Abstract
Regulated intramembrane proteolysis (RIP) is a conserved mechanism that regulates signal transduction across the membrane by recruiting membrane-bound proteases to cleave membrane-spanning regulatory proteins. As the first identified protease that performs RIP, the metalloprotease site-2 protease (S2P) has received extensive study during the past decade, and an increasing number of S2P-like proteases have been identified and studied in different organisms; however, some of their substrates and the related S1Ps remain elusive. Here, we review recent research on S2P cascades, including human S2P, E. coli RseP, B. subtilis SpoIVFB and the newly identified S2P homologs. We also discuss the variation and conservation of characterized S2P cascades. The conserved catalytic motif of S2P and prevalence of amino acids of low helical propensity in the transmembrane segments of the substrates suggest a conserved catalytic conformation and mechanism within the S2P family. The review also sheds light on future research on S2P cascades.
Keywords: intramembrane, proteolysis, site-1 protease, site-2 protease, substrate
Introduction
Regulated intramembrane proteolysis (RIP) is a conserved mechanism to regulate transmembrane signaling. It is carried out by different types of intramembranously cleaving proteases (I-CLiPs), which can be grouped into three or four families on the basis of their catalytic mechanisms, catalytic properties, and substrates. I-CLiPs have been identified in all kingdoms of life except virus.1 Through RIP, they activate or inactivate transcription factors or signaling peptides to coordinate the responses between cellular compartments. These enzymes contain multiple membrane-spanning segments, within which the conserved catalytic residues often reside. They specialize in cleaving peptide bonds that are normally embedded in hydrophobic membranes. One family of I-CLiPs consists of serine proteases, designated as Rhomboid proteases, example as GlpG.2,3 Another family includes two types of aspartyl proteases: Signal peptide peptidases, example as GXGD and Presenilin1 as the catalytic subunit of γ-secretase.4,5 Another includes a large family of metalloproteases, the S2P proteases.6
In this review, we focus on the S2P family, starting with the regulation of S2P, SpoIVFB, RseP and the recently identified S2P family members, then discussing the common properties of S2P cascades and finally touching on promising clues for future investigation.
S2P Cascades in Transmembrane Signaling Processes
S2P in human
As the first member of this family, site-2 protease (S2P) was first identified in humans. It is involved in the feedback regulation of sterol and fatty acid synthesis and uptake by controlling the activity of transcription factors, SREBPs (Sterol regulatory element binding proteins).7–9 Proteins involved in the S2P pathway include site 1 protease (S1P), site 2 protease (S2P), their substrates and other regulatory proteins [Fig. 1(A)]. Before activation, SREBPs reside in the endoplasmic reticulum (ER) membrane while S1P and S2P are located in the Golgi apparatus.7,10,11 This spatial separation prevents SREBPs from unregulated degradation by S1P and S2P. SREBPs are membrane-bound transcription factors responsible for sterol and fatty acid biosynthesis and uptake.9 They are known to directly promote transcription of more than 30 genes required for uptake and synthesis of cholesterol, triglycerides, fatty acids, and phospholipids.12 The N-terminal domain of SREBP is a transcription factor of basic helix-loop-helix leucine zipper (bHLH-Zip). The C-terminus forms a tight complex with the WD repeats domain of Scap (SREBP cleavage-activating protein), which functions as the sterol sensor in this system.13,14 SREBP contains two transmembrane sequences and a short luminal loop.7
Figure 1.
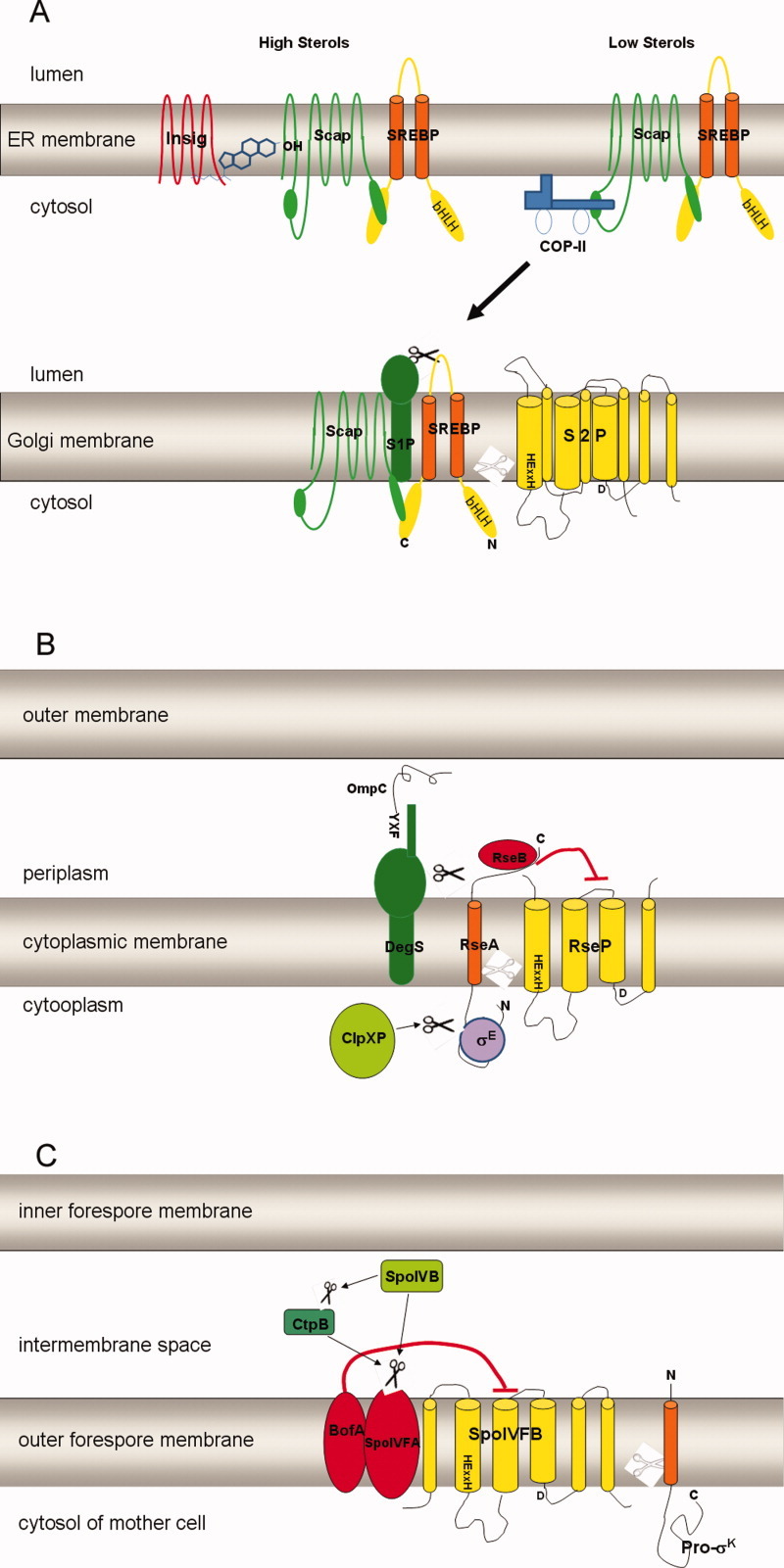
Schematic drawings showing the S2P cascades of human S2P, E. coli RseP and B. subtilis SpoIVFB. Substrates are shown in orange, Site-2 proteases are shown in yellow. The white scissors represent the cleavage by Site-2 proteases while the black scissors represent the cleavage by Site-1 proteases or other proteases. The metal chelating motif that includes HExxH and D is shown in black. A: Under low levels of sterols, human SREBP is transported from the ER membrane to the Golgi to be sequentially cleaved by S1P and S2P. B: In E. coli, RseA is cleaved sequentially by DegS and RseP. C: After activation by SpoIVB, SpoIVFB cleaves pro-σk to release mature σk.
In cells with high sterol, Scap sequesters SREBPs and binds to cholesterol in the ER membrane in such a way that it assumes a conformation that is propitious to bind Insig (insulin-induced gene), a small ER membrane-resident protein.15–17 This binding retains the SREBP-Scap complex in the ER by preventing interaction of Scap with the COPII vesicle-formation proteins at the MELADL motif.17–21 As a consequence, SERBPs are protected from activation by S1P and S2P resident in Golgi and the transcription of its target genes declines.
In sterol depleted cells, conformation changes of Scap and Insig disrupt their binding and the SREBP-Scap complex is sorted in COPII-coated transport vesicles to the Golgi.18 In the Golgi, two sequential proteolytic cleavage events by S1P and S2P finally release the N terminal transcription factor domain of SREBPs from the Golgi membrane.19 S1P, a serine proteases, cleaves after a leucine residue at the consensus sequence RxxL in the luminal loop of SREBPs.22,23 Then the zinc metalloprotease S2P cleaves at a Leu-Cys bond predicted to lie within the membrane.6 This two-stage proteolysis of SREBPs discharges the mature N-terminal bHLH-Zip domain to the nucleus to bind the sterol regulatory element (SRE) sequences in the promoters of target genes, therefore activating transcription of genes involves in sterol and fatty acid synthesis and uptake.7,9 The resultant increase of cholesterol will later feed back to inhibit activation of SREBP through Scap. Through the S2P cascade, the cholesterol feedback pathway in humans is stringently regulated. Cholesterol content in the ER membrane triggers this regulatory machine by mediating the interaction of Scap with Insigs in a switch-like manner.21,24 Thus S2P plays a crucial role in regulating the feedback of cholesterol biosynthesis.
Intriguingly, SREBP is not the only substrate for the S2P pathway. S1P and S2P function in tandem to activate the stress response transcription factor ATF6 in response to unfolded protein in a manner similar to SREBP.25 CREBH was identified as a liver-specific transcription factor that is cleaved sequentially by S1P and S2P upon ER stress and is required to activate expression of acute phase response genes.26 OASIS (old astrocyte specifically induced substance), a transcription factor anchored in ER membrane, modulates the unfolded protein response in astrocytes through the cleavage by S1P and S2P.27,28 Recent results show that Ichthyosis follicularis with atrichia and photophobia (IFAP syndrome) is caused by functional deficiency of S2P (MBTPS2) through uncertain substrates.29 The diversity of substrates indicates that S2P is a pivotal protease and participates in different signaling transduction pathways.
RseP in Escherichia coli
DegS and RseP in E. coli are homologs of human S1P and S2P. DegS is a serine protease anchored to the periplasmic face of the inner membrane and RseP is a multiple transmembrane protease with canonical HEXXH and NPDG motifs and located in the inner membrane.30–34 They successively process RseA, the membrane-bound anti-σE protein. Early studies demonstrated that σE mediates extracytoplasmic stress responses by controlling the expression of genes that facilitate refolding or degradation of misfolded periplasmic proteins.35 Under normal growth conditions, σE is sequestered by RseA through forming a tight complex with the N-terminal cytoplasmic domain of RseA.36–38 When cells are subjected to extracytoplasmic stress which result in unassembled or misfolded proteins signals, RseA is degraded by successive proteolytic events to release σE [Fig. 1(B)]. Misfolded outer membrane porins (OMPs such as ompA or ompC) can interact with the PDZ domain of DegS and initiate the first cleavage of RseA by DegS at its periplasmic domain (Val148-Ser149).33,34,39 This initial, signal-sensing cleavage step is rate-limiting,40 and stimulates the second cleavage of RseA1-148 by RseP. RseP cleaves between Ala108 and Cys109, well inside the predicted TM sequence of RseA.41 Then RseA1–108, the cytoplasmic fragment of RseA, is further degraded by ClpXP or other ATP-dependent proteases to ultimately release σE in cytoplasm.40 Active σE will bind appropriate RNA polymerase complex to modulate expression of target genes.
Many interesting and important studies of the regulation mechanisms have shown complexity and precision of its regulation in nature. As a periplasmic stress sensor, DegS is kept in an inactive state by its PDZ domain under normal conditions.42 When protein folding is compromised in the periplasm, the C-terminus of OMPs bind to the PDZ domains of the trimeric DegS protease and activate its cleavage on RseA.39,43 Crystallographic and biochemical analysis show that DegS is an allosteric enzyme.44 OMPs binding shifts the equilibrium from a nonfunctional state to the functional proteolytic conformation. The ancillary regulatory protein RseB is also involved. Initial studies found that RseP variants with the periplasmic PDZ domain deleted or mutated allowed unregulated cleavage of RseA and consequent σE activation.45 This indicated that the PDZ domain of RseP might be an inhibitor of proteolytic activity.46 A Gln-rich region in the periplasmic domain of RseA was found to be required for the avoidance of the RseP-mediated proteolysis in the absence of site-1 cleavage.45 At the same time, it was found that RseP can cleave intact RseA in a ΔRseB strain and RseB binds to the periplasmic region of RseA strongly with 1:1 stoichiometry.47 These results suggest RseB plays a vital role in inhibiting RseP cleavage of intact RseA; and RseB, the PDZ domain of RseP and the Gln-rich domain of RseA may together exert an inhibitory function. In the ΔRseB strain, RseP cleavage of RseA is impaired by overexpression DegS, which indicates that RseB and DegS each independently inhibit RseP cleavage of intact RseA.47 In vitro RseB inhibits proteolysis by DegS by binding tightly to the periplasmic domain of RseA.36,48 The inhibitory effect may be due to steric hindrance because RseB prevents the substrate from accessing to the active site of DegS. Though RseB inhibit DegS cleavage of RseA in vitro, DegS can cleave RseA in vivo under similar conditions. Maybe other factors interact with RseB to relieve its inhibitory effect.
SpoIVFB in Bacillus subtilis
SpoIVFB from the gram-positive bacterium B. subtilis is another founding member of the S2P metalloprotease family. It is involved in activation of the crucial membrane-associated transcription factors σK during sporulation.49,50 SpoIVFB processes pro-σK to mature and active σK by proteolytic removal of an N-terminal extension of 20 amino acids [Fig. 1(C)].49,51 Sporulation of B. subtilis involves the formation of a polar septum that asymmetrically partitions the cell into a large mother cell and a small forespore. The two cells initially lie side by side, but later the forespore is engulfed by the mother cell to create a cell within a cell.52 As a result of engulfment, the forespore is surrounded by two membranes, the inner forespore membrane (IFM) and the outer forespore membrane (OFM). Gene expression is coordinated between these two chambers in part by a series of sigma factors to drive a series of morphological changes that culminate with lysis of the mother cell to release a dormant spore.50
As inactive precursor, pro-σK associates with the OFM with its N-terminal transmembrane segment (amino acids 1 to 27).53 Proteolytic cleavage by SpoIVFB releases the C-terminal σK into the mother cell for directing transcription of genes involved in spore cortex and coat synthesis.54 Although σK is in charge of the expression of genes in the mother cell, its activation is mediated by signaling proteins from both the mother cell and the forespore cell. Before sporulation, the transmembrane metalloprotease SpoIVFB is held inactive by two other integral-membrane proteins, SpoIVFA and BofA.51,55,56 BofA, SpoIVFA and SpoIVFB are all synthesized in the mother cell. They form a complex in OFM. SpoIVFA plays an essential role in the assembly and localization of the complex while BfoA directly inhibits the activity of SpoIVFB on pro-σK processing probably by supplying a fourth zinc ligand.55,56 The inhibition of SpoIVFB by BofA and SpoIVFA is relieved by two serine proteases, SpoIVB and CtpB.57 In the interspace between IFM and OFM, the signaling serine protease SpoIVB from forespore triggers pro-σK processing by cleaving the extracellular domain of SpoIVFA at multiple sites.58 In vitro, these cleavages do not disrupt the interactions between SpoIVFA, SpoIVFB, and BofA, suggesting that SpoIVB-dependent activation of the processing enzyme results from a conformational change in this complex.59 CtpB, a second serine protease from the mother cell, also triggers pro-σK processing by cleaving SpoIVFA or BofA.57,59,60 From the in vitro data that CtpB appeared to cleave BofA near its C-terminus upon coexpression in E. coli and purified CtpB degraded BofA, a three-step proteolytic cascade was proposed, in which SpoIVB first cleaves SpoIVFA, CtpB then cleaves BofA and finally SpoIVFB cleaves pro-σK.57 Later Campo and Rudner59 proposed another model in which CtpB triggers pro-σK processing by cleaving SpoIVFA. They demonstrated that when SpoIVB is unable to cleave SpoIVFA, it can still activate pro-σK processing through CtpB. Thus a model was proposed in which SpoIVB regulates intramembrane proteolysis through two proteolytic pathways: directly by cleaving the extracellular domain of SpoIVFA and indirectly by cleaving and activating CptB to process SpoIVFA, both of which converge on the same regulator SpoIVFA and release SpoIVFB for processing of pro-σK.
SpoIVFB is unique among S2P family members in that it does not contain PDZ domain and there is no evidence to suggest that pro-σK is cleaved before SpoIVFB. However, regulation of SpoIVFB activity is achieved by a proteolytic cascade involving serine proteases, which are used as S1P protease in other S2P cascades. SpoIVB and CtpB are not involved in Site-1 cleavage of substrate but are required to relieve the inhibition of SpoIVFB by SpoIVFA and BofA.58–60 Recently, Kroos and coworkers61 constructed a biochemical system to validate the direct relationship between spoIVFB and pro-σK. Their results demonstrated that spoIVFB does cleave pro-σK in an ATP-dependent manner.61 This is special in characterized S2P pathways, though more evidence is needed to clarify its regulation mechanism.
RasP in Bacillus subtilis
Another S2P homolog in B. subtilis is RasP. B. subtilis codes for several extracytoplasmic function (ECF) sigma factors including σW. As for σE in E. coli, activity of σW is negatively controlled by a membrane-bound anti-sigma factor, RsiW.62 The σW regulon is induced by different stresses, such as alkaline shock, salt shock, phage infection, and certain antibiotics that damage the cell envelope.63–65 RsiW is cleaved at site-1 and site-2 by PrsW (YpdC) and RasP (YluC), respectively,66–68 and is then further degraded by cytoplasmic Clp peptidases.69 This proteolysis cascade liberates sequestered σW into the cytoplasm to exert its function by ultimately binding to RNA polymerase. Though the molecular mechanism of the initialization and regulation remains elusive at present, a simple profile of its proteolysis process can be drawn. Several peptidases are reported to be involved in trimming of RsiW following PrsW and prior to RasP.70 In a reconstituted E. coli system, PrsW removes 40 amino acids from the C-terminal of RsiW by cleaving between Ala168 and Ser169 of the extracytoplasmic domain, generating RsiW-S1. Then the C-terminus of RsiW-S1 is further processed by Tsp (tail-specific protease, a periplasmic carboxyl terminal processing peptidase) before subsequent RasP proteolysis.70
Another substrate of RasP in B. subtilis has been identified as FtsL, which is involved in division of cells.71 Alignment of FtsL and RsiW and mutant constructs indicate that the conserved RAS motif before, and the AAA motif inside the transmembrane region of FtsL may be involved in recognition and cleavage, respectively (Fig. 3).71 However, the so-called S1P seems unnecessary for FtsL processing. A recent investigation suggested that the ABC transporter EcsAB may be involved in the regulation of RasP cleavage of its substrates in an unknown manner.72
Figure 3.
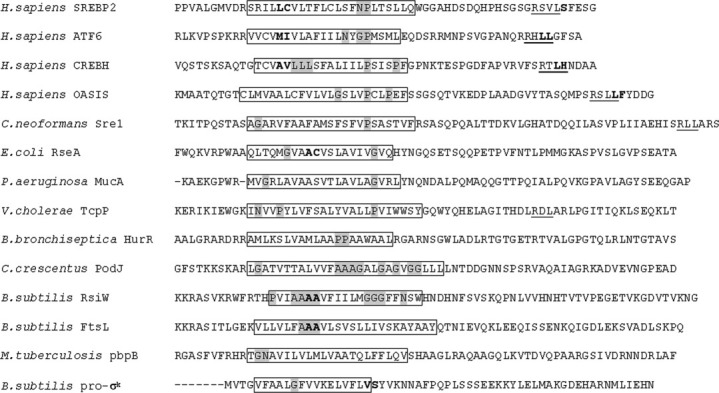
Sequence and properties of S2P substrates. The sequences of the predicted TM segments of 14 identified S2P substrates are shown. The boxes denote the predicted TM domains. Putative helix-destabilizing residues (N, P, G or repeated amino acids) in the TM regions are highlighted in gray. S1P and S2P cleavage sites, where known, are highlighted between residues in bold (outside TM for S1P and inside or close to TM for S2P, respectively). The consensus sequences of the human S1P site (RxxL or RxLx) are underlined. The aligned sequences are: H. sapiens SREBP2 (GI:27477113), H. sapiens ATF6 (GI:2245630), H. sapiens CREBH (GI:14211949), H. sapiens OASIS (GI:21668502), C. neoformans Sre1 (BROAD ID: CNAG_04804), E. coli RseA (GI:89109378), P. aeruginosa MucA (GI:223702393), V. cholerae TcpP (GI:14548354), B. bronchiseptica HurR (GI:33603626), C. crescentus PodJ (GI:221235062), B. subtilis RsiW (GI:62900894), B. subtilis FtsL (GI:1122761), M. tuberculosis pbpB (GI:15609300), B. subtilis pro-σk (GI:133478).
MucP in Pseudomonas aeruginosa
Pseudomonas aeruginosa, a gram-negative bacterium, is an important opportunistic human pathogen causing urinary tract, respiratory system and eye infections, and a variety of systemic infections particularly in individuals with compromised immune defence such as cystic fibrosis (CF).73 During the course of infection in CF, P. aeruginosa colonizers convert from low to high producers of the capsular polysaccharide alginate. Thereby they gain higher tolerance against host immune defence and provide a barrier that allows the pathogen to survive treatment with most common antibiotics.73 The overproduction of alginate is often caused by mutations within mucA. The antisigma factor MucA is a transmembrane protein located in the inner membrane and contains an N-terminal cytoplasmic domain that binds and represses activity of sigma factor σ22 (AlgU/AlgT).74–76 MucA also contains a C-terminal periplasmic region for regulation. In response to extracytoplasmic stress, MucA is sequentially cleaved by AlgW and MucP, the homologs of DegS and RseP in E. coli, respectively. Then MucA releases the sequestered ECF σ22 (AlgU/AlgT) to activate the alginate biosynthesis. Mutations or deletion in mucA found in mucoid strains cause truncations of the C-terminal periplasmic domain of MucA. MucB and MucD are negative regulators for alginate synthesis in the periplasm. Without extracytoplasmic stress, MucB binds tightly to the C-terminal of MucA and strongly inhibits the cleavage by AlgW.77 This easily explains why truncations of the C-terminal periplasmic domain of mucA can induce the mucoid conversion, since it loses the negative regulation by MucB. MucD can eliminate signal proteins that activate AlgW and other proteases to cut MucA.78 The defect in MucD caused MucA instability, suggesting wild type MucD degrades overactive cell wall stress signals, for example, the misfolded periplasmic peptides that can activate AlgW protease even in the absence of stress.79 MucE is the positive regulator of alginate biosynthesis. Three critical amino acid residues at the C terminus of MucE (WVF) were required for mucoid conversion via proteases AlgW and MucP.78 The PDZ domain of AlgW per se is a sensor. It bind MucE C-terminal peptide sequences or other stress peptide signals, which represent the folding/assembly status of proteins in the periplasm.78 And the PDZ domain plays important roles both in repressing proteolytic activity when appropriate peptide signals are absent and in stimulating cleavage when such peptides are present.77 When P. aeruginosa suffers from cell wall stress, such as D-cycloserine, a signal peptide from MucE interacts with the PDZ domain of AlgW to induce the site 1 cleavage of MucA by cleaving the C-terminal periplasmic region between Ala136-Gly137. And then MucP executes its site 2 protease function by cleaving the processed MucA.77 In a study of a revertant of mucoid mutant, the ClpXP and ClpP2 proteins were found to degrade the cytoplasmic portion of the truncated MucA N-terminus to release the sequestered AlgU, which finally drives alginate biosynthesis.80
MmpA in Caulobacter crescentus
Caulobacter crescentus is a dimorphic bacterium that divides asymmetrically and undergoes a characteristic morphological transition as part of its life cycle. During the cell cycle, the polarity determinant PodJ is processed in a S2P cascade. PodJ recruits structural and regulatory proteins required for polar organelle biogenesis to the correct cell pole at a defined time in the cell cycle. The full-length PodJ protein (PodJL) contains 974 amino acids, having a cytoplasmic N-terminal part, a single transmembrane domain and a periplasmic C-terminal region.81 It is synthesized in the early predivisional cell and localizes to the incipient swarmer pole where it recruits PleC histidine kinase/phosphatase and factors required for pili biogenesis.81–83 As cells divide, the periplasmic domain of PodJL is truncated, resulting in a membrane-anchored short form PodJS, which is required for later steps in pole development. Shapiro's group84,85 investigated the proteolytic processing of PodJ in C. crescentus. They found that truncation of PodJL into PodJS requires the periplasmic protease PerP, whose expression is controlled by the DivJ–PleC–DivK monitoring system.84 PodJS is subsequently released from the polar membrane by the S2P homolog MmpA during the swarmer-to-stalked cell transition.85 Eventually, the cytoplasmic part of PodJ is removed by an as-yet-unknown protease. The sequential cleavage of PodJ by PerP and MmpA illustrate how the S2P cascade has been adapted to regulate the asymmetric distribution of PodJ isoforms, which lead to the correct succession of polar organelle development during the cell cycle of C. crescentus.
HurP in Bordetella bronchiseptica
The heme system is one of the three iron retrieval systems in B. bronchiseptica, a gram-negative pathogen of humans and animals that colonizes the respiratory tract. HurP, a S2P family member, is essential for heme-dependent induction of bhuR and downstream genes.86 Expression of the hurIR bhuRSTUV heme utilization operons in B. bronchiseptica is coordinately controlled by the global iron-dependent regulator Fur and the ECF sigma factor HurI. Activation of HurI requires transduction of a heme-dependent signal via HurI, HurR, and BhuR, a three component heme-dependent regulatory system. HurI is sequestered by its antisigma factor, HurR, when iron is sufficient. BhuR is an outer membrane heme receptor protein. These three components are driven by two promoters, PhurI and PbhuR. When iron is abundant, expression of both operons is repressed in a Fur-dependent manner. Upon iron depletion, Fur-derepression at the hurI promoter enables hurIR transcription and read-through transcription of bhuR, resulting in basal production levels of the heme transport proteins.87 Under this condition, BhuR binds heme and initializes the S2P cascade by activating the unknown S1P cleavage of HurR. Then HurP exerts site 2 cleavage on HurR to release the HruI to bind RNA polymerase.86 Finally, this signaling cascade generates the cell response to the iron depletion environment.
S2P homologs in Arabidopsis
EGY1 (ethylene-dependent gravitropism-deficient and yellow-green 1) was identified as the first S2P homolog in plants.88,89 EGY1 is one of the five S2P homologs in Arabidopsis which contain the two canonical motifs HEXXH and NPDG. Located in chloroplast thylakoid membrane, EGY1 is required for development of thylakoid grana and a well-organized lamellae system in chloroplasts. It is also required for the accumulation of chlorophyll and chlorophyll a/b binding (CAB) proteins in chloroplast membranes. Another characterized S2P homologs in Arabidopsis, AraSP, is a chloroplast inner envelop membrane protease and essential for plant development.90 However, the substrates of both EGY1 and AraSP are still under investigation and the relative S1P is elusive.
On the other hand, several membrane-tethered transcription factors (MTF) are reported to be activated by proteolytic release from membrane. Heat stress induces the proteolytic release of the transcription factor domain of MTF AtbZIP28 from ER membrane and results in its redistribution to the nucleus.91 MTF AtbZIP60 is a proteolysis-activated transcription factor involved in ER stress response.92 Processing and relocation to the nucleus of ER MTF AtbZIP17 are important for the up-regulation of salt stress genes.93 AtbZIP17 and AtbZIP28 contain the canonical cleavage site of mammalian S1P as RxxL, and AtS1P responsible for AtZIP17 was indentified.93 Genome-scale screening identified at least 85 membrane-bound transcription factors (MTF) in Arabidopsis.94 They are proposed to be regulated by controlled proteolytic activation in response to various environmental changes.
Other newly identified S2P cascades
In gram-positive bacterium Enterococcus faecalis, antibiotic resistance and virulence determinants can be rapidly spread throughout populations by the transport of mobile genetic elements induced by oligopeptide sex pheromone.95,96 A S2P family member Eep processes the sex pheromone and inhibitor precursors as they pass through the cell membrane. Eep is involved in the production of pheromone cAD1, cPD1, and cCF10, and inhibitor peptide iAD1 and iCF10.97–99 But the substrate recognition and cleavage site remain elusive for Eep. So far no common motif was revealed in the primary sequence of known Eep targets.97 It is proposed that possibly the secondary structure provided by the N terminus of the Eep-recognized polypeptides directs Eep targeting.
Recently, a functionally conserved S2P member Stp1 was characterized in the human fungal pathogen Cryptococcus neoformans. The investigators demonstrated that Stp1 cleaves Sre1, a transcription factor similar to SREBP, within its predicted first transmembrane segment.100 Sre1 pathway is crucial for host adaptation and virulence and is required for both growth and survival in the presence of sterol biosynthesis-inhibiting antifungal drugs,101 thus providing hints for anticryptococcal therapy.
Mycobacterium tuberculosis is an obligate human pathogen threatening global health. In this gram-positive bacteria, knockout mutants of the S2P homolog Rv2869c exhibited altered cell envelope composition and virulence, suggesting S2P mediated RIP as a proximal regulator of cell envelope virulence determinants.102 Later, penicillin-binding protein 3 (PBP3) was found to be cleaved by Rv2869c (MSMEG_2579) in oxidatively stressed cells. Amino acids A102 and A103 of PBP3 are required for Rv2869c mediated cleavage. While Wag31, by virtue of its interaction with PBP3 through amino acid residues 46NSD48, protects it from oxidative stress-induced cleavage.103
Vibrio cholerae is a gram-negative bacterium that causes the severe diarrhoeal disease cholera. Acting in contrast to the common situation of S2P pathway on activating transcription factor, RIP here renders the transcription factors inactive and results in limiting virulence gene expression. TcpP, a membrane-localized virulence activator of V. chelerae, bind promoter DNA and activate gene expression under favorable condition. Under adverse conditions, TcpP is degraded by an unknown S1P and a homolog of YaeL, Vc-YaeL.104
Variation and Conservation of S2P Cascades
As the first known intramembrane protease, mammalian S2Ps have been investigated extensively since Brown and Goldstein discovered their function in regulation of sterol and fatty acid synthesis in 1990s.105 Thus far, homologous proteases have been identified throughout different organisms from prokaryote, archaea to other eukaryote, but some of the substrates and relative S1Ps still remain elusive (Table I). Here, we try to summarize the variation and conservation of S2P cascades, aiming at providing clues for the future study of newly identified members.
Table I.
Currently Characterized S2P Cascades in Different Organisms
| Substrates | S1P | S2P | Location | Inducing signal | Downstream cascade | Ancillary regulators | References | |
|---|---|---|---|---|---|---|---|---|
| Eukaryotic organisms | ||||||||
| Homo sapiens | SREBPs | S1P/SKI-1 | S2P | ER→Golgi | Low levels of membrane cholesterol in ER | Activates cholesterol biosynthesis and uptake | Scap, Insigs, COPII | 7–9, 14, 16–18, 21–23 |
| ATF6 | MBTPS1 | S2P/MBTPS2 | ER→Golgi | Unfolded proteins accumulation as ER stress | Unfolded protein response | BiP/GRP78 | 25, 106, 107 | |
| OASIS | S1P | S2P | ER→Golgi | ER stress | Unfolded protein response in astrocytes | 27 | ||
| CREBH | S1P | S2P | ER→Golgi | ER stress | Acute inflammatory response in liver | 26 | ||
| Drosophila | SREBP | S1P | S2P | Low levels of phosphatidylethanolamine | Activating fatty acid biosynthesis | 108 | ||
| Arabidopsis thaliana | bZIP17 | AtS1P | Unknown | Unknown | 93 | |||
| Unknown | Unknown | EGY1 (At2g35220) | Thylakoid membrane | 88 | ||||
| Unknown | Unknown | AraSP (At2g32480) | Chloroplast inner envelope | 90 | ||||
| Cryptococcus neoformans (fungi) | Sre1 | Unknown | Stp1 | Unknown | Hypoxia, nutrient limiting, presence of sterol synthesis inhibitor | Host adaptation and virulence | 100, 101 | |
| Prokaryotic organisms | ||||||||
| Escherichia coli (G-) | RseA (anti sigma factor of σE) | DegS | RseP (YaeL or EcfE) | CM | Extracytoplasmic stress, accumulation of misfolded outer membrane proteins in the periplasm | Extracytoplasmic stress response, cell envelope biogenesis | RseB, RseC, ClpXP | 30–34, 40, 41, 43, 47, 48, 109 |
| Pseudomonas aeruginosa (G-) | MucA (anti sigma factor of AlgU) | AlgW | MucP | IM/PM | Cell wall stress (induced by e.g. D-cycloserine) | Extracytoplasmic stress response, alginate biosynthesis | MucB, MucD, MucE, Prc, ClpXP | 77–80 |
| Bordetella bronchiseptica (G-) | HurR (anti sigma factor of HurI) | Unknown | HurP | CM/PM | Iron depletion | Activate haem receptor bhuR expression and iron uptake | 86 | |
| Vibrio cholerae (G-) | TcpP | Unknown | Vc-YaeL | CM/PM | Unfavorable conditions | Limiting virulence gene expression | TcpH | 104 |
| Caulobacter crescentus | PodJ (polarity determinant) | PerP | MmpA | CM | In response to the completion of cytokinesis | Polar organelle development | DivJ–PleC–DivK system | 84, 85 |
| Bacillus subtilis (G+) | Pro-σK | No S1P | SpoIVFB | Outer forespore membrane | Signals for sporulation, e.g. nutrient deprivation | σK activates transcription of mother cell genes involved in sporulation | SpoIVFA, BofA, CtpB, SpoIVB | 49, 56, 57, 59, 61 |
| RsiW (anti sigma factor of σW) | PrsW (YpdC) | RasP (YluC) | CM/PM | Stress affecting integrity of cell envelope: alkaline stress, salt stress, phage, antimicrobial peptides | Release of σW to activate extracytoplasmic stress response | EasAB,Tsp or CptB, ClpXP | 64, 66–70, 72 | |
| FtsL (cell division protein) | Unknown | RasP (YluC) | CM | Induced by DNA damage | Cell division | 71 | ||
| Enterococcus faecalis (G+) | cCF10, cAD1, cPD1, iAD1, iCF10 | Unknown | Eep | CM | Release pheromone, signal for conjugation | 96–99, 110 | ||
| Streptococcus uberis (G+) | MtuA | Unknown | Eep | Absence of lipoprotein signal peptidase Lsp | Peptidase cleavage | 111 | ||
| Mycobacterium tuberculosis (G+) | PBP3 (pbpB) | Unknown | Rv2869c | CM/PM | Oxidatively stress | Regulate cell envelope composition and virulence | Wag31 | 102, 103 |
| Archaea | ||||||||
| Methanocaldococcus jannaschii | Unknown | Unknown | mjS2P | 112 | ||||
IM, inner membrane; PM, periplasm membrane; CM, cytoplasmic membrane; G+, gram positive; G-, gram negative.
Various site-1 proteases
As summarized above and in Table I, Site-1 proteases differ markedly from one system to another. They will not be discussed further.
Conserved catalytic motif and mechanism of Site-2 proteases
In contrast to the diverse Site-1 proteases, Site-2 proteases are much more closely related to each other. They all belong to the same family of membrane-embedded, zinc metalloproteases and share the conserved catalytic motif as: HEXXH and NPDG [Fig. 2(A,B)].113 Most HEXXH motifs lie inside the TM helices, while most NPDG motifs are located at the end of or even outside TM helices. Between these helices containing HEXXH and NPDG, usually there is another TM helix, forming a compact three TM structural core (Fig. 4).114,112 Both the HEXXH and NPDG motifs face the cytosolic side, suggesting a conserved active site core domain or conformation to be very close to the membrane surface [Figs. 2(A,B) and 4]. In the studies of substrate recognition and binding by RseP, Koide et al. analyzed the environment of the RseP active site based on the accessibility of various thiol-alkylating reagents to Cys residues introduced around the catalytic residues. The results suggested that the active site of RseP is neither totally embedded in the lipid phase nor exposed to the cytoplasm, but rather located within a folded protein domain partially embedded in the membrane.115
Figure 2.
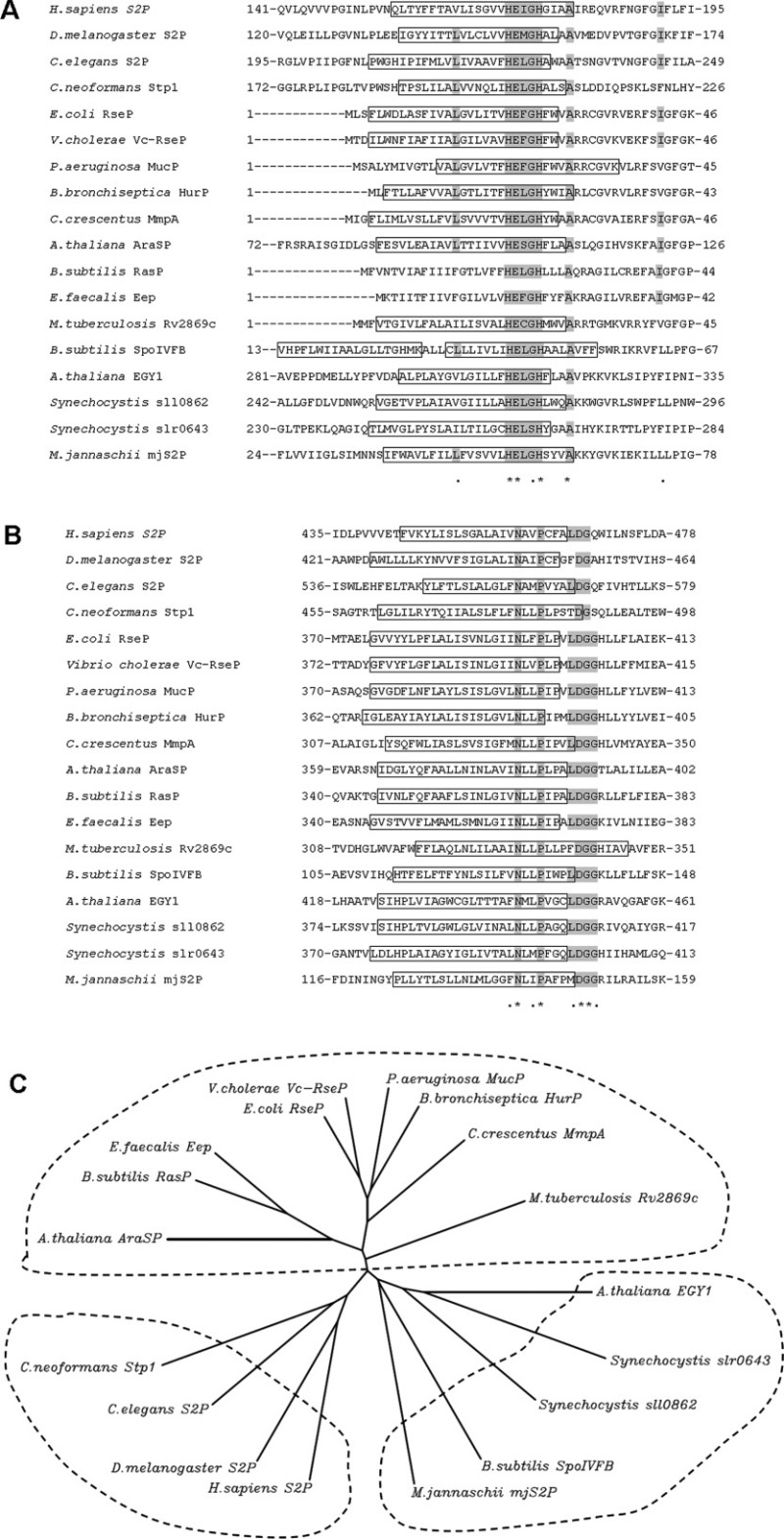
Sequence alignment and relationship among S2P homologs. The sequences of 18 characterized S2P homologs are presented. Only the core TM domains containing the HEXXH and NPDG motifs are shown. A: Sequence alignment in the vicinity of the HEXXH motif. The presumed transmembrane helix (TM) is boxed and the consensus amimo acids are highlighted in gray. B: Sequence alignment for the NPDG motif. C: Unrooted dendrogram of S2P homologs based on the alignment of the HEXXH and NPDG motifs. TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) and PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/) were used to predict TM helix. The unrooted dendrogram was generated by CLUSTALW (http://align.genome.jp/). The aligned sequences are: H.sapiens S2P (GI:6016601), D. melanogaster S2P (GI:19922044), C. elegans S2P (GI:22859116), C. neoformans Stp1 (BROAD ID: CNAG_05742), A. thaliana EGY1 (GI:15238440), Synechocystis sp. PCC6803 sll0862 (GI:16330216), Synechocystis sp. PCC6803 slr0643 (GI:16331565), A. thaliana AraSP (GI:18402981), E.coli RseP (GI:16128169), V. cholerae Vc-RseP (GI:20978850), P. aeruginosa MucP (GI:146448760), B. bronchiseptica HurP (GI:33601589), C. crescentus MmpA (GI:20978837), B. subtilis RasP (GI:20978800), E. faecalis Eep (GI: 256853722), M. tuberculosis Rv2869c (GI:20978863), B. subtilis SpoIVFB (GI:16079849), M. jannaschii mjS2P (GI:2499926).
Figure 4.
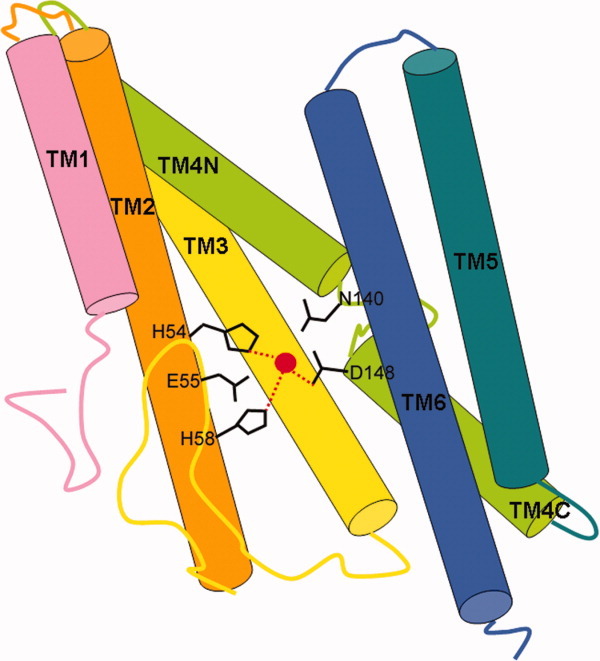
The crystal structure of mjS2P from Methanocaldococcus jannaschii.112 Six transmembrane helices (TM) are shown in different colors. The catalytic zinc atom (red) is coordinated by His54 and His58 on TM2 and Asp148 at the N terminal end of helix TM4C. The conserved Glu55 and Asn140 may contribute to catalysis.
Shi and coworkers have determined the crystal structure of the transmembrane core domain of mjS2P, a putative archaea homolog from Methanocaldococcus jannaschii (Fig. 4).112 They found that mjS2P has six transmembrane segments (TM1–TM6) and zinc is coordinated by three amino acids, His54 and His58 in TM2, and Asp148 in TM4. TM2 and TM4 are stabilized by TM3 and together these three segments constitute a core domain of S2P. These results are consistent with the prior mutational studies. Based on the differences of two mjS2P structures, it was suggested that TM1 and TM6 may function as a lateral gate to allow substrate access to the active site.112,116 But since the active sites are physically located very close to membrane surface and proteases appear to cause significant membrane compression around the enzyme, it cannot be ruled out that the active sites may be exposed outside the membrane to interact with substrate during proteolysis.117
In the alignment of identified S2P homologs, other than the known HEXXH motif, an alanine, three amino acids after HEXXH is found conserved among the S2P homologs analyzed [Fig. 2(A)]. Using the alignment of HEXXH and NPDG sequences, an unrooted dendrogram was developed as Figure 2(C). Three main groups appear to represent the eukaryotes homologs, the prokaryote homologs, and the SpoIVFB group. All the Site-2 proteases contain at lease four hydrophobic and potentially membrane-spanning segments. S2P, ResP, and SpoIVFB are representatives of the three known structural subclasses within the family.117 The structural classification is consistent with the unrooted dendrogram based on the alignment of the catalytic motif [Fig. 2(C)].
Most of the prokaryote homologs, for example, E. coli RseP, V. cholerae Vc-RseP, P. aeruginosa MucP, B. bronchiseptica HurP, C. crescentus MmpA, B. subtilis RasP, E. faecalis Eep, and M. tuberculosis Rv2869c are cataloged in the RseP subclass. They contain only four TM domains and share additional conserved residues GFG 16 amino acids after HEXXH motif [Fig. 2(A)]. The metal-chelating motif HEXXH and NPDG are located within or near the first and third TM segments and face the cytosolic side of the membrane [Fig. 1(B)]. They cleave type II (Nin-Cout) TM helices of membrane-spanning regulatory proteins such as antisigma factors in response to extracellular signals and stresses.118 Among the RseP subclass, Chen et al. reported that MmpA and RseP are functionally interchangeable in heterologous systems.85 Experiments also revealed that the defects in hemin utilization and heme-dependent induction of BhuR were restored when recombinant HurP (or recombinant RseP) was introduced into the mutant. Introduction of HurP into a Vc-yaeL mutant of V. cholerae also complemented its S2P defect.86 These data provided strong evidence that protease activity and cleavage site recognition is conserved in HurP, RseP, and Vc-YaeL. SpoIVFB has additional TM domains before the first and after the fourth TM domains, bringing the total number to six [Fig. 1(C)]. It removes the membrane associated 20aa prosequence of pro-σK. S2P from H. sapiens, D. melanogaster, C. elegans, and Stp1 from C. neoformans are grouped as the eukaryotic S2P homologs [Fig. 2(C)]. Compared with SpoIVFB, they have either one or several additional TM domain at the N-terminal [Fig 1(A)]. They cleave type II TM helices.
It is intriguing to find that two homologs from Arabidopsis, AraSP, and EGY1 are classified in different groups [Fig. 2(C)]. AraSP actually has four TM helices, conserved GFG residues and a PDZ domain,90 similar with other homologs in RseP subclass. While EGY1 and its cyanobacterium homologs, sll0862 and slr0643 from Synechocystis sp. PCC6803 do not contain PDZ domain and have several additional TM helices. Alignment here suggests they are closer to the SpoIVFB group. Chloroplasts are though to have derived from a cyanobacterium ancestor by endosymbiosis. The high similarity between EGY1 and sll0862 indicates the nuclear gene of EGY1 may be transferred from the genome of ancestral cyanobacterium. Further investigation of these new members, especially regarding their substrate, relative S1P, and biological function may reveal their relationship clearly.
Substrate recognition
In most S2P cascades, substrates are membrane-bound transcription factors or antisigma factors that sequestrate sigma factors. These transcription factors or sigma factors are responsible for expression of genes that respond to perturbation of the extracytoplasmic or cytoplasmic environment.118 S2P cascades liberate the mature form of transcription factors or sigma factors from membrane by sequential cleavages, which enables them to bind RNA polymerase (RNAP). The complete assembly of the RNAP complex will trigger the expression of responsive genes. Generally this cascade is negatively controlled to prevent the cells from constitutively expressing unnecessary genes under normal conditions and it will be initiated immediately once the cells suffer from abnormal environmental stresses.
Interestingly, more than one substrate has been identified for several S2P cascades and some substrates share sequence similarity at the cleavage site. For example, SREBP2, ATF6, CREBH, and OASIS are substrates of human S1P and S2P. They share a similar S1P cleavage site designated as RxLx or RxxL (Fig. 3).26 RsiW and FtsL are substrates of RasP in Bacillus subtilis and they have the same S2P cleavage site with sequence AAAV (Fig. 3).71 When comparing all the S2P substrates identified so far, it is obvious that there is no consensus sequence at the substrate cleavage site. However, the substrates all contain a transmembrane helix or membrane associated sequence, within which the S2P cleavage site reside inside or near the membrane (Fig. 3). They all contain amino acids of low helical propensity, for example, Pro (P), Gly (G), and Asn (N), in the TM segment (Fig. 3). The resultant helix destabilization may facilitate the cleavage process.
RseP-substrate recognition
Intensive investigations have been conducted on RseP to reveal its substrate recognition. In vivo and in vitro studies show that RseP can cleave transmembrane sequences of some model membrane proteins that are unrelated to RseA, provided that the transmembrane region contains residues of low helical propensity. These results show that RseP has proteolytic activity against a broad range of TM sequences and is dependent on residues of low helical propensity in the transmembrane segment of substrates.41 An α-helix is generally resistant to proteolytic cleavage, because the conformation makes the amide bonds inaccessible to the protease active site; helix-destabilizing residues may make the substrate polypeptide backbone more susceptible to hydrolysis. Helix-destabilizing residues in the TM sequence of a substrate were also reported to promote cleavage by Rhomboid proteases and the signal peptide peptidase.119–121
Through coimmunoprecipitation assays, Koide et al. showed that helix-destabilizing residues in a substrate transmembrane segment, which were previously shown to be required for efficient proteolysis of the substrate by RseP, stabilized the substrate-RseP interaction.122 The third transmembrane region of RseP is also involved in substrate binding. Asn-394(N) and Pro-397(P) are conserved amino acid in TM3 of RseP and might play a role in the substrate binding.122 Inaba et al. isolated deregulated RseP mutants which suggested that the proteolytic function is controlled by ligand binding to two tandem PDZ domains in RseP, especially the first one.123
Other than SpoIVFB, most site-2 proteases cut following the site-1 cleavage. Several investigations have explored why Site-1 cleavage must precede Site-2 cleavage. It was found that cleavage by DegS may remove the Gln-rich domain of RseA and eliminate the RseB mediated three-component inhibitory system on RseP (Fig. 1).45,46 Recently in the reconstituted sequential cleavages of the E. coli RseA by DegS (S1P) and RseP (S2P), Shi and coworkers found that Val148 of RseA, the newly exposed carboxyl-terminal hydrophobic amino acid is required for the cleavage of RseA by RseP following DegS cleavage. Its mutation to a charged or dissimilar amino acid abolished the Site-2 cleavage. Structural analysis revealed that the putative peptide-binding groove in the second, but not the first, PDZ domain of RseP is poised for binding a single hydrophobic amino acid. Based on the structural evidence, they suggested that after DegS cleavage, the newly exposed carboxyl terminus of RseA Val148 interacts with the second PDZ domain of RseP to facilitate its cleavage.124 This finding parallels observations on the role of ectodomain shedding in γ-secretase.125 A common model indicates that ectodomain shedding by S1P frees a single terminal residue of substrates to interact with extracytoplasmic domain of I-CliPs and activate the second cleavage.125
SpoIVFB-substrate recognition
Prince et al. found a small N-terminal segment (amino acids 1 to 27) was sufficient for membrane localization of a pro-σK-GFP chimera.53 Longer segments, however, are required for RIP. It was found that nearly half of pro-σK (amino acids 1 to 117) was required for RIP of pro-σK-GFP chimeras in sporulating B. subtilis. Likewise, pro-σK-hexahistidine chimeras demonstrated that the N-terminal 117 amino acids of pro-σK are sufficient for RIP, although the N-terminal 126 amino acids allowed much better accumulation of the chimeric protein in sporulating B. subtilis and more efficient processing by SpoIVFB in E. coli.53
S2P-Subtrate recognition
Shen and Prywes found that short luminal domains of ATF6 result in S1P-independent S2P cleavage.106 The addition of artificial irrelevant domains onto these short ATF6 luminal domains restored the S1P dependence of S2P cleavage, suggesting that dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6 but not the specific sequences in the luminal domain.106
Perspective
As one of the families executing intramembrane cleavage, S2P and its homologs are involved in RIP events that control stress responses,33,34 cell division,71 bacterial mating,98 pathogenesis,1 and polar organelle biogenesis,85 among other activities. The nearly universal presence of S2P homologs in different organisms and the diverse functions they play suggest that they perform fundamental functions that convey advantages to cells. Though responsive to different signals and initiated by various S1Ps, the S2P cascades still exhibit common properties. S2P homologs share the conserved HEXXH and NPDG motifs and are easier to identify through a bioinformatics approach.113,114 The prevalence of amino acids of low helical propensity in the TM segment of substrates may suggest a conserved catalytic conformation and mechanism in these S2P homologs. Further structural analysis of the S2Ps and their substrates may help to answer this question. The absence of identified substrates is the major bottleneck for protease research because it may hamper further progress in understanding the molecular basis of how these proteases function. Some substrates and relative regulator proteins have been identified through forward genetic approaches that look for suppressors of protease mutant phenotypes. Differential proteomics, protein arrays and yeast two-hybrid screening are useful tools for substrate identification. While the protease cleavage specificity and its subcellular location might help to select candidate substrates. And amino acids of low helical propensity in the TM segment of substrates may help to identify substrates of new S2P homologs.
References
- 1.Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nature Rev Microbiol. 2009;7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urban S. Taking the plunge: integrating structural, enzymatic and computational insights into a unified model for membrane-immersed rhomboid proteolysis. Biochem J. 2010;425:501–512. doi: 10.1042/BJ20090861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman M. Rhomboid proteases and their biological functions. Ann Rev Gen. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 4.Fluhrer R, Steiner H, Haass C. Intramembrane proteolysis by signal peptide peptidases: a comparative discussion of GXGD-type aspartyl proteases. J Biol Chem. 2009;284:13975–13979. doi: 10.1074/jbc.R800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy JV, Twomey C, Wujek P. Presenilin-dependent regulated intramembrane proteolysis and gamma-secretase activity. Cell Mol Life Sci. 2009;66:1534–1555. doi: 10.1007/s00018-009-8435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 7.Sakai J, Duncan EA, Rawson RB, Hua XX, Brown RS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 8.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang XD, Sato R, Brown MS, Hua XX, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 10.Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 11.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein—evidence from in vivo competition studies. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda M, Korn BS, Hammer RE, Moon YA, Komuro R, Horton JD, Goldstein JL, Brown MS, Shimomura I. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng Y, Schwarz EJ, Lazar MA, Genin A, Spinner NB, Taub R. Cloning, human chromosomal assignment, and adipose and hepatic expression of the CL-6/INSIG1 gene. Genomics. 1997;43:278–284. doi: 10.1006/geno.1997.4821. [DOI] [PubMed] [Google Scholar]
- 16.Brown AJ, Sun LP, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 17.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 18.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci USA. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawson RB. The SREBP pathway—insights from insigs and insects. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 20.Sun LP, Li L, Goldstein JL, Brown MS. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 21.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espenshade PJ, Cheng D, Goldstein JL, Brown MS. Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:22795–22804. doi: 10.1074/jbc.274.32.22795. [DOI] [PubMed] [Google Scholar]
- 23.Duncan EA, Brown MS, Goldstein JL, Sakai J. Cleavage site for sterol-regulated protease localized to a Leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- 24.Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 26.Zhang KZ, Shen XH, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 27.Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, Iseki K, Wanaka A, Imaizumi K. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol. 2005;7:186–U112. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- 28.Murakami T, Kondo S, Ogata M, Kanemoto S, Saito A, Wanaka A, Imaizumi K. Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J Neurochem. 2006;96:1090–1100. doi: 10.1111/j.1471-4159.2005.03596.x. [DOI] [PubMed] [Google Scholar]
- 29.Oeffner F, Fischer G, Happle R, Konig A, Betz RC, Bornholdt D, Neidel U, Boente MD, Redler S, Romero-Gomez J, Salhi A, Vera-Casano A, Weirich C, Grzeschik KH. IFAP syndrome is caused by deficiency in MBTPS2, an intramembrane zinc metalloprotease essential for cholesterol homeostasis and ER stress response. Am J Hum Gen. 2009;84:459–467. doi: 10.1016/j.ajhg.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dartigalongue C, Loferer H, Raina S. EcfE, a new essential inner membrane protease: its role in the regulation of heat shock response in Escherichia coli. EMBO J. 2001;20:5908–5918. doi: 10.1093/emboj/20.21.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alba BM, Zhong HJ, Pelayo JC, Gross CA. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma(E) activity. Mol Microbiol. 2001;40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 32.Kanehara K, Akiyama Y, Ito K. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene. 2001;281:71–79. doi: 10.1016/s0378-1119(01)00823-x. [DOI] [PubMed] [Google Scholar]
- 33.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli sigma(E) regulon. J Biol Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 36.Collinet B, Yuzawa H, Chen T, Herrera C, Missiakas D. RseB binding to the periplasmic domain of RseA modulates the RseA: sigma(E) interaction in the cytoplasm and the availability of sigma(E)center dot RNA polymerase. J Biol Chem. 2000;275:33898–33904. doi: 10.1074/jbc.m006214200. [DOI] [PubMed] [Google Scholar]
- 37.Tam C, Collinet B, Lau GKK, Raina S, Missiakas D. Interaction of the conserved region 4.2 of sigma(E) with the RseA anti-sigma factor. J Biol Chem. 2002;277:27282–27287. doi: 10.1074/jbc.M202881200. [DOI] [PubMed] [Google Scholar]
- 38.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigma(E) with the cytoplasmic domain of its anti-sigma RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 39.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 40.Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA. Design principles of the proteolytic cascade governing the sigma(E)-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 2007;21:124–136. doi: 10.1101/gad.1496707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasselblatt H, Kurzbauer R, Wilken C, Krojer T, Sawa J, Kurt J, Kirk R, Hasenbein S, Ehrmann M, Clausen T. Regulation of the sigma E stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress. Genes Dev. 2007;21:2659–2670. doi: 10.1101/gad.445307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 44.Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 45.Kanehara K, Ito K, Akiyama Y. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J. 2003;22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bohn C, Collier J, Bouloc P. Dispensable PDZ domain of Escherichia coli YaeL essential protease. Mol Microbiol. 2004;52:427–435. doi: 10.1111/j.1365-2958.2004.03985.x. [DOI] [PubMed] [Google Scholar]
- 47.Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Fine-tuning of the Escherichia coli sigma(E) envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cezairliyan BO, Sauer RT. Inhibition of regulated proteolysis by RseB. Proc Natl Acad Sci USA. 2007;104:3771–3776. doi: 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus-subtilis enhances processing of the sigma-factor precursor pro-sigma(K) in the absence of other sporulation gene-products. J Bacteriol. 1995;177:1082–1085. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Ann Rev Gen. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 53.Prince H, Zhou RB, Kroos L. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis Pro-sigma(K) J Bacteriol. 2005;187:961–971. doi: 10.1128/JB.187.3.961-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cutting S, Oke V, Driks A, Losick R, Lu SJ, Kroos L. A forespore checkpoint for mother cell gene-expression during development in Bacillus-subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 55.Zhou RB, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-sigma(K) in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-sigma(K) processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou RB, Kroos L. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-sigma(K) during Bacillus subtilis development. Mol Microbiol. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong TC, Cutting SM. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-sigma(K) in Bacillus subtilis. Mol Microbiol. 2003;49:1425–1434. doi: 10.1046/j.1365-2958.2003.03651.x. [DOI] [PubMed] [Google Scholar]
- 59.Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 60.Pan Q, Losick R, Rudner DZ. A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor UK in Bacillus subtilis. J Bacteriol. 2003;185:6051–6056. doi: 10.1128/JB.185.20.6051-6056.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou RB, Cusumano C, Sui DX, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc Natl Acad Sci USA. 2009;106:16174–16179. doi: 10.1073/pnas.0901455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshimura M, Asai K, Sadaie Y, Yoshikawa H. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology-Sgm. 2004;150:591–599. doi: 10.1099/mic.0.26712-0. [DOI] [PubMed] [Google Scholar]
- 63.Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Mol Microbiol. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- 64.Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma(W)-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 65.Pietiainen M, Gardemeister M, Mecklin M, Leskela S, Sarvas M, Kontinen VP. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology-Sgm. 2005;151:1577–1592. doi: 10.1099/mic.0.27761-0. [DOI] [PubMed] [Google Scholar]
- 66.Schobel S, Zellmeier S, Schumann W, Wiegert T. The Bacillus subtilis sigma(W) anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol. 2004;52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 67.Heinrich J, Wiegert T. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol Microbiol. 2006;62:566–579. doi: 10.1111/j.1365-2958.2006.05391.x. [DOI] [PubMed] [Google Scholar]
- 68.Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006;20:1911–1922. doi: 10.1101/gad.1440606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zellmeier S, Schumann W, Wiegert T. Involvement of Clp protease activity in modulating the Bacillus subtilis sigma(W) stress response. Mol Microbiol. 2006;61:1569–1582. doi: 10.1111/j.1365-2958.2006.05323.x. [DOI] [PubMed] [Google Scholar]
- 70.Heinrich J, Hein K, Wiegert T. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol Microbiol. 2009;74:1412–1426. doi: 10.1111/j.1365-2958.2009.06940.x. [DOI] [PubMed] [Google Scholar]
- 71.Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol Microbiol. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- 72.Heinrich J, Lunden T, Kontinen VP, Wiegert T. The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiol-Sgm. 2008;154:1989–1997. doi: 10.1099/mic.0.2008/018648-0. [DOI] [PubMed] [Google Scholar]
- 73.Govan JRW, Deretic V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schurr MJ, Yu H, MartinezSalazar JM, Boucher JC, Deretic V. Control of AlgU, a member of the sigma(E)-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie ZD, Hershberger CD, Shankar S, Chakrabarty AM. Sigma factor anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma(22) for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu DR, Eisinger VM, Rowen DW, Yu HWD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wood LF, Ohman DE. Use of cell wall stress to characterize sigma(22) (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 80.Qiu DR, Esinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiol-Sgm. 2008;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol Microbiol. 2006;59:301–316. doi: 10.1111/j.1365-2958.2005.04935.x. [DOI] [PubMed] [Google Scholar]
- 82.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 84.Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 86.King-Lyons ND, Smith KF, Connell TD. Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. J Bacteriol. 2007;189:6266–6275. doi: 10.1128/JB.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vanderpool CK, Armstrong SK. Integration of environmental signals controls expression of Bordetella heme utilization genes. J Bacteriol. 2004;186:938–948. doi: 10.1128/JB.186.4.938-948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen G, Bi YR, Li N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 2005;41:364–375. doi: 10.1111/j.1365-313X.2004.02308.x. [DOI] [PubMed] [Google Scholar]
- 89.Guo D, Gao XR, Li H, Zhang T, Chen G, Huang PB, An LJ, Li N. EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Mol Biol. 2008;66:345–360. doi: 10.1007/s11103-007-9273-5. [DOI] [PubMed] [Google Scholar]
- 90.Bolter B, Nada A, Fulgosi H, Soll J. A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett. 2006;580:789–794. doi: 10.1016/j.febslet.2005.12.098. [DOI] [PubMed] [Google Scholar]
- 91.Gao HB, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iwata Y, Fedoroff NV, Koizumi N. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell. 2008;20:3107–3121. doi: 10.1105/tpc.108.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim SG, Lee S, Seo PJ, Kim SK, Kim JK, Park CM. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics. 2010;95:56–65. doi: 10.1016/j.ygeno.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 95.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 96.Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci USA. 2005;102:15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol. 2008;190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Antiporta MH, Dunny GM. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol Microbiol. 2009;74:672–690. doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 102.Makinoshima H, Glickman MS. Regulation of mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature. 2005;436:406–409. doi: 10.1038/nature03713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mukherjee P, Sureka K, Datta P, Hossain T, Barik S, Das KP, Kundu M, Basu J. Novel role of Wag31 in protection of mycobacteria under oxidative stress. Mol Microbiol. 2009;73:103–119. doi: 10.1111/j.1365-2958.2009.06750.x. [DOI] [PubMed] [Google Scholar]
- 104.Matson JS, DiRita VJ. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc Natl Acad Sci USA. 2005;102:16403–16408. doi: 10.1073/pnas.0505818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J Lipid Res. 2009;50:S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen JS, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279:43046–43051. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- 107.Shen JS, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 108.Matthews KA, Kunte AS, Tambe-Ebot E, Rawson RB. Alternative processing of sterol regulatory element binding protein during larval development in Drosophila melanogaster. Genetics. 2009;181:119–128. doi: 10.1534/genetics.108.093450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alba BM, Gross CA. Regulation of the Escherichia coli sigma(E)-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 110.An FY, Clewell DB. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J Bacteriol. 2002;184:1880–1887. doi: 10.1128/JB.184.7.1880-1887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Denham EL, Ward PN, Leigh JA. Lipoprotein signal peptides are processed by lsp and eep of Streptococcus uberis. J Bacteriol. 2008;190:4641–4647. doi: 10.1128/JB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Feng L, Yan HC, Wu ZR, Yan N, Wang Z, Jeffrey PD, Shi YG. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 113.Lewis AP, Thomas PJ. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence hornologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koide K, Maegawa S, Ito K, Akiyama Y. Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J Biol Chem. 2007;282:4553–4560. doi: 10.1074/jbc.M607339200. [DOI] [PubMed] [Google Scholar]
- 116.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr Opin Struct Biol. 2008;18:432–441. doi: 10.1016/j.sbi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ha Y. Structure and mechanism of intramembrane protease. Sem Cell Dev Biol. 2009;20:240–250. doi: 10.1016/j.semcdb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heinrich J, Wiegert T. Regulated intramembrane proteolysis in the control of extracytoplasmic function sigma factors. Res Microbiol. 2009;160:696–703. doi: 10.1016/j.resmic.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 119.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–1037. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 120.Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 121.Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 122.Koide K, Ito K, Akiyama Y. Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. J Biol Chem. 2008;283:9562–9570. doi: 10.1074/jbc.M709984200. [DOI] [PubMed] [Google Scholar]
- 123.Inaba K, Suzuki M, Maegawa K, Akiyama S, Ito K, Akiyama Y. A pair of circularly permutated PDZ domains control RseP, the S2P family intramembrane protease of Escherichia coli. J Biol Chem. 2008;283:35042–35052. doi: 10.1074/jbc.M806603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li XC, Wang BY, Feng LH, Kang H, Qi Y, Wang JW, Shi YG. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc Natl Acad Sci USA. 2009;106:14837–14842. doi: 10.1073/pnas.0903289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dries DR, Yu G. Rip exposed: how ectodomain shedding regulates the proteolytic processing of transmembrane substrates. Proc Natl Acad Sci USA. 2009;106:14737–14738. doi: 10.1073/pnas.0908124106. [DOI] [PMC free article] [PubMed] [Google Scholar]


