type at 12 hours, P<0.05. Results are mean ± s.e.m.
DISCUSSION
Daam1, as a formin protein, contains characteristic FH1 and FH2 domains, which are pivotal in mediating actin polymerization (Goode and Eck, 2007). Previous work has suggested that Daam1 is an important signaling protein mediating the non-canonical Wnt/PCP pathway (Habas et al., 2001), which governs cell polarity and alignment during tissue morphogenesis and organogenesis (Zallen, 2007). The potential impact of Daam1 on Drosophila and Xenopus development has been evaluated, although the mutant phenotypes were discrepant (Habas et al., 2001; Matusek et al., 2006). This may be partially attributed to the evolutionary distance between species. The biological function of mammalian Daam1 has not heretofore been analyzed. Here, we generated a Daam1-deficient mouse model and examined the potential role for Daam1 during tissue morphogenesis, in particular, heart morphogenesis. The majority of Daam1gt/gt mice died in utero, exhibiting a range of heart defects, including ventricular noncompaction, DORV and VSD. Genetic reversion of the Daam1gt/gt allele via a cardiovascular lineage-restricted Cre recombinase (Nkx2-5Cre) results in viable animals with normal hearts, confirming that Daam1gt/gt mice died from cardiac defects and reinforcing the theory that Daam1 directly impacts cardiac morphogenesis. As Daam1 has been shown to possess the capacity of directly mediating actin polymerization in vitro (Higashi et al., 2008; Moseley et al., 2006), we closely examined potential alterations of cytoskeletal organization in Daam1gt/gt mice. Our work demonstrates that there is a significant reduction of F-actin in Daam1gt/gt myocardia and aberrant sarcomeric organization, cell adhesion and alignment in Daam1gt/gt cardiomyocytes. Interestingly, Daam1 was also found to be important for proper lamellipodia formation and related cellular functions (e.g. adhesion and migration) in fibroblasts. Collectively, these observations corroborate the ability of Daam1 to regulate actin organization in vivo and reveal its essential role in tissue morphogenesis. Tissue morphogenesis and organogenesis are accomplished through a series of actin-based cellular activities, such as coordinated cell movement and alignment. In heart, correct alignment of the great arteries, closure of the ventricular septum and compaction of ventricular trabecular myocardium all require proper movement and alignment of cardiomyocytes. Thus, the cardiac defects seen in Daam1gt/gt mice very probably derive from the aberrant cytoskeletal architecture and function in Daam1gt/gt cardiomyocytes.
It has been proposed that Daam1 regulates cytoskeletal organization through modulating RhoA activity (Habas et al., 2001; Jenny and Mlodzik, 2006). On the contrary, several in vitro and in vivo studies have demonstrated that Daam1 acts downstream of RhoA (Aspenstrom et al., 2006; Matusek et al., 2006). Our biochemical analyses performed on the Daam1gt/gt cardiomyocytes agree with these latter studies. No impact was observed on the activities of the small GTPases (RhoA, Rac1 and Cdc42), nor on the phosphorylation of their downstream targets in Daam1gt/gt hearts. It has been suggested that Rac1 mediates a parallel signaling pathway in regulating the cytoskeleton. Both Rac1 and Daam1 have been shown to be immediately downstream of Dishevelled (Liu et al., 2008). However, our data suggest that Rac1, and also RhoA and Cdc42, do not respond in a compensatory manner to the disruption of the cytoskeleton by Daam1 deficiency.
Previous studies using loss-of-function mouse mutants targeting potential Wnt/PCP pathway upstream components (e.g. Wnt5a, Wnt11, and Dishevelled2) have suggested that the non-canonical Wnt/PCP pathway plays a crucial role in cardiac morphogenesis (Hamblet et al., 2002; Schleiffarth et al., 2007; Zhou et al., 2007). Interestingly, these mutants exhibited similar cardiac defects to those observed in Daam1gt/gt mice: DORV and VSD. Furthermore, similar alterations in cytoskeletal architecture and function found in Daam1-deficient mice, including reduced myocardial F-actin levels, disrupted cell-cell adhesion and poor cardiomyocyte alignment, are also observed in Wnt11 knockout mice and Scrib (a PCP gene) mutant mice (Phillips et al., 2007). In addition, the activity of Daam1 has been shown to be regulated by Wnts and Dishevelled2 in the Wnt/PCP pathway (Habas et al., 2001; Liu et al., 2008). Thus, our data suggest that Daam1 potentially plays an effector role for the Wnt/PCP signaling during heart morphogenesis.
It is of interest to note that about 10% of Daam1 mutants survived to adulthood. Given that the Daam1gt allele encodes low levels of wild-type Daam1 transcripts, it is likely that Daam1 expression in some mutant animals exceeded a threshold required for viability. It is also possible that there are modifier genes compensating for the Daam1-deficiency in the mutant survivors, as the mice are maintained in a mixed 129Sv/C57 background. The mutant survivors appeared grossly normal, and showed relatively normal systolic cardiac function, despite the presence of isolated ventricular noncompaction. The Daam1gt/gt survivors are thus rather unique, as the majority of other existing ventricular noncompaction mouse mutants exhibit other cardiac defects and die in utero or at birth (Chen et al., 2009). The etiology of ventricular noncompaction in humans is complex and is probably caused by multi-genetic factors. The results obtained here suggest Daam1 is a novel candidate gene linked to ventricular noncompaction clinically.
In summary, this study demonstrates that the formin protein Daam1 is essential for cardiac morphogenesis. At the subcellular level, Daam1 is crucial for the regulation of actin and its derivative structures such as sarcomeres and cell junctions, and consequently for normal cytoskeletal architecture and function. Our study highlights a critical role for Daam1 in regulating the actin cytoskeleton and tissue morphogenesis.
Supplementary Material
Acknowledgments
We thank Dr Terry Yamaguchi (NIH, Frederick) for kindly sending us the Daam1 in situ probe plasmid, Dr Mary Dinauer (IU, Indianapolis) for generously providing us with the Rac1 antibody, Dr Lei Wei (IU, Indianapolis) for generously providing the phospho-MYPT1 and phospho-LIMK1 antibodies, and Dr Anthony Firulli (IU, Indianapolis) for providing the Nkx2-5 and Gata4 plasmids. We also thank Dr R. Mark Payne and Dr Xin Zhang for valuable discussions. This work was supported, in whole or in part, by National Institutes of Health Grants HL81092 (to W.S.), HL70259 (to W.S.) and HL85098 (to W.S. and L.J.F.), and by Riley Children's Foundation (to W.S., L.S.H., R.J.C., M.R. and L.J.F.). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.055566/-/DC1
References
- Aspenstrom P., Richnau N., Johansson A. S. (2006). The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp. Cell Res. 312, 2180-2194 [DOI] [PubMed] [Google Scholar]
- Bishop G. A., Hsing Y., Hostager B. S., Jalukar S. V., Ramirez L. M., Tomai M. A. (2000). Molecular mechanisms of B lymphocyte activation by the immune response modifier R-848. J. Immunol. 165, 5552-5557 [DOI] [PubMed] [Google Scholar]
- Campellone K. G., Welch M. D. (2010). A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shi S., Acosta L., Li W., Lu J., Bao S., Chen Z., Yang Z., Schneider M. D., Chien K. R., et al. (2004). BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219-2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang W., Li D., Cordes T. M., Mark Payne R., Shou W. (2009). Analysis of ventricular hypertrabeculation and noncompaction using genetically engineered mouse models. Pediatr. Cardiol. 30, 626-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot A. C., Bartelings M. M., Deruiter M. C., Poelmann R. E. (2005). Basics of cardiac development for the understanding of congenital heart malformations. Pediatr. Res. 57, 169-176 [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J. (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593-627 [DOI] [PubMed] [Google Scholar]
- Habas R., Kato Y., He X. (2001). Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843-854 [DOI] [PubMed] [Google Scholar]
- Hallett M. A., Dagher P. C., Atkinson S. J. (2003). Rho GTPases show differential sensitivity to nucleotide triphosphate depletion in a model of ischemic cell injury. Am. J. Physiol. Cell Physiol. 285, C129-C138 [DOI] [PubMed] [Google Scholar]
- Hamblet N. S., Lijam N., Ruiz-Lozano P., Wang J., Yang Y., Luo Z., Mei L., Chien K. R., Sussman D. J., Wynshaw-Boris A. (2002). Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development 129, 5827-5838 [DOI] [PubMed] [Google Scholar]
- Higashi T., Ikeda T., Shirakawa R., Kondo H., Kawato M., Horiguchi M., Okuda T., Okawa K., Fukai S., Nureki O., et al. (2008). Biochemical characterization of the Rho GTPase-regulated actin assembly by diaphanous-related formins, mDia1 and Daam1, in platelets. J. Biol. Chem. 283, 8746-8755 [DOI] [PubMed] [Google Scholar]
- Jenny A., Mlodzik M. (2006). Planar cell polarity signaling: a common mechanism for cellular polarization. Mt Sinai J. Med. 73, 738-750 [PubMed] [Google Scholar]
- Kida Y. S., Sato T., Miyasaka K. Y., Suto A., Ogura T. (2007). Daam1 regulates the endocytosis of EphB during the convergent extension of the zebrafish notochord. Proc. Natl. Acad. Sci. USA 104, 6708-6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. H., Han J. K. (2007). Essential role for beta-arrestin 2 in the regulation of Xenopus convergent extension movements. EMBO J. 26, 2513-2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhu X., Chen M., Cheng L., Zhou D., Lu M. M., Du K., Epstein J. A., Parmacek M. S. (2005). Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc. Natl. Acad. Sci. USA 102, 8916-8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Gundersen G. G. (2008). Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9, 860-873 [DOI] [PubMed] [Google Scholar]
- Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L. W., Habas R. (2008). Mechanism of activation of the Formin protein Daam1. Proc. Natl. Acad. Sci. USA 105, 210-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Takahashi K., Nariai T., Ito T., Takatani T., Fujio Y., Azuma J. (2005). N-cadherin-mediated cell adhesion determines the plasticity for cell alignment in response to mechanical stretch in cultured cardiomyocytes. Biochem. Biophys. Res. Commun. 326, 228-232 [DOI] [PubMed] [Google Scholar]
- Matusek T., Djiane A., Jankovics F., Brunner D., Mlodzik M., Mihaly J. (2006). The Drosophila formin DAAM regulates the tracheal cuticle pattern through organizing the actin cytoskeleton. Development 133, 957-966 [DOI] [PubMed] [Google Scholar]
- Matusek T., Gombos R., Szecsenyi A., Sanchez-Soriano N., Czibula A., Pataki C., Gedai A., Prokop A., Rasko I., Mihaly J. (2008). Formin proteins of the DAAM subfamily play a role during axon growth. J. Neurosci. 28, 13310-13319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Maiti S., Goode B. L. (2006). Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 406, 215-234 [DOI] [PubMed] [Google Scholar]
- Moses K. A., DeMayo F., Braun R. M., Reecy J. L., Schwartz R. J. (2001). Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31, 176-180 [DOI] [PubMed] [Google Scholar]
- Nakaya M. A., Habas R., Biris K., Dunty W. C., Jr, Kato Y., He X., Yamaguchi T. P. (2004). Identification and comparative expression analyses of Daam genes in mouse and Xenopus. Gene Expr. Patterns 5, 97-105 [DOI] [PubMed] [Google Scholar]
- Noorman M., van der Heyden M. A., van Veen T. A., Cox M. G., Hauer R. N., de Bakker J. M., van Rijen H. V. (2009). Cardiac cell-cell junctions in health and disease: electrical versus mechanical coupling. J. Mol. Cell. Cardiol. 47, 23-31 [DOI] [PubMed] [Google Scholar]
- Olson M. F. (2008). Applications for ROCK kinase inhibition. Curr. Opin. Cell Biol. 20, 242-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Kitchen S. M., West R. A., Sigler R., Eisenmann K. M., Alberts A. S. (2007). Myeloproliferative defects following targeting of the Drf1 gene encoding the mammalian diaphanous related formin mDia1. Cancer Res. 67, 7565-7571 [DOI] [PubMed] [Google Scholar]
- Phillips H. M., Rhee H. J., Murdoch J. N., Hildreth V., Peat J. D., Anderson R. H., Copp A. J., Chaudhry B., Henderson D. J. (2007). Disruption of planar cell polarity signaling results in congenital heart defects and cardiomyopathy attributable to early cardiomyocyte disorganization. Circ. Res. 101, 137-145 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Cooper J. A. (2009). Actin, a central player in cell shape and movement. Science 326, 1208-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992). The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401-410 [DOI] [PubMed] [Google Scholar]
- Rodriguez L. G., Wu X., Guan J. L. (2005). Wound-healing assay. Methods Mol. Biol. 294, 23-29 [DOI] [PubMed] [Google Scholar]
- Sarmiento C., Wang W., Dovas A., Yamaguchi H., Sidani M., El-Sibai M., Desmarais V., Holman H. A., Kitchen S., Backer J. M., et al. (2008). WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J. Cell Biol. 180, 1245-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffarth J. R., Person A. D., Martinsen B. J., Sukovich D. J., Neumann A., Baker C. V., Lohr J. L., Cornfield D. N., Ekker S. C., Petryk A. (2007). Wnt5a is required for cardiac outflow tract septation in mice. Pediatr. Res. 61, 386-391 [DOI] [PubMed] [Google Scholar]
- Schonichen A., Geyer M. (2010). Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta 1803, 152-163 [DOI] [PubMed] [Google Scholar]
- Skarnes W. C., Auerbach B. A., Joyner A. L. (1992). A gene trap approach in mouse embryonic stem cells: the lacZ reported is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 6, 903-918 [DOI] [PubMed] [Google Scholar]
- Skwarek-Maruszewska A., Hotulainen P., Mattila P. K., Lappalainen P. (2009). Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J. Cell Sci. 122, 2119-2126 [DOI] [PubMed] [Google Scholar]
- Weaver A. M., Hussaini I. M., Mazar A., Henkin J., Gonias S. L. (1997). Embryonic fibroblasts that are genetically deficient in low density lipoprotein receptor-related protein demonstrate increased activity of the urokinase receptor system and accelerated migration on vitronectin. J. Biol. Chem. 272, 14372-14379 [DOI] [PubMed] [Google Scholar]
- Weber K. T., Sun Y., Tyagi S. C., Cleutjens J. P. (1994). Collagen network of the myocardium: function, structural remodeling and regulatory mechanisms. J. Mol. Cell. Cardiol. 26, 279-292 [DOI] [PubMed] [Google Scholar]
- Yang C., Czech L., Gerboth S., Kojima S., Scita G., Svitkina T. (2007). Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5, e317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A. (2007). Planar polarity and tissue morphogenesis. Cell 129, 1051-1063 [DOI] [PubMed] [Google Scholar]
- Zhou F., Leder P., Zuniga A., Dettenhofer M. (2009). Formin1 disruption confers oligodactylism and alters Bmp signaling. Hum. Mol. Genet. 18, 2472-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lin L., Majumdar A., Li X., Zhang X., Liu W., Etheridge L., Shi Y., Martin J., Van de Ven W., et al. (2007). Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat. Genet. 39, 1225-1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Soonpaa M. H., Chen H., Shen W., Payne R. M., Liechty E. A., Caldwell R. L., Shou W., Field L. J. (2009). Acute doxorubicin cardiotoxicity is associated with p53-induced inhibition of the mammalian target of rapamycin pathway. Circulation 119, 99-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.