Abstract
Shikimic acid can be transformed into monovalent and multivalent glycomimetics that target different members of the C-type lectin class, including DC-SIGN, a dendritic cell lectin that facilitates HIV transmission.
Carbohydrates act in conjunction with carbohydrate-binding proteins (lectins) to govern numerous cellular processes, including cell adhesion, recognition, and signaling.1 Despite the importance of lectins, ligands that can be used to interrogate or mitigate their function are scarce.2–4 In principle, carbohydrates themselves can serve as probes, but they have liabilities. First, they tend to bind with low affinity. Second, optimizing carbohydrate leads can be difficult because it typically requires the production of complex oligosaccharide analogs via multistep chemoenzymatic or chemical synthesis routes. Finally, since the specificity of lectins can overlap, natural oligosaccharides often bind multiple lectins, thereby complicating their use as probes of a target lectin in the presence of others.5 Non-carbohydrate antagonists are an appealing alternative, but few have been reported.2–4 Although screens of large compound libraries are beginning to yield inhibitors,6–10 general approaches to block lectins are lacking. Here, we describe a privileged glycomimetic scaffold that can give rise to effective and selective inhibitors of the C-type lectin class. Moreover, we show that such glycomimetics can be incorporated into multivalent displays to generate potent inhibitors. To date, the targeting of lectins with multivalent glycomimetics is underexplored;11–13 our data indicate it can serve as a powerful strategy.
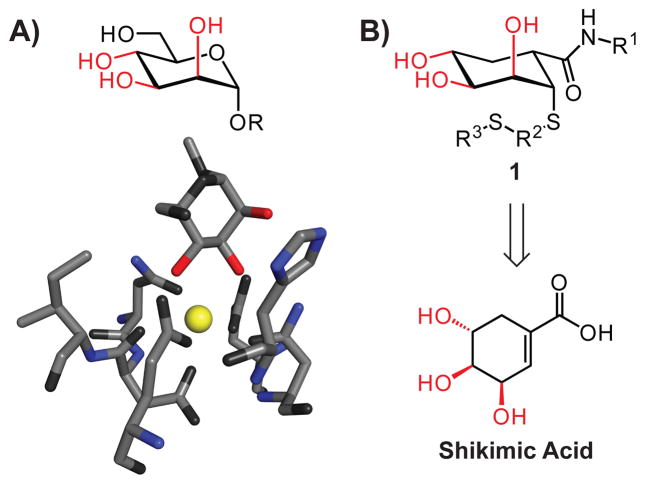
C-type lectins are a large class of proteins that are integral to immune system function; they mediate pathogen recognition and processing as well as cell–cell interactions.14 C-type lectins, which are named for their dependence on calcium ions for carbohydrate complexation, often bind mannosides. In these complexes, the 2-, 3- and 4-hydroxyl groups of the sugar contribute to binding (Fig. 1A).15 We therefore hypothesized that scaffold 1, which mimics the arrangement of the D-mannose 2-, 3-, and 4-hydroxyl groups (Fig. 1B), could afford glycomimetic probes of carbohydrate function. We demonstrated that the natural product shikimic acid16 could be transformed into compounds with the necessary arrangement of hydroxyl groups.18 From the resulting collections, inhibitors were identified of a prototype C-type lectin, mannose-binding protein A (MBP-A).
Fig. 1.
Strategy for inhibitor design. A) D-Mannose (top) and a substructure of the binding site of a complex of mannose and MBP-A (bottom) (PDB accession code 1kwy17). The hydroxyl groups necessary for lectin recognition and binding are shown in red, Ca2+ is yellow. B) Glycomimetics 1 resemble mannose and can be obtained from the natural product shikimic acid.
A key feature of the aforementioned approach is that it has the potential to be general. Specifically, compounds represented by 1 might have the requisite attributes to bind C-type lectins other than MBP-A. One attractive target for testing this possibility is dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN). DC-SIGN resides on the surface of dendritic cells, which are critical antigen-presenting cells.19 DC-SIGN is involved in pathogen recognition and facilitates dendritic cell–T cell interactions, but it is its involvement in the dissemination of infectious human pathogens that led us to seek inhibitors. DC-SIGN can interact with viruses, such as HIV-1 or Ebola virus, and bacterial species, such as Mycobacterium tuberculosis, to facilitate infection.20 Compounds that bind DC-SIGN and thereby prevent it from interacting with pathogens could serve as therapeutic leads. Moreover, several pathogens that bind DC-SIGN subvert normal immune system function, and DC-SIGN ligands could probe the underlying mechanisms. MBP-A and DC-SIGN are both mannose-binding C-type lectins; therefore, our objective—to generate agents that block DC-SIGN selectively—serves as a challenging test of our design strategy.
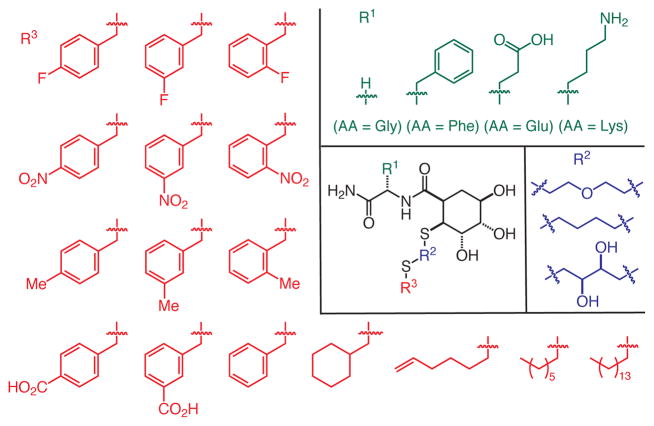
DC-SIGN binds weakly to monosaccharide ligands such as N-acetyl mannosamine (ManNAc, Kd = 8.7 mM) and L-fucose (Kd = 6.7 mM).21 The affinity for oligosaccharides is marginally higher (Kd = 0.21 mM for Man9GlcNAc).21 We hypothesized that our strategy could yield effective glycomimetics with improved activity. To this end, we used solid-phase synthesis to assemble a collection of putative mannose mimics that vary at three positions (Fig. 2).
Fig. 2.
Building blocks used in the synthesis of the glycomimetic library targeting DC-SIGN.
Aside from the triol-substituted 6-membered ring that we anticipate would mimic mannose, the glycomimetic scaffold differs structurally from the natural ligands. We envisioned substituents at the points of variation could endow ligands with lectin affinity and specificity. Accordingly, we wanted to test a range of functionality at each variable position. Our synthetic approach was designed to utilize building blocks that are readily accessible (e.g., either commercially available or synthesized in a few steps). For example, we varied the amino acid substituent to explore how changes in R1 influence binding. Glycine serves as a small, flexible amino acid, while phenylalanine is larger, more hydrophobic, and the aryl group can engage in a range of interactions. Glutamic acid and lysine were chosen to test the influence of anionic or cationic substituents, respectively. The R3 substituent was varied using a collection of alkylating agents. We tested some aliphatic R3 groups, but we focused on benzyl substituents because aromatic side chains often line carbohydrate binding sites.22, 23 Accordingly, aromatic rings with a range of functional groups were introduced, including those bearing halides, hydrogen bond donors or acceptors, and electron donating or withdrawing groups. The dithiol linkers (R2) used are commercially available and capable of positioning the R3 substituents near the carbohydrate or secondary binding sites.
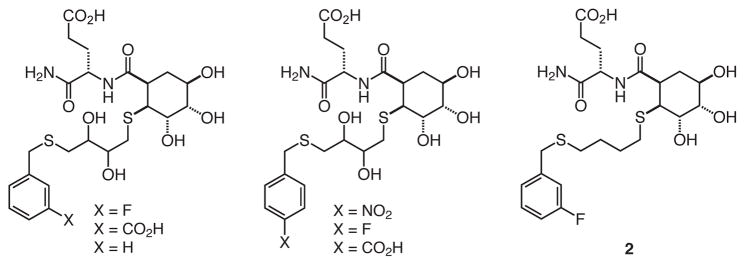
These building blocks were employed18 to synthesize 192 compounds. The compounds were cleaved from resin and screened without further purification to get an estimate of their activity. The screen involved a fluorescence-based high-throughput competition assay that assessed the ability of compounds to compete against immobilized mannan for binding to the fluorophore-labeled tetrameric extracellular domain of DC-SIGN (DC-SIGN/ECD).10, 21 In this way, we identified several compounds that block DC-SIGN (Fig. 3). Trends in the inhibition data were readily apparent. The identity of the R1 moiety affected binding most significantly; all of the hits contain glutamic acid at this position, but compounds with lysine at this position were largely ineffective. At the R2 position, the DTT-derived linker was most prevalent, followed by the butanedithiol-derived linker. At the R3 position, there was a preference for aromatic groups with electron withdrawing substituents. To confirm the validity of the inhibitory values upon which the trends were based, we purified several library members and determined their IC50 values. The relative values parallel those determined from the initial screen.
Fig. 3.
The most potent hits identified from library screen.
Based on its activity in the initial screen, compound 2 (Fig. 3), was chosen for further characterization. To determine its IC50 value, we measured its ability to compete with probe, a fluorescein-labeled bovine serum albumin displaying 20–25 copies of mannose, for the immobilized extracellular domain of DC-SIGN.10 The IC50 value of 2 (3.2 ± 0.6 mM) is superior to that of N-acetylmannosamine (IC50 = 11.2 ± 0.7 mM). The MBP-A inhibitors identified previously were no more potent than the monosaccharide ligand α-methyl-D-mannopyranoside.18 Thus, our data indicate that this strategy can give rise to DC-SIGN inhibitors that are superior to known monosaccharide ligands. The results, therefore, provide a starting point for optimizing ligand potency. Most significantly, these results support the generality of our design—the same scaffold can be used to generate ligands for two lectins with different binding sites.
We envisioned augmenting the potency of our selective glycomimetic through multivalency. DC-SIGN is tetrameric, and multiple copies of the protein are displayed on the dendritic cell surface.21 Its ability to detect antigens likely depends upon multivalent binding. As with other protein-carbohydrate interactions,24–27 DC-SIGN could exploit multivalent binding to achieve the necessary functional affinity.13, 28–31 The vast majority of multivalent lectin inhibitors consist of scaffolds decorated with the mono-or oligosaccharide binding epitopes recognized by the target lectin.32 In these cases, selectivity is a concern because the carbohydrate epitope can interact with many lectins, but glycomimetics can display higher selectivity for their target lectin.5 5For this reason, multivalent glycomimetics may serve as useful probes,13 but few have been described.
Our strategy for multivalent ligand synthesis involves the application of ring-opening metathesis polymerization (ROMP).5 This method can yield polymers of defined lengths that function as highly effective biological probes.33–38 Additionally, ROMP-generated polymers can promote receptor clustering,39 and this property could facilitate investigations of DC-SIGN-mediated internalization. Finally, ROMP can give rise to polymers of defined length with polydispersity indices (PDIs) near unity. These values are comparable to those obtained for the functionalization of higher generation dendrimers. In summary, ROMP affords products with valuable attributes.
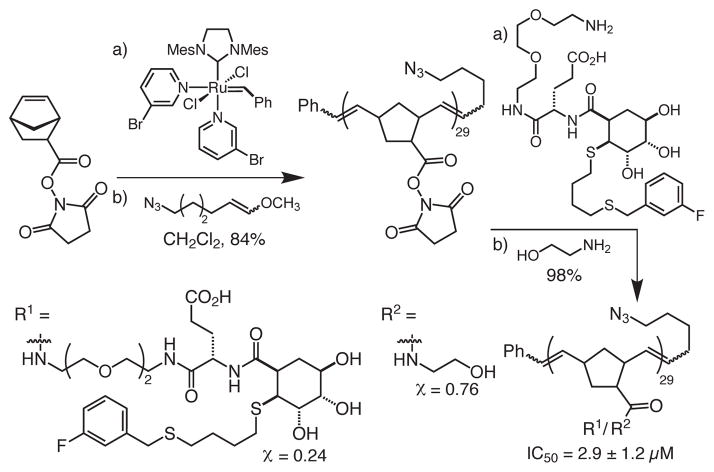
The multivalent glycomimetics were generated from polymer backbones bearing succinimide esters, which could be modified post-polymerization40 to append the glycomimetic epitopes (Scheme 1). To generate polymers that bind avidly to DC-SIGN, the length of the polymer must be sufficient for it to engage in multivalent binding, i.e., either occupy multiple binding sites within the tetrameric DC-SIGN or cluster multiple copies of DC-SIGN on the cell surface. We used the structure of the monomeric DC-SIGN carbohydrate-recognition domain (pdb accession code 1s1441) to estimate the distances between monomers of DC-SIGN. The width of the DC-SIGN CRD is roughly 40 Å. When extended, we estimate that each monomer within a ROMP-derived polymer spans approximately 5 Å.42 For polymers to engage multiple copies of DC-SIGN, our analysis indicates that they must have a minimum length of 40 Å. We generated polymers with a length of 25 monomeric units, as these should be capable of multivalent binding. Accordingly, the degree of polymerization (29) was controlled by using a ratio of ruthenium initiator to norbornene monomer of 1:25.43
Scheme 1.
Synthetic route to ROMP-derived multivalent scaffold 3, with a degree of polymerization of 29. The ratio of ethanolamine spacer to DC-SIGN inhibitor is approximately 1:3.
The ratio of glycomimetic to ethanolamine was chosen to balance epitope density, which has been shown to be important for biological function,44 with water solubility. The glycomimetic was conjugated to the polymer through reaction of the amine-terminated ethylene glycol linker with the succinimidyl ester moiety on each backbone unit. This process afforded a multivalent inhibitor displaying approximately seven glycomimetics per polymer. Like the corresponding monomer, the polymer also inhibits DC-SIGN/ECD binding; thus, the linker did not impair ligand binding. Intriguingly, the IC50 value for the polymer is 2.9 ± 1.2 μM, which indicates the polymer is 1000-fold more potent than the monomeric inhibitor. These results illustrate the utility of the glycomimetic strategy for generating potent multivalent ligands for lectins.
In summary, the shikimic acid-derived glycomimetic scaffold can be used to generate ligands for C-type lectins. This approach can yield inhibitors that are selective and potent. Both MBP-A and DC-SIGN possess significantly different binding sites, revealing the general utility of our approach for targeting diverse lectins. Additionally, our conversion of a lead glycomimetic into a multivalent ligand yields a highly potent non-carbohydrate inhibitor of DC-SIGN. Moreover, it demonstrates that multivalency can be exploited to generate highly effective non-carbohydrate inhibitors of lectins. Thus, the strategy we have outlined will be useful in developing probes not only of DC-SIGN function but also of the many other C-type lectins whose function and dysfunction are critical for human health.
Supplementary Material
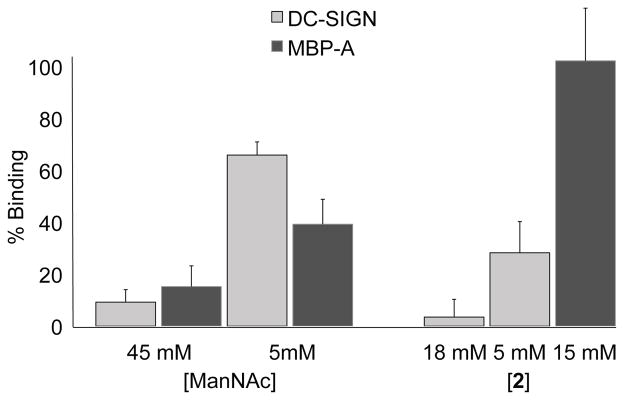
Fig. 4.
Percent probe binding to DC-SIGN/ECD and MBP-A in the presence of ManNAc or 2 determined in fluorescence assay.
Acknowledgments
This research was supported by the NIGMS (R01 GM049975). We thank M. J. Borrok for preliminary studies, J. Grim for helpful conversations, Prof. J. Frost and Dr. K. Draths for a donation of shikimic acid, and Prof. K. Drickamer for supplying the DC-SIGN expression vector. KCAG was supported by a National Science Foundation Graduate Research Fellowship and the NIH Chemistry-Biology Interface Training Grant (T32 GM008505), EEC was supported by the NIH Biotechnology Training Program (T32GM08349), and KW was a Hilldale Undergraduate Research Fellow.
Footnotes
Electronic Supplementary Information (ESI) available: Additional figures, including binding curves, and detailed biochemical and synthetic experimental procedures.
Notes and references
- 1.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst B, Magnani JL. Nat Rev Drug Discov. 2009;8:661–677. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardi A, Cheshev P. Chem Eur J. 2008;14:7434–7441. doi: 10.1002/chem.200800597. [DOI] [PubMed] [Google Scholar]
- 4.Kaila N, Thomas BE. Med Res Rev. 2002;22:566–601. doi: 10.1002/med.10018. [DOI] [PubMed] [Google Scholar]
- 5.Sears P, Wong CH. Angew Chem Int Ed. 1999;38:2300–2324. [PubMed] [Google Scholar]
- 6.Slee DH, Romano SJ, Yu J, Nguyen TN, John JK, Raheja NK, Axe FU, Jones TK, Ripka WC. J Med Chem. 2001;44:2094–2107. doi: 10.1021/jm000508c. [DOI] [PubMed] [Google Scholar]
- 7.Ohta S, Inujima Y, Abe M, Uosaki Y, Sato S, Miki I. Inflamm Res. 2001;50:544–551. doi: 10.1007/PL00000232. [DOI] [PubMed] [Google Scholar]
- 8.Kaila N, Janz K, DeBernardo S, Bedard PW, Camphausen RT, Tam S, Tsao DHH, Keith JC, Nickerson-Nutter C, Shilling A, Young-Sciame R, Wang Q. J Med Chem. 2007;50:21–39. doi: 10.1021/jm0602256. [DOI] [PubMed] [Google Scholar]
- 9.Schon MP, Krahn T, Schon M, Rodriguez ML, Antonicek H, Schultz JE, Ludwig RJ, Zollner TM, Bischoff E, Bremm KD, Schramm M, Henninger K, Kaufmann R, Gollnick HP, Parker CM, Boehncke WH. Nat Med. 2002;8:366–372. doi: 10.1038/nm0402-366. [DOI] [PubMed] [Google Scholar]
- 10.Borrok MJ, Kiessling LL. J Am Chem Soc. 2007;129:12780–12785. doi: 10.1021/ja072944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angyalosi G, Grandjean C, Lamirand M, Auriault C, Gras-Masse H, Melnyk O. Bioorg Med Chem Lett. 2002;12:2723–2727. doi: 10.1016/s0960-894x(02)00486-9. [DOI] [PubMed] [Google Scholar]
- 12.Shamay Y, Paulin D, Ashkenasy G, David A. J Med Chem. 2009;52:5906–5915. doi: 10.1021/jm900308r. [DOI] [PubMed] [Google Scholar]
- 13.Sattin S, Daghetti A, Thepaut M, Berzi A, Sanchez-Navarro M, Tabarani G, Rojo J, Fieschi F, Clerici M, Bernardi A. ACS Chem Biol. 2010;5:301–312. doi: 10.1021/cb900216e. [DOI] [PubMed] [Google Scholar]
- 14.Cambi A, Koopman M, Figdor CG. Cell Microbiol. 2005;7:481–488. doi: 10.1111/j.1462-5822.2005.00506.x. [DOI] [PubMed] [Google Scholar]
- 15.Weis WI, Taylor ME, Drickamer K. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 16.Knop DR, Draths KM, Chandran SS, Barker JL, von Daeniken R, Weber W, Frost JW. J Am Chem Soc. 2001;123:10173–10182. doi: 10.1021/ja0109444. [DOI] [PubMed] [Google Scholar]
- 17.Ng KKS, Kolatkar AR, Park-Snyder S, Feinberg H, Clark DA, Drickamer K, Weis WI. J Biol Chem. 2002;277:16088–16095. doi: 10.1074/jbc.M200493200. [DOI] [PubMed] [Google Scholar]
- 18.Schuster MC, Mann DA, Buchholz TJ, Johnson KM, Thomas WD, Kiessling LL. Org Lett. 2003;5:1407–1410. doi: 10.1021/ol0340383. [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek TBH, van Vliet SJ, Engering A, t Hart BA, van Kooyk Y. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 20.van Kooyk Y, Geijtenbeek TBH. Nat Rev Immunol. 2003;3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell DA, Fadden AJ, Drickamer K. J Biol Chem. 2001;276:28939–28945. doi: 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- 22.Weis WI, Drickamer K. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 23.Kiessling LL, Pohl NL. Chem Biol. 1996;3:71–77. doi: 10.1016/s1074-5521(96)90280-x. [DOI] [PubMed] [Google Scholar]
- 24.Lee YC, Lee RT. Acc Chem Res. 1995;28:321–327. [Google Scholar]
- 25.Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Collins BE, Paulson JC. Curr Opin Chem Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Kiessling LL, Splain RA. Annu Rev Biochem. 2010;79:619–653. doi: 10.1146/annurev.biochem.77.070606.100917. [DOI] [PubMed] [Google Scholar]
- 28.Rojo J, Delgado R. J Antimicrob Chemother. 2004;54:579–581. doi: 10.1093/jac/dkh399. [DOI] [PubMed] [Google Scholar]
- 29.Adams EW, Ratner DM, Bokesch HR, McMahon JB, O’Keefe BR, Seeberger PH. Chem Biol. 2004;11:875–881. doi: 10.1016/j.chembiol.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Wang SK, Liang PH, Astronomo RD, Hsu TL, Hsieh SL, Burton DR, Wong CH. Proc Natl Acad Sci U S A. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Avila O, Bedoya LM, Marradi M, Clavel C, Alcami J, Penades S. Chem Bio Chem. 2009;10:1806–1809. doi: 10.1002/cbic.200900294. [DOI] [PubMed] [Google Scholar]
- 32.Kiessling LL, Strong LE, Gestwicki JE. Annu Rep Med Chem. 2000;35:321–330. [Google Scholar]
- 33.Courtney AH, Puffer EB, Pontrello JK, Yang ZQ, Kiessling LL. Proc Natl Acad Sci U S A. 2009;106:2500–2505. doi: 10.1073/pnas.0807207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baessler KA, Lee Y, Sampson NS. ACS Chem Biol. 2009;4:357–366. doi: 10.1021/cb900013d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borrok MJ, Kolonko EM, Kiessling LL. ACS Chem Biol. 2008;3:101–109. doi: 10.1021/cb700211s. [DOI] [PubMed] [Google Scholar]
- 36.Kolonko EM, Kiessling LL. J Am Chem Soc. 2008;130:5626–5627. doi: 10.1021/ja8001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rawat M, Gama CI, Matson JB, Hsieh-Wilson LC. J Am Chem Soc. 2008;130:2959–2961. doi: 10.1021/ja709993p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith D, Pentzer EB, Nguyen ST. Polym Rev. 2007;47:419–459. [Google Scholar]
- 39.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 40.Strong LE, Kiessling LL. J Am Chem Soc. 1999;121:6193–6196. [Google Scholar]
- 41.Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI, Drickamer K. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- 42.Kanai M, Mortell KH, Kiessling LL. J Am Chem Soc. 1997;119:9931–9932. [Google Scholar]
- 43.Choi TL, Grubbs RH. Angew Chem Int Ed. 2003;42:1743–1746. doi: 10.1002/anie.200250632. [DOI] [PubMed] [Google Scholar]
- 44.Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. J Am Chem Soc. 2002;124:1615–1619. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.