Abstract
Independent component analysis (ICA) methods have received growing attention as effective data-mining tools for microarray gene expression data. As a technique of higher-order statistical analysis, ICA is capable of extracting biologically relevant gene expression features from microarray data. Herein we have reviewed the latest applications and the extended algorithms of ICA in gene clustering, classification, and identification. The theoretical frameworks of ICA have been described to further illustrate its feature extraction function in microarray data analysis.
INTRODUCTION
With advances in DNA microarray technology, it is now possible to quantify expression levels of thousands of genes in parallel. In fact, in various human disease studies, the technology has become a mature gene expression profiling tool. Implicit in the work of gene expression analysis today is the assumption that no gene in the human genome is expressed completely independently of other genes. Therefore, to understand the synergistic effects of multiple genes, researchers, as part of the process, extract the underlying features from any multivariable microarray data set to reduce dimensionality and redundancy.
To permit effective extraction of these features, however, microarray technology must address the issue of noise in the array systems themselves. Due to the inherent noise in most array systems, single control and experiment designs are not always reliable, making it difficult to understand the functionality of the complicated correlations among the large numbers of genes present in the genome.
Overcoming all of these challenges requires mathematical and computational methods that are versatile enough to capture the underlying biological features and simple enough to be applied efficiently to large data sets. Two kinds of unsupervised analysis methods for microarray data, principal component analysis (PCA) and independent component analysis (ICA), have been developed to accomplish this task. PCA projects the data into a new space spanned by the principal components. Each successive principal component is selected to be orthonormal to the previous ones and to capture the maximum variance that is not already present in the previous components. The constraint of mutual orthogonality of components implied by classical PCA, however, may not be appropriate for biomedical data. Moreover, since PCA features all use second-order statistics technique to achieve the correlation learning rules and only covariances between the observed variables are used in the estimation, these features are only sensitive to second-order statistics. The failure of correlation-based learning algorithms is that they reflect only the amplitude spectrum of the signal and ignore the phase spectrum. Extracting and characterizing the most informative features of the signals, however, require higher-order statistics.
In contrast to PCA, the goal of ICA is to find a linear representation of non-Gaussian data so that the components are statistically independent, or as independent as possible. Such a representation can be used to capture the essential structure of the data in many applications, including feature extraction and signal separation. ICA is already widely used for performing blind source separation (BSS) in signal processing and is considered a very important research topic in many areas, such as biomedical engineering, medical imaging, speech enhancement, remote sensing, communication systems, exploration seismology, geophysics, econometrics, data mining, etc. (1). BSS techniques principally do not use any training data and do not assume a priori knowledge about the parameters of filtering and mixing systems. BSS, therefore, has been successfully used in processing electroencephalograms (EEG), magnetoencephalograms (MEG), and functional magnetic resonance (fMRI) images (2–5).
In the first phase of ICA biomedical applications, Hori and colleagues first proposed an ICA-based gene classification method to effectively classify yeast gene expression data into biologically meaningful groups (6,7). Liebermeister proposed an ICA method for microarray data analysis to extract expression modes whereby each mode represents a linear influence of a hidden cellular variable (8). Following these developments, ICA has been widely used in microarray data analysis for feature extraction, gene clustering, and classification. In biomedical data mining, ICA has been shown to outperform PCA.
METHODOLOGY
Theory and Model of ICA/BSS
The technique of ICA was first used in 1982 for analyzing a problem pertaining to neurophysiology (9). In the middle of the 1990s, after the term ICA was first coined by Comon (10), ICA received wide attention and growing interest when many efficient approaches were put forward, such as the infomax principle by Bell and Sejnowski (11), natural gradient-based infomax by Amari (12), and the fixed-point (FastICA) algorithm by Hyvärinen (13–16). Due to ICA’s computational efficiency, it was widely used in many large-scale problems. ICA and its various extended theories, algorithms, and applications have subsequently become a very important research topic in many areas.
ICA, as a specific embodiment of BSS, has been further developed in the last decade. BSS solves the problem of recovering statistically independent signals from the observations of an unknown linear mixture. Although BSS and ICA refer to the same or similar models and can solve problems with similar algorithms under the assumption of mutually independent primary sources, the terms are often interchangeably used in the literature. However, BSS and ICA have somewhat different objectives. The objective of BSS is to estimate the original source signals even if they are not all statistically independent, while the objective of ICA is to determine a data transformation that assures that the output signals are as independent as possible. For this purpose, ICA methods often employ higher-order statistics (HOS) to extract a higher-order structure of signals in feature extraction (10,11).
The BSS model can be described as follows:
Let s(t) = [s_1(t), s_2(t), …, s_M(t)]T, where m represents mutually independent random processes whose exact distribution functions are unknown, and t is a discrete time variable. Suppose that we have n sensors to provide observed signals x(t) = [x_1(t), x_2(t), …, x_N(t)]T, with xi(t), i = 1, 2, …, n, which measure a linear combination of the n signals. This can be expressed as:
| [Eq. 1] |
It can also be rewritten in the matrix format as:
| [Eq. 2] |
where A is an unknown n × m full-column rank linear matrix that represents the mixing system. In this system, X is the information that is available, while A and S are unknown. The objective of BSS is, in general, to recover all source signal S by using the assumption of mutual independence among source signals. In most cases, to simplify BSS problems, the number of source signals are always assumed to be equal to the number of observed signals, n = m. Then, the mixing matrix A becomes a n × n square matrix.
Without knowing the source signals and the mixing matrix, we want to recover the original signals from the observations X by the following linear transform:
| [Eq. 3] |
where Y denotes the vector of [y_1(t), y_2(t), …, y_N(t)]T. Y is the estimation of the source signal S, and W is a n × n linear matrix.
Since the mixing matrix A and the source vector S are unknown, it is impossible to obtain the original sources in an exact order and amplitude. Nevertheless, it is possible to obtain scaled and permutated sources. In this case, the de-mixing matrix W is written as:
| [Eq. 4] |
where D is a nonsingular diagonal matrix and P is a permutation matrix.
ICA can be used to blindly separate mixtures mixed by several mutually independent sources in BSS problems, in which we focus on the independent components in S as the original sources. In addition, together with higher-order statistical techniques, it can also be applied to extract features from sources, in which the basis vectors in matrix A and the projection coefficients S are both concerned. In applying ICA on a feature extraction field, we use the same model as BSS but with different denotations for the variables. In ICA, X is the original signal instead of the mixture (as in BSS), the columns of A are denoted as the basis vectors or latent vectors of the ensemble of signals, and the rows of S are the mutual independent projection coefficients. ICA assumes the observation X to be the linear mixture of the independent components in S. In ICA feature extraction, the number of independent components is always assumed to be the same as the number of input signals, implying that one can extract as many features as the number of input signals. The objective of ICA feature extraction then is to extract the basis vectors when the projection coefficients are the most statistically independent from one another.
ICA Modeling of Gene Expression
Recently, ICA has been used by biomedical scientists as an unsupervised approach to explore gene expression features and discover novel underlying biological information from large microarray data sets. Gene expression data provided by microarray technology is considered a linear combination of independent components having specific biological interpretations.
Let the n × k matrix X denote microarray gene expression data with k genes under n samples or conditions. xij in X is the expression level of the jth gene in the ith sample. Generally speaking, the number of genes k is much larger than that of the samples n, k≫n. Suppose that the data have been preprocessed and normalized (i.e., each sample has zero mean and standard deviation); then the ICA model for gene expression data is the same as Equation 2, X = AS. In some publications, the k × n matrix X is used to denote k genes under n samples. In this case, the transform, XT is used in the ICA model: XT = AST. So, XT here denotes the same n × k matrix as used in the ICA model.
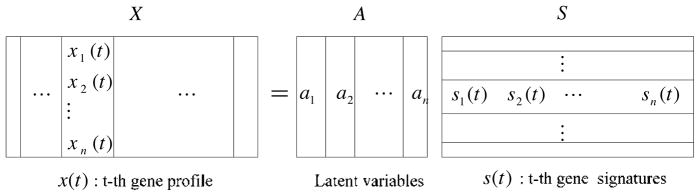
In ICA modeling of microarray data, the columns of A = [a1, a2,…, an] are the n × n latent vectors of the gene microarray data, S denotes the n × k gene signature matrix or expression mode, in which the rows of S are statistically independent to each other, and the gene profiles in X are considered to be a linear mixture of statistically independent components S combined by an unknown mixing matrix A. Figure 1 presents the vector framework of the ICA model. References 17 and 18 give detailed analysis for matrices S and A. The ith row matrix A contains the weights with which the n expression modes of the k genes contribute to the ith observed expression profile. Hence the assignment for the observed expression profiles with different classes is valid for the rows of A, and each column of A can be associated with one specific expression mode. For an example of two classes, suppose one of the independent expression modes s is characteristic of a putative cellular gene regulation process. In this case, it should contribute substantially to one of the class experiments, whereas its contribution to another class experiment should be less, or vice versa. Since the jth column of A contains the weights with which s contributes to all observations, this column should show large or small entries according to the class labels. After such characteristically latent variables have been obtained, the corresponding elementary modes can be identified, which yields useful information for classification. Also, the distribution of gene expression levels generally features a small number of significantly overexpressed or under-expressed genes which form biologically coherent groups and may be interpreted in terms of regulatory pathways (6–8,18,19).
Figure 1.
ICA vector model of microarray gene expression data.
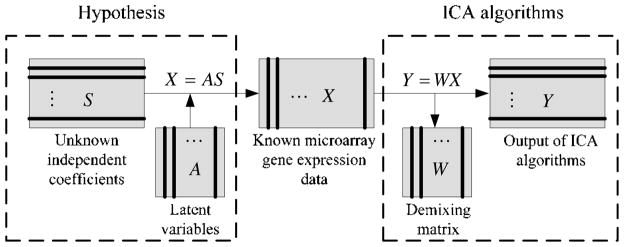
To obtain S and A, the demixing model can be expressed the same as Equation 3, Y = WX, where W is an n × n demixing matrix. Figure 2 shows the processing of ICA algorithms on microarray gene expression data.
Figure 2.
Theoretical framework of ICA algorithms on microarray gene expression data.
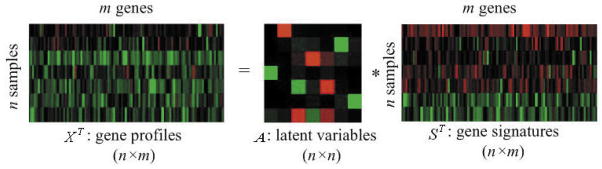
Figure 3 gives us a small example of applying ICA, in this case to microarray gene expression data of seven hippocampal tissue samples from individuals with severe Alzheimer’s disease [data provided by Blalock and colleagues (20)]. In Figure 3, X denotes the Alzheimer’s disease microarray data set of 7 samples and 101 selected genes, A is the 7 × 7 latent variables matrix, and S represents the gene signatures in a 7 × 101 matrix whose rows are mutual statistically independent coefficients.
Figure 3.
ICA model of severe Alzheimer’s disease microarray gene expression data.
Typical ICA algorithms are infomax, minimum mutual information (MMI) (11), FastICA and maximum non-Gaussianity (13–16). Due to its high speed and effectiveness, FastICA [proposed by Hyvärinen, (13)] is frequently used in performing ICA for microarray data. In FastICA, maximizing negentropy is used as the contrast function since negentropy is an excellent measurement of nongaussianity (13). Using the concept of differential entropy, the negentropy J is defined by
| [Eq. 5] |
where yG is a Gaussian random vector of the same covariance matrix as vector y. Mutual information, I, is known as a natural measure of the independence of random variables; it is widely used as the criterion in ICA algorithms. The mutual information I between the n variables yi, i = 1,…, n, can be expressed by negentropy:
| [Eq. 6] |
It is known that the independent components are determined when mutual information I is minimized. From Equation 6, it is clearly shown that minimizing the mutual information I is equivalent to maximizing the negentropy J(y). To estimate the negentropy of yi = wTx, an approximation to identify independent components one by one is designed as follows:
| [Eq. 7] |
where G can be practically any non-quadratic function, E{·} denotes the expectation, and ν is a Gaussian variable of zero mean and unit variance. In Reference 13, Hyvärinen proved the consistency of the vector w obtained by maximizing JG, and analyzed the asymptotic variance for choosing the function G to be used in the contrast function. Robustness as a very attractive property was also discussed by Hyvärinen: for a bounded function G, the estimation of w is rather robust against outliers, or one should at least choose a function G(yi) that does not grow very fast when |y| grows. Hyvärinen offered three nonlinear functions of G for a different probability density distribution of output signal y. They are
| [Eq. 8] |
| [Eq. 9] |
and
| [Eq. 10] |
where 1 ≤ a1 ≤ 2 and a2 ≈ 1 are constants For these nonlinear functions, G1 is a good general-purpose contrast function, G2 is suggested to be used when the independent components are high super-Gaussian or when robustness is very important, and G3 can be used for sub-Gaussian independent components with no outliers. It is demonstrated by Lee et al. (17) that the distributions of gene signatures are typically super-Gaussian, therefore, the nonlinear function G1 and G2 are suitable in a FastICA algorithm for analyzing gene microarray data.
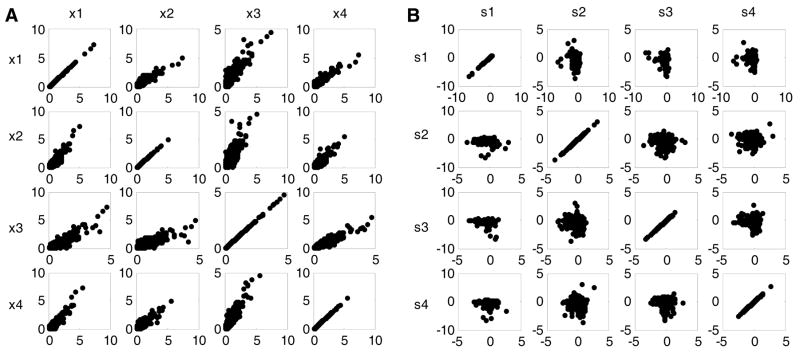
Figure 4 shows an example of the joint distributions between samples of the severe Alzheimer’s disease gene expression data set before and after FastICA transformation. Figure 4A shows strong correlations between the pairs of the original gene microarray data since the distributions are concentrated along a linear line for every pair: (x1, x2), (x1, x3), (x1, x4), (x2, x3), (x2, x4), and (x3, x4). However, Figure 4B displays no such correlations between the pairs of gene signatures, especially the pairs (s2, s3), (s2, s4) and (s3, s4). The gene signatures were captured using the FastICA algorithm with as the nonlinear function in the contrast function. The mutually independent gene signatures obtained from FastICA combined with the joint distributions shown in Figure 4 confirmed that ICA extracts the underlying statistically independent information from the mutually dependent gene microarray data. The FastICA programs we ran in MATLAB 7.0 can be downloaded freely from Hyvärinen’s homepage (www.cis.hut.fi/projects/ica/fastica).
Figure 4. Joint distributions between pairwise gene expression data and gene signatures.
(A) The joint distributions between four original severe Alzheimer’s disease genes: x1, x2, x3, and x4. (B) The joint distributions between four corresponding gene signatures s1, s2, s3, and s4 captured by FastICA.
Number Estimation of Latent Vectors
The estimation of the number of latent vectors is very challenging since it depends on how to find an efficient measurement method to detect the functional significance of each latent variable. To this aim, Martoglio and colleagues presented a variational Bayesian framework of the ICA method for reducing a high-dimensional DNA array data to a smaller set of latent variables. They reported that the number of latent variables can be assessed after the learning process rather than having to pre-set the number of possible clusters (21).
While using the basic ICA model (Equation 1), we have taken note that the number of latent variables in the framework described above is exactly the same as the number of samples or conditions given in gene expression data. In many cases, the size of the latent variables n (which are equal to the number of microarray samples or experiments) is much smaller than the number of genes. In that case, the number of latent variables can be selected and reduced by (i) selecting all the latent variables if their measurement contributions are all of the similar significance (we will discuss the methods of measurement later in this section), and (ii) selecting latent variables by setting a threshold of the cumulated contribution of the eigenvalues when using PCA as pre-processing before performing ICA (22,23). In other words, after PCA projection, we can select smaller principal components (eigenvectors) corresponding to the largest eigenvalues based on cumulated contribution over a threshold (for example, 95% is a threshold determined by the assumption that the selected principal components can catch most of the information of the original inputs if the cumulative contribution of their corresponding eigenvalues is over 95%). In particular, the latent variables obtained by the FastICA deflation approach (estimating ICs one by one) can be reduced by simply discarding the last latent variables, since more and more noise and useless information remain in the later ICs. Therefore, the FastICA deflation approach is sometimes effective at eliminating counterproductive influences from the weak components that might not capture essential information (7).
In the case of large sizes of microarray experiments, several ways of obtaining the number of latent variables can be summarized:
To evaluate the significance of latent variables, Lee et al. (24) presented the L2-norm (||a(i)||, where a(i) is the ith column vector of A) to calculate the contributions from the columns of matrix A. By reordering the basis vectors using L2-norm and their coefficient variance respectively, they showed that those two ordering methods provided almost the same order. Then, the top significant basis vectors could be selected by removing the basis vectors with negligible L2-norm and variance.
Hansen et al. used dynamic decor-relation to estimate probabilities of competing hypotheses for ICA (25). They used an approximate Bayesian framework for computing relative probabilities of the relevant hypotheses. The method showed efficiency in detecting the content of dynamic components, selecting the number of independent components, and testing whether a given number of components are significant relative to white noise.
Another value of the Bayesian framework that should be stated here is the work of Teschendorff et al., who in 2005 (26) presented a variational Bayesian Gaussian mixture model to estimate the number of clusters as an external parameter within an independent component that had been inferred using MLICA and compared with the accuracy of the cluster number estimation using the EM-BIC. The experiments on tumor microarray data showed that the variational Bayesian method was more accurate in finding the true number of clusters and avoided the shortcoming of specifying the number of clusters in advance in k-means and self-organizing maps (SOM) methods.
In other work from Teschendorff et al. published in 2007, a method of feature selection was presented (27) that suggested finding genes that are differentially activated by setting a threshold. The threshold can be set typically two or three standard deviations from the mean, and genes are selected whose absolute weights exceed this threshold.
Application of ICA to Microarray Data
ICA can decompose input microarray data into components so that each component is statistically as independent from the other as possible and can reveal the underlying gene expression patterns. The statistical analysis of microarray data will make the detection of gene clustering patterns, the identification of cancer and oncogenic pathways, and the identification of their associated gene network more effective and accurate.
Application of ICA to Gene Clustering and Classification
In 2001, ICA was first applied to a blind gene classification by Hori et al. (6,7), who defined the columns of the mixing matrix A and matrix ST as “model profile” and “controlling factors,” respectively. Then the ICA model was known as linear combinations of model profiles weighted by the contributions of controlling factors. By using ICA on gene expression data of yeast during sporulation, the method automatically identified typical gene profiles that were similar to average profiles of biologically meaningful gene groups, and the authors classified yeast genes based on the obtained typical profiles. By comparing the average profiles, they validated that the genes DIT1, SPR28, SSP1, and SPR3 were related to spore wall formation, whereas the genes MEP2, ERG6, ACS2, and IDH1 were associated with metabolism.
Independently, Liebermeister et al. applied ICA to gene expression data in 2002 (8). The expression of each gene was modeled as a linear function of the expression modes based on hidden variables termed “expression modes.” They showed that (i) cell cycle-related gene expression in yeast was related to distinct biological functions, such as phases of the cell cycle or the mating response; and that (ii) the gene expression in human lymphocytes revealed modes that were related to characteristic differences between cell types. The expression modes and their influences can be used to visualize the samples and genes in low-dimensional space.
Lee et al. did excellent work in 2003 to analyze the performance of different kinds of ICA algorithms (17). They applied linear and nonlinear ICA to project microarray data into statistically independent components and to cluster genes according to over- or underexpression in each component. They tested six linear ICA methods: natural-gradient maximum-likelihood estimation (NMLE) (28,29), joint approximate diagonalization of eigenmatrices (JADE) (30), FastICA with three decorrelation and nonlinearity approaches (31), and extended infomax (32). They also tested two variations of nonlinear ICA: Gaussian radial basis function (RBF) kernel (NICAgauss) and polynomial kernel (NICApoly). Overall, the linear ICA algorithms consistently performed well in all data sets. The nonlinear ICA algorithms performed best in the small data sets but were unstable in the two largest data sets. They also showed that ICA outperforms PCA, k-means clustering, the Plaid model, and the construction of functionally coherent clusters on microarray data sets from Saccharomyces cerevisiae, Caenorhabditis elegans, and humans.
In the same year, Suri et al. compared the performance of ICA with PCA for detecting coregulated gene groups in microarray data collected from different stages of the yeast cell cycle (33). The experiments demonstrated that PCA did not find periodically regulated gene groups. In contrast, ICA revealed two gene groups that are apparently regulated within the cell cycle. This analysis of the same data with PCA and ICA suggested that ICA was more successful in finding coregulated gene groups.
Najarian et al. in 2004 used FastICA to discover biological gene clusters from gene expression data obtained by using DNA microarrays, and scored the genes based on the protein interactions within a cluster (34). Specifically, they used the mutually independent coefficients generated by FastICA for determining the proximity measure in gene clustering and for establishing the biological gene clusters that are co-expressed and that interact with the protein products.
FastICA was also used to supervise gene classification in the yeast functional genome by Lu et al. in 2005 (35). They first preprocessed the gene expression data sets with FastICA to reduce the noise and obtain the gene expression data with hidden patterns (GEHP), then trained the supervised classifications using the GEHP, and labeled the tested genes with the class of the output of the classification method. Experimental results showed that their method can recognize the hidden patterns of the gene expression profiles and efficiently reduce noise.
In 2006, using leukemia and colon cancer data sets, Zhu et al. (36) applied ICA in gene dimension reduction and identified informative genes prior to the clustering.
In the same year, Huang et al. used ICA and a multivariate penalized regression model to classify four sets of DNA microarray data involving various human normal and tumor tissue samples (37). For determining the number of latent variables, the sequential floating forward selection (SFFS) technique was used to find the most discriminating ICA feature. They showed that their method was effective and efficient in discriminating normal and tumor tissue samples and the results can hold up under re-randomization of the samples.
Application of ICA to Cancer or Oncogenic Pathway Identification
The goal of many microarray analyses is to identify genes in cell lines or tissue samples that are differentially transcribed under different biological conditions. In 2002, Liao and colleagues (38) used FastICA to recognize the functional identities of known genes. Based on the ICA work with gene expression profiles of yeast cells exposed to different stimuli, the partial least-squares (PLS) method was used as a regression model to identify the correlation between the multi-dimensional gene expression profile and the drug activity pattern. The result shows that a group of 15 genes are important.
In the same year, Wang et al. developed a statistically-principled neural computation approach to the independent component imaging for disease signatures (39). The features can identify an informative index subspace over which mixed imaginary sources can be separated by the infomax algorithm. They discussed the theoretical roadmap of the approach and its applications to partial volume correction in cDNA microarray expression data and to neurotransporter binding separation data collected by positron emission tomography (PET) imaging.
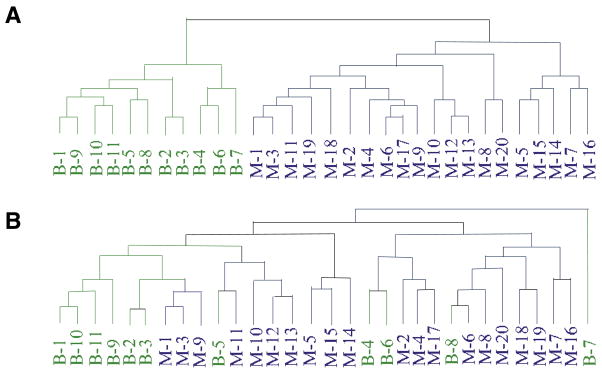
In 2004, Saidi et al. compared ICA to the established methods of PCA, Cyber-T and significance analysis of microarrays (SAM) to identify coregulated genes associated with endometrial cancer (40). In contrast to PCA, ICA generated patterns that clearly characterized the malignant samples studied. The hierarchical clustering results of ICA and PCA outputs in classifying the benign (B1–B11) and malignant (M1–M20) endometrial samples are shown in Figure 5. To save space, only the superimposed hierarchical cluster dendrogram is presented here; more details can be seen in Reference 40. Figure 5 shows that ICA outperformed PCA in achieving distinct separation between benign (B1–B11) and malignant (M1–M20) endometrial samples. Moreover, several genes involved in lipid metabolism that were differentially expressed in endometrial carcinoma were found only through the ICA method.
Figure 5. The superimposed hierarchical dendrogram in clustering benign (B1–B11) and malignant (M1–M20) endometrial samples by using ICA and PCA outputs in Saidi’s work (40).
(A) ICA output with superimposed hierarchical cluster dendrogram. The hierarchical clustering was applied to the columns of matrix A and showed a distinct separation into benign (green lines) and malignant (blue lines) groups. (B) PCA output with superimposed hierarchical cluster dendrogram based on 15 principal components.
Also in 2004, Chiappetta et al. performed ICA on two data sets: breast cancer data and Bacillus subtilis sulfur metabolism data. They found that some of the obtained gene families correspond to well known families of coregulated genes (19). In the same year, Berger et al. (41) used ICA on DNA microarray data of breast cancer with the goal of identifying distinct, biologically relevant functions from gene expression data. They uncovered these functions based on observed gene expression by selecting outlier values of gene influence from the ICA estimates and examining their corresponding gene annotations.
FastICA was used by X.W. Zhang et al. (42) in colon and prostate cancer data sets to extract specific diagnostic patterns of normal and tumor tissues (2005). They proposed a method of biomarker selection by designing discriminant vectors and calculating the Euclidean distance between test vectors and discriminant vectors, and then classified the samples into normal or tumor samples. The genes that can achieve any user-predefined classification accuracy can be selected as biomarkers.
To compare the results based on correlated expression and gene clusters with those significantly enriched gene ontology categories, Frigyesi and colleagues in 2006 (43) showed that ICA can achieve higher resolution. The component characteristics for molecular subtypes and for tumors with specific chromosomal translocations were identified. ICA also identified more than one gene cluster that was significant for the same ontology categories and that disclosed a higher level of biological heterogeneity even within coherent groups of genes.
Journée and Teschendorff et al. gave a review of some standard ICA algorithms and the link between the ICA approach and the theory of geometric optimization (44), discussing three fundamental contrast functions, the matrix manifold, and the optimization process. To validate the ICA approach on a breast cancer microarray data set, four ICA algorithms [JADE (45), RADICAL (46), Kernel-ICA (47), and FastICA (13)] and PCA were applied on one of the largest breast cancer microarray data sets. After evaluating the biological significance of the results, ICA achieved a more realistic representation of the gene expression data than PCA when comparing results with known oncogenic and cancer-signaling pathways.
In follow-up studies, Teschendorff and Journée et al. did more detailed research (27) in which the four ICA algorithms of JADE, RADICAL, Kernel-ICA and FastICA were also applied to six breast cancer microarray data sets to identify cancer signaling and oncogenic pathways and regulatory modules that play a prominent role in breast cancer. They then related the differential activation patterns to breast cancer phenotypes. Some novel associations linking immune response and epithelial-mesenchymal transition pathways with estrogen receptor status and histological grade, respectively, were found in the results. In addition, associations linking the activity levels of biological pathways and transcription factors with clinical outcome in breast cancer were found. They showed that ICA results in a more biologically relevant interpretation of genome-wide transcriptomic data.
Extended ICA Algorithm for Microarray Data
To better understand and extract the underlying independent biological expression profiles in microarray data, some further studies and new algorithms were introduced into the ICA methods. In 2003, the independent feature subspace analysis (IFSA) method was proposed by Kim et al. to achieve a linear transformation such that feature subspaces become independent but components in a feature subspace are allowed to be dependent (48). IFSA can extract phase- and shift-invariant features. Based on a membership scoring method, the experiment showed the usefulness in grouping functionally related genes in the presence of time-shift and expression phase variance. In the same year, Wang et al. (49) presented a partial ICA technique in tissue heterogeneity correction to estimate the parameters of a linear latent variable model that performs a blind decomposition of gene expression profiles from mixed cell populations.
The gene signatures S and the latent variables A obtained from any ICA algorithm contain both positive and negative entries. Miskin et al. showed that enforcing the latent variables to be positive does not result in an increase of the evidence. On the other hand, enforcing positivity of the gene signatures seems to result in a significant increase in the evidence for encoding the gene signatures as a whole. However, for any individual gene signature, the outcome could be worse (21,50). For this reason, a better approach would encode the fact that some gene signatures can be all positive (such as the housekeeping genes) and some should be both positive and negative.
Recently, the non-negative matrix factorization (NMF) technique was improved and combined with ICA to enable further understanding of microarray data because non-negative sources are meaningful in microarray profiling. Zhang et al. (2003) presented a method of blind non-negative source separation to factorize the observation matrix into a non-negative mixing matrix and a non-negative source matrix where the microarray sources may be dependent (51). To solve the tissue heterogeneity problem, the method was applied to partial volume correction of real microarray expression profiles. Experiments showed that the non-negative ICA method can be used to decorrelate observations and recover sources without any limitation so that the sources must be statistically independent or distributed in a non-Gaussian manner. This finding indicates a superior performance compared with the normal ICA approach. In 2006, non-negative ICA as a gene clustering approach was presented for microarray data analysis by Gong and his colleagues (52). Their method better suits the corresponding putative biological processes due to the positive nature of gene expression levels. In conjunction with a visual statistical data analyzer, they showed that the significant enrichment of gene annotations within clusters can be obtained. In the simulation experiments of the NMF method, however, an inevitable problem of not converging to a stationary point existed in the common NMF approach (46,53). One reason is that the basis of a space may not be unique theoretically; therefore, separate runs of NMF lead to different results. Another reason may come from the algorithm itself, such that the cost function is sometimes stuck in a local minimum during its iteration. Stationarity is important as it is a necessary condition for solving local minimum and optimization problems. A popular approach to solve NMF optimization problems is a multiplicative update algorithm by Lee and Seung (54). Lee and Seung proved that the update causes the function value to be nonincreasing, even though there is no proof showing that any limit point is stationary. Some projection and gradient descend approaches have been performed to improve the convergence and optimize uniqueness. In a 2007 publication, Lin (53) discussed the difficulty of proving the convergence and proposed minor modifications of existing updates that proved their convergence.
To address the problem that most ICA algorithms return the number of components specified without any indication as to which ones are the most stable, Chiappetta et al. (55) constructed consensus components by rerunning the FastICA algorithm with random initializations and by only including components that passed certain stability criteria in the final analysis. Himberg et al. (56) also introduced re-sampling of ICA components and used estimated centrotypes as representatives of ICA components.
DISCUSSION
ICA Within Information Theory Frames
Information theory is one approach to finding a suitable code that depends on the statistical properties of data. Here, we formulate an ICA model using information theory on coding. From the above publications (18,27,28,33), researchers showed that ICA can extract valid latent variables from complex and noisy measurement environments. This property of sparse coding, a method for finding a description of data in which only a small number of the representations are significantly nonzero, is the reason that noise and independent patterns unrelated to the subject of investigation can be removed. ICA transfer is an adaptive process in which representation is estimated from the data so that the independent components are as sparse as possible. For this property, super-Gaussianity or leptokurtosis are equivalent to sparsity. Lee et al. (17) found that the underlying biological processes (latent variables) are more super-Gaussian than original inputs from microarray gene data, since only a few relevant genes are significantly affected, and the majority of genes are left relatively unaffected.
Several methods have been presented to measure the significance and select genes from each independent component. In the work of Lee et al. (17), genes were selected by the loads within each independent component under the hypothesis that genes showing relatively high or low expression levels within the component are the most important for the biological process. Then, two clusters for each component were created: one cluster containing C% of all genes with larger loads, and one cluster containing C% of genes with smaller loads, where C was an adjustable coefficient. Similarly, the selection methods of sorting significant genes within a range of largest or smallest values of independent component were presented in the works of Zhu (36) and Gong (52). Moreover, as described in Reference 18, new expression modes were generated by using positive parts of rows of, S, , and the absolute values of negative parts of rows of S, . Then, the genes whose expression level exceeds a given threshold can be selected.
Frigyesi et al. (44) selected genes associated with specific components with the highest absolute loading on a given component, and tested the remaining data to fit a normal distribution using the Lilliefors’ test. This was repeated until the remaining data converged to a normal distribution.
Kong et al. (57–59) showed another method, in this case applied to speech signals, by selecting activities from each ICA-transformed vector by using linear or nonlinear shrinkage functions estimated by a maximum likelihood method. They assumed that the activities with small absolute values are noise or redundant information and set them to zero, retaining just a few components with large activities.
Future ICA Developments and Concluding Remarks
For the application of ICA in gene microarray data analysis, future developments will need to address two central challenges.
First, to extract valid gene signatures that can identify or classify the inherent pathophysiological processes, nonlinear ICA algorithms should be considered in order to represent interactions among biological processes that are not necessarily linear. Examples of nonlinear interactions in gene regulatory networks include some complex logic units, toggle switch or oscillatory behavior, and multiplicative effects resulting from expression cascades (60–62). Nonlinear ICA methods like kernel ICA may be appropriate in such cases, because nonlinear and kernel methods have been shown to extract independent components from nonlinear correlative speech and image signals (63–66). The key to nonlinear ICA algorithms is to construct an appropriate nonlinear feature space between gene signatures and gene microarray data; thereby, a kernel method based on radial basis function (RBF) and polynomial can be considered.
Second, it will be necessary to evaluate the biological significance of the independent components (ICs) from different microarray gene data sets. The IC number selection discussed in the previous sections is mainly based on mathematical analysis. However, it may be necessary to take more biological and physiological information into account. With the connection of mathematical informatics theory and bioinformatics knowledge related to etiology and nosology, the functional significance of each gene signature can be better understood, allowing the analysis of microarray gene expression data to be performed more easily and effectively.
In summary, ICA-based methods can successfully identify and cluster both the known biological signaling pathways or regulatory modules and the novel correlations involved with gene expression levels and pathways. The ability of ICA to extract latent and biologically relevant gene expression information from microarray data, despite the presence of significant noise, underscores its importance as a data-mining tool for microarray and other biomedical data analysis.
Acknowledgments
We were saddened by the passing of Dr. Hiromi Gunshin during the preparation of this manuscript. Her warmth, intelligence, and devotion to science will always be remembered.
This work was supported by the Research Foundation of Shanghai Municipal Education Commission (nos. 06FZ012 and 2008098) and the research funds of the BWH Radiology Department. It was partially supported by the NIA/NIH (grant no. 5R21AG028850, to X.H.) and the Alzheimer’s Association (grant no. IIRG-07-60397, to X.H.). We thank Ms. Kim Lawson at the BWH Radiology Department for her extremely helpful comments and editing of our manuscript. This paper is subject to the NIH Public Access Policy.
Footnotes
To purchase reprints of this article, contact: Reprints@BioTechniques.com
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
References
- 1.Vigário R, Jaakko S, Veikko J. Independent component approach to the analysis of EEG and MEG recordings. IEEE Trans Biomed Eng. 2000;47:589–593. doi: 10.1109/10.841330. [DOI] [PubMed] [Google Scholar]
- 2.James CJ. Independent component analysis for biomedical signals. Physiol Meas. 2005;26:R15–R39. doi: 10.1088/0967-3334/26/1/r02. [DOI] [PubMed] [Google Scholar]
- 3.Hong B, Pearlson GD, Calhoun VD. Source-density driven independent component analysis approach for FMRI data. Hum Brain Mapp. 2005;25:297–307. doi: 10.1002/hbm.20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigário R, Jaakko S, Veikko J. Independent component approach to the analysis of EEG and MEG recordings. IEEE Trans Biomed Eng. 2000;47:589–593. doi: 10.1109/10.841330. [DOI] [PubMed] [Google Scholar]
- 5.Potter M, Kinsner W. Competing ICA techniques in biomedical signal analysis. Canadian Conference on Electrical and Computer Engineering, 2001. 2001;2:987–992. [Google Scholar]
- 6.Hori G, Inoue M, Nishimura S, Nakahara H. Blind gene classification based on ICA of microarray data. International Conference on Independent Component Analysis and Signal Separation, ICA 2001; 2001. pp. 332–336. [Google Scholar]
- 7.Hori G, Inoue M, Nishimura SI, Nakahara H. Blind gene classification—an application of a signal separation method. Genome Informatics. 2001;12:255–256. [Google Scholar]
- 8.Liebermeister W. Linear modes of gene expression determined by independent component analysis. Bioinformatics. 2002;18:51–60. doi: 10.1093/bioinformatics/18.1.51. [DOI] [PubMed] [Google Scholar]
- 9.Jutten C. Source separation: from dusk till dawn. Proceedings of the International Workshop on Independent Component Analysis and Blind Source Separation, ICA 2000; 2000. pp. 15–26. [Google Scholar]
- 10.Comon P. Independent component analysis, a new concept? Signal Process. 1994;36:287–314. [Google Scholar]
- 11.Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- 12.Amari SI, Cichocki A, Yang HH. A new learning algorithm for blind source separation. Adv Neural Inf Process Syst. 1996;8:757–763. [Google Scholar]
- 13.Hyvärinen A, Oja E. A fast fixed-point algorithm for independent component analysis. Neural Comput. 1997;9:1483–1492. doi: 10.1142/S0129065700000028. [DOI] [PubMed] [Google Scholar]
- 14.Hyvärinen A. A family of fixed-point algorithms for independent component analysis. Proceedings of the 1997 IEEE International Conference on Acoustics, Speech and Signal Processing; 1997. pp. 3917–3920. [Google Scholar]
- 15.Hyvärinen A. Survey on Independent Component Analysis. Neural Comput Surv. 1999;2:94–128. [Google Scholar]
- 16.Hyvärinen A, Karhunen J, Oja E. Independent Component Analysis. In: Haykin SH, editor. A Volume in the Wiley Series on Adaptive and Learning Systems for Signal Processing, Communications, and Control. John Wiley & Sons, Inc; New York, NY: 2001. pp. 1–481. [Google Scholar]
- 17.Lee SI, Batzoglou S. Application of independent component analysis to microarrays. Genome Biology. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schachtner R, Lutter D, Theis FJ, Lang EW, Schmitz G, Tomé AM, Vilda P Gómez. How to extract marker genes from microarray data sets. Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2007; 2007. pp. 4215–4218. [DOI] [PubMed] [Google Scholar]
- 19.Chiappetta P, Roubaud MC, Torrésani B. Blind source separation and the analysis of microarray data. J Comput Biol. 2004;11:1090–1109. doi: 10.1089/cmb.2004.11.1090. [DOI] [PubMed] [Google Scholar]
- 20.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martoglio AM, Miskin JW, Smith SK, Mackay DJC. A decomposition model to track gene expression signatures: preview on observer-independent classification of ovarian cancer. Bioinformatics. 2002;18:1617–1624. doi: 10.1093/bioinformatics/18.12.1617. [DOI] [PubMed] [Google Scholar]
- 22.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misra J, Schmitt W, Hwang D, Hsiao L, Gullans S, Stephanopoulos G, Stephanopoulos G. Interactive exploration of microarray gene expression patterns in a reduced dimensional space. Genome Res. 2002;12:1112–1120. doi: 10.1101/gr.225302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J-H, Jung H-Y, Lee TW, Lee S-Y. Speech feature extraction using independent component analysis. Proceedings of the 2000 IEEE International Conference on Acoustics, Speech, and Signal Processing; 2000. pp. 1631–1634. [Google Scholar]
- 25.Hansen LK, Larsen J, Kolenda T. Blind detection of independent dynamic components. Proceedings of the 2001 IEEE International Conference on Acoustics, Speech, and Signal Processing; 2001. pp. 3197–3200. [Google Scholar]
- 26.Teschendorff AE, Wang YZ, Barbosa-Morais NL, Brenton JD, Caldas C. A variational bayesian mixture modeling framework for cluster analysis of gene-expression data. Bioinformatics. 2005;21:3025–3033. doi: 10.1093/bioinformatics/bti466. [DOI] [PubMed] [Google Scholar]
- 27.Teschendorff AE, Journee M, Absil PA, Sepulchre R, Caldas C. Elucidating the altered transcriptional programs in breast cancer using independent component analysis. PLOS Comput Biol. 2007;3:e161. doi: 10.1371/journal.pcbi.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amari SI. Natural gradient works efficiently in learning. Neural Comput. 1998;10:251–276. [Google Scholar]
- 29.Bell AJ, Sejnowski TJ. Learning the higher-order structure of a nature sound. Netw: Comput Neural Syst. 1996;7:261–266. doi: 10.1088/0954-898X/7/2/005. [DOI] [PubMed] [Google Scholar]
- 30.Cardoso JF. High-order contrasts for independent component analysis. Neural Comput. 1999;11:157–192. doi: 10.1162/089976699300016863. [DOI] [PubMed] [Google Scholar]
- 31.Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans Neural Netw. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- 32.Lee TW, Girolami M, Sejnowski TJ. Independent component analysis using an extended Infomax algorithm for mixed subgaussian and supergaussian sources. Neural Comput. 1999;11:417–441. doi: 10.1162/089976699300016719. [DOI] [PubMed] [Google Scholar]
- 33.Suri RE. Application of independent component analysis to microarray data. International Conference on Integration of Knowledge Intensive Multi-Agent Systems; 2003; 2003. pp. 375–78. [Google Scholar]
- 34.Najarian K, Kedar A, Paleru R, Darvish A, Zadeh RH. Independent component analysis and scoring function based on protein interactions. Proceedings of the 2004 Second International IEEE Conference on Intelligent Systems; 2004. pp. 595–599. [Google Scholar]
- 35.Lu X-G, Lin Y-P, Yue W, Wang H-J, Zhou S-W. ICA based supervised gene classification of microarray data in yeast functional genome. Proceedings of the Eighth International Conference on High-Performance Computing in Asia-Pacific Region; 2005. pp. 633–638. [Google Scholar]
- 36.Zhu L, Tang C. Microarray sample clustering using independent component analysis. Proceedings of the 2006 IEEE/SMC International Conference on System of Systems Engineering; 2006. pp. 112–117. [Google Scholar]
- 37.Huang DS, Zheng CH. Independent component analysis-based penalized discriminant method for tumor classification using gene expression data. Bioinformatics. 2006;22:1855–1862. doi: 10.1093/bioinformatics/btl190. [DOI] [PubMed] [Google Scholar]
- 38.Liao XJ, Dasgupta N, Lin SM, Carin L. ICA and PLS modeling for functional analysis and drug sensitivity for DNA microarray signals. Proceedings of the 2002 IEEE International Conference on Acoustics, Speech, and Signal Processing; 2002. pp. 3880–3883. [Google Scholar]
- 39.Wang Y, Zhang JY, Huang K, Khan J, Szabo Z. Independent component imaging of disease signatures. Proceedings of 2002 IEEE International Symposium on Biomedical Imaging; 2002. pp. 457–460. [Google Scholar]
- 40.Saidi SA, Holland CM, Kreil DP, MacKay D, Charnock-Jones DS. Independent component analysis of microarray data in the study of endometrial cancer. Oncogene. 2004;23:6677–6683. doi: 10.1038/sj.onc.1207562. [DOI] [PubMed] [Google Scholar]
- 41.Berger JA, Mitra SK, Edgren H. Studying DNA microarray data using independent component analysis. Proceedings of the 2004 First International Symposium on Control, Communications and Signal Processing; 2004. pp. 747–750. [Google Scholar]
- 42.Zhang XW, Yap YL, Wei D, Chen F, Danchin A. Molecular diagnosis of human cancer type by gene expression profiles and independent component analysis. Eur J Hum Genet. 2005;13:1303–1311. doi: 10.1038/sj.ejhg.5201495. [DOI] [PubMed] [Google Scholar]
- 43.Frigyesi A, Veerla S, Lindgren D, Hoglund M. Independent component analysis reveals new and biologically significant structures in micro array data. BMC Bioinformatics. 2006;7:290–301. doi: 10.1186/1471-2105-7-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Journée M, Teschendorff AE, Absil P-A, Tavaré S, Sepulchre R. Geometric optimization methods for the analysis of gene expression data. In: Gorban AN, Kégl B, Wunsch DC, Zinovyev A, editors. Principal Manifolds for Data Visualization and Dimension Reduction. Springer; New York, NY: 2006. pp. 6–27. [Google Scholar]
- 45.Cardoso JF. High-order contrasts for independent component analysis. Neural Comput. 1999;11:157–192. doi: 10.1162/089976699300016863. [DOI] [PubMed] [Google Scholar]
- 46.Learned-Miller EG, Fisher JW., III ICA using spacings estimates of entropy. J Mach Learn Res. 2003;4:1271–1295. [Google Scholar]
- 47.Bach FR, Jordan MI. Kernel independent component analysis. J Mach Learn Res. 2002;3:1–48. [Google Scholar]
- 48.Kim H, Choi S, Bang S-Y. Membership scoring via independent feature subspace analysis for grouping co-expressed genes. Proceedings of the International Joint Conference on Neural Networks; 2003; 2003. pp. 1690–1695. [Google Scholar]
- 49.Wang Y, Zhang JY, Khan J, Clarke R, Gu ZP. Partially-independent component analysis for tissue heterogeneity correction in microarray gene expression analysis. 2003 IEEE 13th Workshop on Neural Networks for Signal Processing; 2003. pp. 23–32. [Google Scholar]
- 50.Miskin JM. PhD thesis. Department of Physics, University of Cambridge; Cambridge, UK: 2002. Ensemble learning for independent component analysis; pp. 1–212. [Google Scholar]
- 51.Zhang JY, Wei L, Wang Y. Computational decomposition of molecular signatures based on blind source separation of non-negative dependent sources with NMF. 2003 IEEE 13th Workshop on Neural Networks for Signal Processing; 2003. pp. 409–418. [Google Scholar]
- 52.Gong T, Zhu Y, Xuan JH, Li H, Clarke R, Hoffman EP, Wang Y. Latent variable and nICA modeling of pathway gene module composite. 28th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2006. pp. 5872–5875. [DOI] [PubMed] [Google Scholar]
- 53.Lin CJ. On the convergence of multiplicative update algorithms for nonnegative matrix factorization. IEEE Trans Neural Netw. 2007;18:1589–1596. doi: 10.1109/TNN.2010.2076831. [DOI] [PubMed] [Google Scholar]
- 54.Lee DD, Seung HS. Advances in Neural Information Processing Systems. Vol. 13. MIT Press; Cambridge, MA: 2001. Algorithms for non-negative matrix factorization; pp. 556–562. [Google Scholar]
- 55.Chiappetta P, Roubaud MC, Torrésani B. Blind source separation and the analysis of microarray data. J Comput Biol. 2004;11:1090–1109. doi: 10.1089/cmb.2004.11.1090. [DOI] [PubMed] [Google Scholar]
- 56.Himberg J, Hyvärinen A, Esposito F. Validating the independent components of neuroimaging time-series via clustering and visualization. Neuroimage. 2004;22:1214–1222. doi: 10.1016/j.neuroimage.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 57.Kong W, Zhou Y, Yang J. An extended speech denoising method using GGM-based ICA feature extraction. Lecture Notes Comput Sci. 2004;3287:296–302. [Google Scholar]
- 58.Yang B, Kong W. Efficient feature extraction and de-noising method for Chinese speech signals using GGM-based ICA. Lecture Notes Comput Sci. 2005;3773:925–932. [Google Scholar]
- 59.Kong W, Yang B. Higher-order feature extraction of non-gaussian acoustic signals using GGM-based ICA. Lecture Notes Comput Sci. 2006;3972:712–718. [Google Scholar]
- 60.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 61.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 62.Savageau MA. Design principles for elementary gene circuits: elements, methods, and examples. Chaos. 2001;11:142–159. doi: 10.1063/1.1349892. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X, Yan W, Zhao X, Shao H. Nonlinear on-line process monitoring and fault detection based on kernel ICA. International Conference on Information and Automation; 2006; 2006. pp. 222–227. [Google Scholar]
- 64.Bai L, Xu A, Guo P, Jia Y. Kernel ICA feature extraction for spectral recognition of celestial objects. IEEE International Conference on Systems, Man and Cybernetics; 2006; 2006. pp. 3922–3926. [Google Scholar]
- 65.Almeida MSC, Almeida LB. Separating nonlinear image mixtures using a physical model trained with ICA. Proceedings of the 2006 16th IEEE Signal Processing Society Workshop on Machine Learning for Signal Processing; 2006. pp. 65–70. [Google Scholar]
- 66.Zhang Y, Li H. Linear and nonlinear ICA based on mutual information. International Symposium on Intelligent Signal Processing and Communication Systems; 2007; 2007. pp. 770–773. [Google Scholar]