Abstract
Export of mRNA occurs via nuclear pores, large nano-machines with diameters of roughly 120 nm that are the only link between nucleus and cytoplasm1. Hence, mRNA export occurs over distances smaller than the optical resolution of conventional light microscopes. There is extensive knowledge on the physical structure and composition of the NPC2–7, but transport selectivity and dynamics of mRNA export at nuclear pores remain unknown8. We developed a super-registration approach using fluorescence microscopy that can overcome the current limitations of colocalization by means of measuring intermolecular distances of chromatically different fluorescent molecules with nm precision. With this method we achieve 20 ms time- and at least26 nm spatial precision, rendering the capture of highly transient interactions in living cells possible. With this method we were able to spatially resolve the kinetics of mRNA transport and present a three step model consisting of docking (80ms), transport (5–20ms) and release (80ms), totalling 180 ± 10 ms. Importantly, the translocation through the channel was not the rate-limiting step, mRNAs can move bi-directionally in the pore complex and not all pores are equally active.
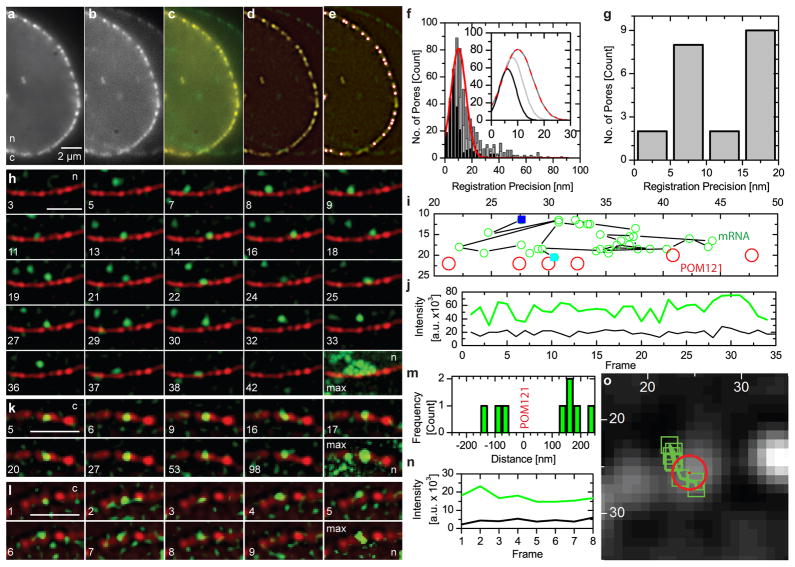
We generated a stable cell line, derived from a transgenic mouse, where all β-actin mRNA is labelled by yellow fluorescent protein (YFP) fused to a MS2-protein tag9,10 (Methods, Fig. S1&2). β-actin mRNA is an essential gene with an estimated size of less than 25 nm, being diffraction limited even if the MS2 sequence should be extended (SI). To ensure sufficient labelling of mRNAs at low expression levels of MS2-protein, we enriched the tag in the nucleus by adding a nuclear localization signal which does not interfere with mRNA export (Methods, SI & Fig. S3). To allow simultaneous imaging of mRNAs and NPCs, POM121 was labelled with tandem Tomato (Fig. S1). POM121 exists in at least eight copies per NPC and is part of the NPC scaffold11,12. Using a high numerical aperture objective single NPCs in the equatorial plane of the nucleus were resolved and showed a distribution of the number of labelled POM121 per NPC (SI & Fig. S4). We simultaneously took high speed movies of NPCs and mRNAs using their distinct fluorescence tags on two precisely registered cameras (Fig 1, SI & Fig. S1C). Rapid imaging was possible because amplification of the transcribed MS2 motif led to excellent signal-to-noise ratios for mRNAs (Figs. 1 & S5), even in cells expressing only low levels of the MS2-YFP tag (Fig. 1, SI and movies). We found that mRNA export events for an individual pore occurred infrequently, beneficial for single molecule observations (SI Movie 1). An immediate observation in our movies was that mRNAs scan multiple pores (Fig. 1H, SI Movie 1). Together with a frequency analysis of mRNA-NPC interactions we concluded that not all pores are equally active in mRNA export at any given time (SI & Fig. S6F). Because s-actin mRNA represents ~0.1% of all molecules passing through NPCs during this time, possibly pore scanning represents a waiting phenomenon. To obtain the spatial precision capable of locating the mRNA relative to NPC dimensions, we developed a method for super-registration of the detection channels below the diffraction limit by registering two cameras within 10 nm (Fig. 1F&G & SI). The fluorescence of POM121-tdTomato was used to acquire inherent dual channel registration markers for each cell imaged.
Fig. 1.
Super-Registration Precision and Detection of Nuclear mRNA. a–g) the registration precision achieved in this experiment was based on imaging nuclear pores on two cameras immediately before data acquisition (SI). Data from both cameras (a red, b green), merged image (c) after registration. A filtered merged image (d) with 21 nuclear pores, white circles outlined in red (e). f) Co-registration precision between the best aligned 6 (black bars and black line in inset), 10 (light grey) and 15 (dark grey) nuclear pores. Fit (inset), Gaussian fit to the ‘15 pore’ data set: registration= 10±1 nm, 13±1 nm FWHM. g) Distances between pores in e). Peak= 7.5 nm. h) mRNAs interacted with nuclear pores infrequently and not all interactions resulted in export of mRNAs from the nucleus (frame numbers in SI Movie 1, mRNA scanning pores, t = 40 ms. i) full length traces h) and SI Movie 1; first (blue), last (cyan). j) Intensity trace (green),tracked mRNA, background (black). k) slow export images from SI Movie 2 (frame indicated). l) fast export (SI Movie 3). m) Distances between mRNA and pore from l) colocalization precision, 26 nm total (SI). Nucleoplasmic mRNA (+) cytoplasmic mRNA (−) (SI & Fig. S5). n) Intensity mRNA signal (green) vs. background. o) mRNA positions (green boxes) and pores (red circle) overlaid on nuclear pore from l. Bars = 2 μm, ‘n/c’ = nucleus/cytoplasm, ‘max’ =maximum intensity projection, i) & o) axis pixels (= 64 nm). h–o LoG filtered (ImageJ, D. Sage).
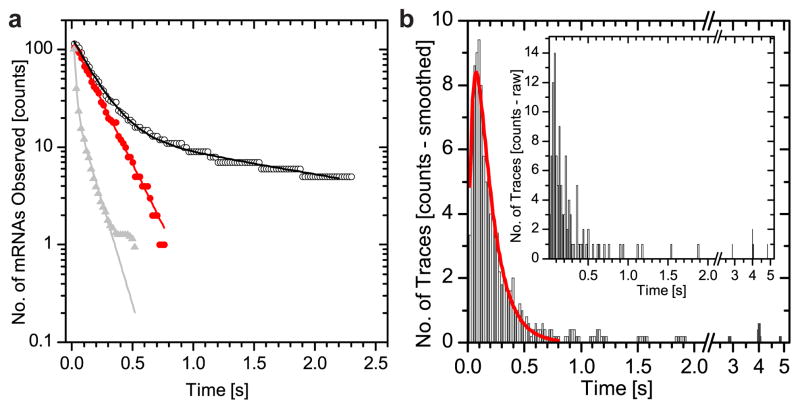
Next we observed the dwell times of mRNAs interacting with NPCs (Fig. 1) compared to those in an equivalent observation volume in the nucleoplasm. Kinetics were much faster for nucleoplasmic diffusion (τ=15±1 ms) than for NPC interaction (τ=172±3 ms, Fig. 2A & Table S2, SI & Fig. S7). During transport, mRNAs were co-registered with NPCs for durations of milliseconds to seconds (Fig. 2 & Table S2). Dwell times at the NPC showed bi-exponential decay kinetics (Fig. 2A). 10% of slow events could be segmented from the total dwell time distribution using a threshold of 800 ms (Fig. 2B & Table S3), whereupon the decay plot became mono-exponential (Fig. 2A). This indicated that the bi-exponential dwell time distribution resulted from two transport species rather than from two kinetic steps in the transport process. The trace duration histogram showed an initial increase followed by a decrease of observed traces per time bin (Fig. 2B) indicating that the fast transport process was a convolution of at least two kinetic steps. We fit the data to y = A · (e(−k2x) − e(−k1x)) with k1 and k2 being rate constants (red line Fig. 2B) suggesting that the observed co-registration of mRNAs with NPCs contains two or more rate-limiting transitions (Fig. 2 & SI)13.
Fig. 2.
Dwell times of β-actin mRNA at the NPC. mRNA co-localized with NPCs, no. frames as milliseconds. Histogram = observed mRNAs per time bin of 20 ms. a) Fit of dwell time of cumulative trace length distribution27 (black circles). First bin = total number of observed traces. Fast transport events (<0.8 s) show mono-exponential decay (red circles). Dwell time = 172±3 ms, (red line, first component black line). Second time constant = 2000±120 ms is needed to fit complete data set (black line). mRNA in the nucleoplasm (grey line), dwell time = 15±1 ms (90%) and 104±6 ms (10%). Data normalized. b) Data from a) (black circles) re-plotted as trace duration histogram (black bars). Cut-off (SI &Table S3, adjacent averaging width = 5 bins). Inset = unprocessed raw data. Two-step convolution model (red line) reveals two kinetic rates28, dwell times kfast = 43±1 ms and kslow = 139±10 ms. Identifying export = two observations = 40 ms. Result consistent with multistep process containing at least two rate constants, total time = 180ms.
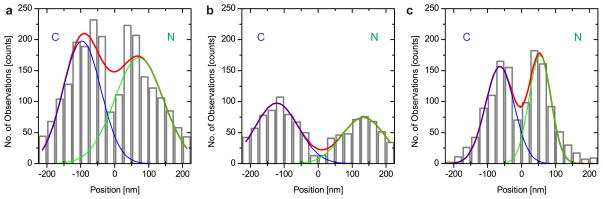
Next we identified export events by identifying nucleoplasmic (+) or cytoplasmic (−) locations of mRNAs (SI & Fig. S8). A change in sign indicated a transport event within a trace. We observed 765 traces within 225 nm of a NPC many showing mRNAs traveling along the nuclear border without engaging nuclear pores (SI Movie 1 & 4). We identified 115 transport traces, containing more than 2300 positional mRNA observations in 33 cells. This translates into a transport efficiency of 15% for this class of mRNAs. Transport traces that showed slow exporting mRNAs contributed ~60% of the positional data (Table S2). Three transport traces showed import of mRNA and 46 traces (40%) showed more than one directional change supporting the principle of reversibility of the translocation step through the central channel14,15. Transport traces were further analyzed by calculating the distance between each observed mRNA position and the closest NPC (Fig. 3A). The resultant ‘binding site’ histograms displayed symmetric distributions with peaks on both pore surfaces. Faster exporting mRNAs (Fig. 3B & Table 1) showed broader binding peaks than slower transporting mRNAs (Fig. 3C) and both were rarely observed within the central channel, arguing for a translocation time below our imaging rate. Within the 50 nm central channel (±25 nm from the POM121-tdT), fast transporting mRNAs accounted for 2.5%, while slow transporting mRNAs accounted for 12.8% of the observations contained in the binding site histograms (Fig. 3B&C). Observation frequencies of mRNA can be linearly correlated to the transit time at any given point along the NPC axis, resulting in transit times of 4.25 ms across the central channel for faster mRNAs. Slower exporting mRNAs might not be interpreted in this manner due to multiple back and forth movements through the pore (SI Movie 2). The similarity of the binding site distributions for fast and slow transiting mRNAs emphasizes that functional interaction sites exist in the NPC outside of the core structure and central channel. Binding to the cytoplasmic or nuclear surfaces of the NPC accounted for the majority of observations of transporting mRNAs (Fig. 3). The kinetic analyses gave a total transport time of ~180 ms (Fig. 2). The binding site analysis combined with the channel translocation time argues for a three-step transport mechanism that involves nucleoplasmic docking (~80 ms), a fast translocation through the central channel (5–20 ms) and a cytoplasmic release step (~80 ms) (Fig. 4). The symmetry in the nuclear and cytoplasmic binding frequencies argues for similar kinetics on both sides of the pore.
Fig. 3.
‘Binding Sites’ of mRNAs at Nuclear Pores. Distances between mRNA and POM121-tdT (zero position) bin widths = 25 nm. (−) = cytoplasmic (blue C), (+) = nucleoplasmic position (green N). Red lines are global fits, green and blue lines are fits to cytoplasmic and nucleoplasmic binding distributions. a) Histogram of all observed transport events at NPCs (b + c). b) Histogram for fast transported mRNAs (90% translocation). c) Histogram for slow mRNAs, observed for extended times at NPC.
Table 1.
Binding site for mRNA at the NPC
| mRNA | xcyto (nm) | xnux (nm) | wcyto (nm) | wnux (nm) | Frcyto (%) | Frnux (%) |
|---|---|---|---|---|---|---|
| All | −97±17 | 71±22 | 56±14 | 72±22 | 54 | 46 |
| Fast | −122±4 | 134±6 | 64±5 | 63±7 | 57 | 43 |
| Slow | −64±6 | 54±4 | 42±6 | 32±4 | 47 | 53 |
Peaks in Fig 3 have been fitted to A1exp(−(x–xc1)/2w12)+A2exp(− (x–xc2)/2w22) to determine the peak positions (xi) and width (wi) as reported in this Table. The amplitudes (Ai) from the fit are normalized to relative fractions (Fri). cyto, cytoplasmic localization; nux, nucleoplasmic localization.
Fig. 4.
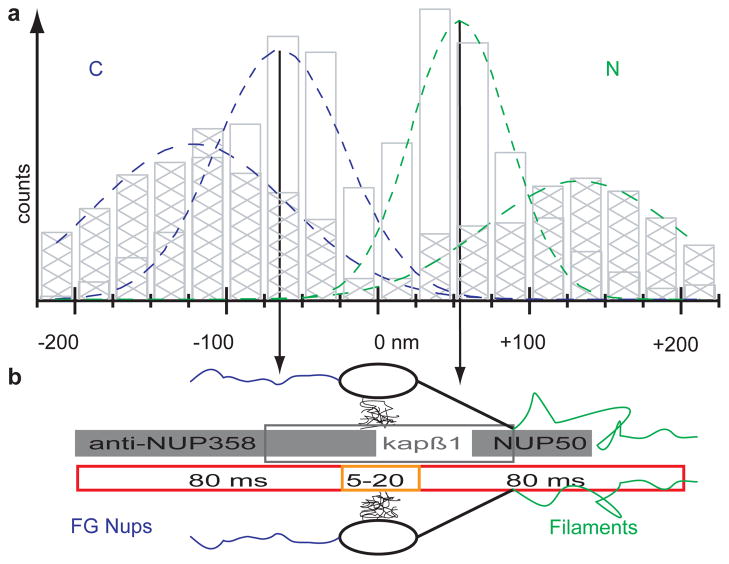
NPC Topography of mRNA Export. Results from Fig. 3B&C (hatched & open bars) plotted (a) to scale with known NPC dimensions (b)4. mRNA export timescale (red = kslow; orange = kfast) along NPC axis combined with single molecule data (grey bars) of Nup35827, import factors29 and import release site30. Nuclear peak position of slow transporting mRNAs located between binding sites for import factors and import release site (Fig. 4 & Table 1). Length of grey bars = FWHM of binding site distributions.
The widths of the binding sites were in the range of ~60 nm (Table 1). The combined cytoplasmic positions from fast and slow mRNAs led to a narrower width of the fit but on the nuclear side, the width of the combined datasets broadens (Fig. 3A & Table 1, p<0.05). This could be interpreted as the existence of one narrow release site on the cytoplasmic surface of the NPC, but a larger target for mRNA binding on the nuclear face. The binding site for slow transporting mRNAs is located closer to the central channel (proximal) than for fast transporting mRNAs (distal). This could be consistent with this inner binding site functioning as a checkpoint, e.g. resembling the Nup98/Gle1 interaction with TAP or CRM116,17. The cytoplasmic peak could be related as a release step, e.g. triggered by DBP5 as suggested by structural data18. Fast transporting mRNAs showed interactions outside of the NPC structure (Fig. 4). These locations may be coincident with nuclear filaments of the NPC (as have been described in EM studies19,20) and cytoplasmic Nups1,21,22. While export of most mRNAs is mediated by the Tap/p15 transport factor complex and is independent of RAN-GTP levels, it depends on available ATP in the cell18. Short term energy depletion assays led to the observation of a narrow peak at 79 nm on the nucleoplasmic side of the pore and resulted in an extended dwell time for exporting cargo arguing for an energy dependent step in transport outside the central channel (Fig. S9).
Several models for providing selectivity in nucleocytoplasmic transport have been described3,23,24. It has been proposed that a channelling effect, called ‘reduction in dimensionality’ results in a fast transport across the pore, once the cargo gains access to the central channel25. Regarding the translocation step, the existing models either formulate the central channel as the major barrier and ‘de facto sorter’ (selective phase model) or an entropic gate made of disordered FG rich filaments that is overcome by receptor mediated binding to the pore (virtual gating hypothesis)3,5.
Here we were able to follow the interaction of single cargos and pores during export and resolved individual transient steps of the export process and their rate constants, which were previously undefined. Despite the large size of the mRNP (SI), the transport time through the central channel is surprisingly fast (~5–20 ms). We calculate the 1D diffusion coefficient for a 5 ms transport time through the central channel to be 0.5 μm2/s, which is in the lower range of mobility found for our mRNPs in the nucleus (Fig S7). Extrapolation of on rates of cargos using artificial nucleoporin gels predicts longer dwell times for the transit step but is limited by missing off rates5. We favour a model where the central channel does not impose a rate-limiting step. Our data demonstrate that the major interaction sites are located at the NPC surfaces rather than within the central channel. Therefore the rate limiting step for mRNA transport is not the transition through the central channel, but rather access to and release from the NPC (Fig. 4).
Three advances have made these observations possible. First, we observed labelled endogenous mRNA molecules (modified with the MS2 tag) in their undisturbed native environment, forgoing the usual caveats concerning reporter genes that, in most cases are over-expressed and non-physiological. Second, an internal reference based “super-registration” allows studying events that regulate mRNA transport in real time in living cells on length scales below the diffraction limit. “Super-registration” is to be distinguished from super-resolution where a large photon flux is used to describe the exact position of an emitting molecule. In contrast, our approach registers two spectral sources of photons with subdiffraction precision relative to each other by utilizing marker signals that pass through the same optical path used to collect the single molecule data. Importantly, this protocol is designed for use in vivo, minimizing photo damage using light fluxes of only a few hundred μW total input power. Finally, combining sensitive high-speed cameras with high signal to noise labelling methods, observations can be made with a time resolution of 20 ms.
This approach is likely to be applicable to other cellular structures, such as DNA “factories”, interaction of nuclear RNA in “speckles” or Cajal bodies or mRNA degradation in “P-bodies”26. The classical use of colocalization in fluorescence microscopy suffers from possible misinterpretations concerning the actual proximity of components due to intrinsic errors in registration. The method of “super-registration” described here provides an order of magnitude greater resolution and hence a more rigorous criterion for the interaction of any two spectrally distinguishable components at the molecular level.
Methods Summary
An immortalized cell line was generated from a homozygous mouse carrying the MS2 stem-loop cassette in the endogenous β-actin gene so that all β-actin mRNA will be labelled by a genetically expressed fluorescent YFP-MS2 tag9,10. This cell line was modified to express the NPC marker POM121-tandem Tomato (Fig. S1), allowing for simultaneous imaging. The cell line showed no growth defects (Fig. S2 & Table S1). To visualize NPCs and mRNA with sufficient time resolution (50 Hz frame rates) and field of view (21.5 μm diameter) two EMCCDs were employed. For magnification adjustment, fine tuning of excitation energies and illumination field, maximal light transition and enabling of precise mechanical pre-alignment of the two cameras, we setup a microscope based on an IX71 microscope stand (Olympus) using a 1.45 N.A. 150x oil objective lens (Fig. S1). All other components were replaced with custom parts, for a detailed description of the setup see SI. Synchronization (nsec) of the cameras was achieved by triggering one camera to a TTL pulse generated by the other camera. Super-registration uses an inherent dual channel marker, here the high signal state of POM121-tdTomato. Prior to data acquisition, the emission signal and the surface reflection of the splitting dichroic mirror are imaged in both channels at the same time. These pore signals are used to register images post-experiment, taking into account inhomogeneities of cover glass thickness and aberrations attributed to optical distortions in living cells. A detailed description of the super-registration assay is in the SI.
Methods
Setup ‘for super-registration’ microscopy
‘Super-registration’ refers to the ability to generate an internal registration signal from the sample, e.g. each cell imaged, that can be used to register spectrally different channels relative to each other to achieve spatial precision below the optical resolution limit. Image series were acquired on a customized dual channel setup (see Fig. S1C) using an Olympus 150x 1.45 N.A. oil immersion objective lens. The right side port of an IX 71 (Olympus) was modified by removing the tube lens. Outside of the stand we placed a 514.5 nm notch filter (Semrock), a 300 mm focal length lens, followed by a 568 nm notch filter (Semrock) that was rotated by 17 degrees to the normal to achieve blocking of 561 nm scattered light. The effective magnification of the optical system was 250x resulting in a pixel size of 64 nm. A dichroic mirror (z543rdc, Chroma) was used to split the fluorescence onto two EMCCDs (Andor iXon, Model DU897 BI). A combination of mirrors and CCD supports (x,y,z, ϕ-and θ-angle) was used to physically pre-align both CCDs to optimize ‘super-registration’ after image processing. A resolution standard (Gellermicro), focal check beads (Invitrogen) and diffraction limited multi-color beads (Invitrogen) were used for pre-alignment. Using two cameras it is possible to adjust their focal plane to account for small axial chromatic shifts. ‘Super-registration’ is achieved by combination of precise mechanical alignment and image processing using transformations based on the registration signal that is detected on both cameras. CCDs were synchronized by a start signal generated by one CCD that was directly delivered to the second CCD. The offset between the two CCDs was determined to be three orders of magnitude below our integration time (2.1 ± 0.2 ns/frame/ms). For excitation of fluorescent proteins an Argon laser with 514.5 nm emission (Melles Griot) and a 561 nm laser line (Cobolt) were merged into a mono mode optical fiber (Qioptiq). The output of the fiber was collimated and delivered through the back port of the IX71 stand and reflected towards the objective by a dichroic mirror (z514-561-1064rpc, Chroma). Alignment onto the optical axis of the objective was achieved with a 4 axis controlled support for the collimator. An adjustable size iris was used to restrict the illumination to an area of approximately 25 μm in diameter. The intensity profile in this area had a flatness of about 5%. Each laser had a shutter (Uniblitz) that was controlled from the imaging software. To allow reasonably fast switching (100 ms) between high and low power settings with the 561 nm line a motorized filter wheel with appropriate neutral density filters was placed behind the shutter but before the merging dichroic of the laser module. The microscope was equipped with a heated stage inset (Warner Scientific) and an objective heater (Bioptechs). During the experiment the stage was covered by a 100 mm cell culture dish wrapped with aluminum foil to exclude stray light. Heating devices were run overnight before an experiment. One hour prior to an experiment three small dishes with a few ml of water were placed on the stage inset to provide humidity. Cells were imaged in a closed dish.
Image Acquisition
Simultaneous imaging of nuclear pores and mRNA enabled a relative measurement of distances (drifts are accounted for by the tracking of both entities) and hence overcomes a limitation in earlier work on imaging nucleocytoplasmic transport, namely missing information on the exact position of the nearest nuclear pore during the acquisition of the cargo signal. To achieve this goal with both, sufficient spatial and temporal resolution EMCCDs, laser shutters and the filter wheel were controlled from the camera software using customized scripts written in Andor Basic. Using sub-frames (~2/3 of each chip, 330 x 330 pixel) on both cameras we were able to observe whole nuclei at a frame rate of 50 Hz equaling a time resolution of 20 ms for tracking single mRNAs. The effective integration time was 19.92 ms. A frame rate of 20 ms was chosen to gain sufficient tracking resolution. Test experiments at 50 ms frame rates showed blurring of mRNA signals while 20 ms frame rates offered adequate signal accumulation to “freeze” the RNAs with a positional accuracy sufficient for tracking, see section on image processing and Eq. 2. To generate the ‘super-registration’ signal used for post experimental, computaional fine alignment of the two detectors the following imaging protocol was implemented: Potential cells of interest were selected and brought into focus (equatorial plane) at very low power settings (0.5 W/cm2) in the red channel using maximal gain on the camera, by avoiding excitation at 514.5 nm bleaching in the green channel was minimized. Next, an automated protocol was used to image NPCs only at 561 nm laser using ‘high’ power setting (180 W/cm2) for 50 frames, followed by a 100 ms break to save data, switch gain settings and filter wheel position, followed by 400 frames with both laser lines (514.5 nm used at 15W/cm2, 561 nm used at 18 W/cm2). While the green channel CCD was used with 1000x gain during both imaging cycles the gain on the red channel CCD was adjusted between 450 for the first cycle and 1000x for the second cycle. The first imaging cycle generated a detectable signal from the NPC staining on both cameras, due to surface reflection on the dichroic mirror between the cameras. The front surface reflection was more pronounced than the back surface reflection and could be detected well enough to use an average time projection of the 50 images collected in the first imaging cycle as a reference for image alignment. (Fig. S1). Power measurements were done using an objective power meter (Carpe). Stage drifts during data acquisition were minimal and as the nuclear pores and the mRNA were imaged simultaneously no extrinsic drift control was needed.
Image Processing
The image information of the mRNA and NPC signals needed to be fine registered post experimentally. For each cell imaged, two data sets per channel were collected as described above. The first set contained signal from the nuclear pore label, POM121-tdT, which was recorded on both cameras. Time projection of the average signal yielded an image that identified single NPCs. Original image stacks were divided into two sub-stacks with only half the area but still retained the same number of images to achieve better registration because of non monotonic distortion over the field of view. Time projected images from both cameras were registered using ‘projective’ transformation in MatLab. The individual transformation matrixes were applied to the second movie from the red channel of each data set to overlay NPCs with the mRNA signal. The signal of the NPC label in the second movie was much lower due to bleaching during the recording of the registration data. To improve the SNR a sliding average of 15 – 25 frames was calculated for the second movie and used to fit the NPC positions during the experiment. This averaging resulted in a reduced time resolution for the NPC signal. As nuclear pores are relatively immobile at least 6 nuclear pores per cell from 15 different cells were tracked for at least 150 frames in these averaged movies to estimate the localization precision of our nuclear pore signal. Based on the mean error of the localized position of these NPC we achieved 15 nm localization precision. This value is an underestimation, as cellular movement will contribute to the error source for localization over this time range. The drift of an average NPC was 1.1 ± 0.2 nm between subsequent frames (20 ms integration time).
The image registration precision was tested by fitting NPC positions on the green channel registration data set and the registered red channel data set for nine cells. The resulting registration precision was better than 10 nm (see Fig. 1 and following text). Determination of the absolute colocalization precision in living cells by our method is limited by the available signal in the green channel. As photons contributing to this image are reflected off the glass surface of a dichroic that is designed to transmit light at this wavelength the SNR in the green channel is clearly worse than in the red channel. Compensation could be reached by longer imaging at high laser intensities, but at the cost of losing the capability to track nuclear pores during acquisition of export movies in the green channel. The applied transformation matrix is based on four pores that have been identified in both images. We hence tested our co-registration precision by calculating the distances between 6, 10 and 15 nuclear pores in both images for a total of 21 registered nuclei from two of three experimental sets (a total of 33 cells, see Fig. 1). Each registered image series contained an expected number of 40 to 60 nuclear pores, depending on the size of the nucleus. Based on the differences in SNR between the two registration images we argue that 10 nuclear pores are a fair sub-sample to estimate registration precision, leading to a registration precision of 8±1 nm. Six pores might be too few as the number is almost identical with the number of pores used for ‘super-registration’, while 20 pores would introduce a co-registration uncertainty that would be majorly determined by the SNR of nuclear pores imaged in the green channel. As can be seen in Fig. 1 the resulting registration precision is 10±1 nm if we apply a 15 pore criteria. NPC and mRNA signals were evaluated by Gaussian fitting. While the localization precision for nuclear pores could be determined experimentally within our data sets to be 15 nm, the localization precision for our mRNA signal was estimated from the number of detected photons and the FWHM of the Gaussian fit by Eq. 11.
| Eq. 1 |
The number of photons (N) was calculated from the counts detected by the camera and reported by the fitting routine using the manufacturer’s calibration data for each camera, taking into account the EM gain, electrons generated per A/D count, quantum efficiency of the CCD and the energy of a photon at the center emission wavelength. The factor ‘s’ is the standard deviation of the Gaussian approximation of the point-spread function. It is determined by fitting a steady signal repeatedly and calculating the distances between identical positions in different frames. Our mRNA is moving and hence we need to estimate this value for use in Eq. 1. One consequence of an inherent mobility of the signal is that it will spread and be less bright than an immobilized equally labeled sample. We used the following assumption: a signal that can be fitted has to have one brightest pixel. The brightest pixel will be a lower approximation for the true position of the mRNA. Hence ‘s’ can be approximated as ‘a’ The pixel size ‘a’ was 64 nm, and the background b was estimated form our data sets. The resulting localization precision for our mRNA signal was 19 nm. The colocalization precision between NPC and mRNA signal is given by Eq. 2.
| Eq. 2 |
The precision of mRNA signal is σmRNA= 19 nm, nuclear pores are localized with σNPC= 15 nm and the registration between the channels is σregistration= 10 nm. The overall colocalization precision that equals our achieved ‘super-registration’ is calculated to be 26 nm. All our numbers for registration precision between cameras, localization of mRNAs and nuclear pores are the average of our data. While such an average is a reliable and well defined measure, we argue that such a number might be of limited relevance for the biological problem. In detail, the observed kinetics of transient interactions in living cells would be heavily biased if traces would be cut short because in individual frames during the total interaction time the signal of one of the observed entities drops below the threshold value for registration precision. Accordingly, selection of data points based on the localization precision, as used in single molecule based super resolution techniques, is not an option for tracking in living cells. The data presented here present a break-through in spectrally resolved super-registration microscopy as they are mostly limited by the detection precision of the mRNA signal, not the pore signal or the channel registration precision. Gaussian fits were preformed with two routines. One routine included automated particle identification and nearest neighbor tracking as described by Thompson et al.31. The other routine was analogous to Kubitscheck et al.32 but implemented in a semi-automated way. Upon ‘clicking’ of a signal the brightest spot in a ten pixel environment is found and a center of mass algorithm delivers the start point for the Gaussian fit. A number of control checks was used to validate the fit. All fit parameters are immediately reported to the user to allow direct appreciation of the fit. A graphical help was also implemented to disallow for confusion of particles. This routine was used to fit all signals within a 10–15 pixel distance of the nuclear envelope. This allowed visual identification of signals and manual tracking. As the focal thickness of our observation volume was small, due to the high N.A. of the objective, manual tracking allowed better control of ‘blinking’ events. Both routines used raw data to perform the fitting. Localization precisions are based on fits performed according to Thompson et al.31
Cells
Immortalized Mouse Embryo Fibroblast cells (MEF) from a homogeneous transgenic knock-in mouse for β-actin-24-MBS were infected with a lentivirus coding for NLS-MCP-YFP protein. The mouse develops normally having all β-actin transcripts tagged with the 24x MBS repeats. This stable cell line was FACS sorted for low expression levels of NLS-MCP-YFP and infected with a lentivirus coding for POM121-tandem-Tomato (POM121-tdT). Cells were FACS sorted for double positive signals in the green and red channels. Successive FACS analysis was used to separate cells with homogeneous NLS-MCP-YFP and POM121-tdT expression. Growth curves of the immortalized MEFs, MEFs derived from the β-actin 24 MBS mouse, β-actin MEFs with either NLS-MCP-YFP or MCP-GFP expression and β-actin MEFs with additional POM121-tdT expression were collected (Fig. S2). Cells were seeded at 3000 cells/ml density in 60 mm dishes. A total of 30 dishes for each cell line were seeded and up to four dishes a day were harvested and counted. A hemacytometer (Fisher) was used for counting and at least four samples from each dish were counted. All five cell lines grew with the same doubling times (Table S1), suggesting that neither the MCP label for the RNA nor the POM121 label for the NPC have major effects on cellular metabolism. The labeling ratio of NPCs is discussed in more detail in the SI in Fig. S4.
Cells were grown in DMEM (Cellgro, Mediatech) containing 10% FBS (Sigma) under 5% CO2 atmosphere. 24 – 36 hours prior to imaging, cells were split into glass bottom dishes (MatTek). Shortly before imaging, cells were washed with PBS (Sigma) and transferred into DMEM w/o Phenol Red, containing 10% FBS and 25 mM HEPES (Gibco). Each dish was imaged at 37°C for less than 60 min.
Supplementary Material
Acknowledgments
Thanks to T. Dange, K. Czaplinski, T. Lionnet and X. Meng for help with cell lines and cloning. To A. Gennerich, H.Y. Park and D. Entenberg for help on data analysis and programming. S.M. Shenoy for developed of alignment software for detector pre-registration and his additional support. D. Larson, I. Lepper and U. Kubitscheck for providing fitting routines. A. Wells for FISH and TIRF data, proof reading and discussion. Lentiviral vector was a gift from G. Mostoslavsky and R.C. Mulligan. Supported by DFG 3388/1 to D.G.. Supported by NIH EB2060 and GM86217 to RHS. The authors declare no competing interests.
Footnotes
Authors Contributions D.G. designed and performed experiments, established cell lines, performed data analysis and built microscopy equipment in consultation with RHS. D.G. and R.H.S. discussed data and wrote the manuscript.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Stoffler D, et al. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J Mol Biol. 2003;328 (1):119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 2.Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. Journal of Cell Science. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 3.Rout MP, et al. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148 (4):635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306 (5700):1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 5.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130 (3):512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450 (7170):695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 7.Lim RY, et al. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318 (5850):640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 8.Mor A, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12 (6):543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 9.Stockley PG, et al. Probing sequence-specific RNA recognition by the bacteriophage MS2 coat protein. Nucleic Acids Res. 1995;23 (13):2512–2518. doi: 10.1093/nar/23.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Current Biology. 2003;13 (2):161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122 (3):513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronshaw JA, Krutchinsky AN, Zhang WZ, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158 (5):915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildiz A, et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5–nm localization. Science. 2003;300 (5628):2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 14.Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci U S A. 1999;96 (17):9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopito RB, Elbaum M. Reversibility in nucleocytoplasmic transport. Proc Natl Acad Sci U S A. 2007;104 (31):12743–12748. doi: 10.1073/pnas.0702690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachi A, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. Rna. 2000;6 (1):136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty P, Satterly N, Fontoura BM. Nuclear export assays for poly(A) RNAs. Methods. 2006;39 (4):363–369. doi: 10.1016/j.ymeth.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122 (12):1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke WW, Scheer U. The ultrastructure of the nuclear envelope of amphibian oocytes: a reinvestigation. II. The immature oocyte and dynamic aspects. J Ultrastruct Res. 1970;30 (3):317–327. doi: 10.1016/s0022-5320(70)80065-x. [DOI] [PubMed] [Google Scholar]
- 20.Franke WW, Scheer U. The ultrastructure of the nuclear envelope of amphibian oocytes: a reinvestigation. I. The mature oocyte. J Ultrastruct Res. 1970;30 (3):288–316. doi: 10.1016/s0022-5320(70)80064-8. [DOI] [PubMed] [Google Scholar]
- 21.Bastos R, Pante N, Burke B. Nuclear pore complex proteins. Int Rev Cytol. 1995;162B:257–302. doi: 10.1016/s0074-7696(08)62619-4. [DOI] [PubMed] [Google Scholar]
- 22.Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136 (3):515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20 (6):1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65(4):570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6 (5):421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 26.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4 (8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 27.Kubitscheck U, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168 (2):233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clemen AE, et al. Force-dependent stepping kinetics of myosin-V. Biophys J. 2005;88 (6):4402–4410. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dange T, Grunwald D, Grunwald A, Peters R, Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J Cell Biol. 2008;183 (1):77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun C, Yang W, Tu LC, Musser SM. Single-molecule measurements of importin alpha/cargo complex dissociation at the nuclear pore. Proc Natl Acad Sci U S A. 2008;105 (25):8613–8618. doi: 10.1073/pnas.0710867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82 (5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubitscheck U, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168 (2):233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.