Abstract
The mature protease from Group N human immunodeficiency virus Type 1 (HIV-1) (PR1N) differs in 20 amino acids from the extensively studied Group M protease (PR1M) at positions corresponding to minor drug-resistance mutations (DRMs). The first crystal structure (1.09 Å resolution) of PR1N with the clinical inhibitor darunavir (DRV) reveals the same overall structure as PR1M, but with a slightly larger inhibitor-binding cavity. Changes in the 10s loop and the flap hinge propagate to shift one flap away from the inhibitor, whereas L89F and substitutions in the 60s loop perturb inhibitor-binding residues 29–32. However, kinetic parameters of PR1N closely resemble those of PR1M, and calorimetric results are consistent with similar binding affinities for DRV and two other clinical PIs, suggesting that minor DRMs coevolve to compensate for the detrimental effects of drug-specific major DRMs. A miniprecursor (TFR1–61-PR1N) comprising the transframe region (TFR) fused to the N-terminus of PR1N undergoes autocatalytic cleavage at the TFR/PR1N site concomitant with the appearance of catalytic activity characteristic of the dimeric, mature enzyme. This cleavage is inhibited at an equimolar ratio of precursor to DRV (∼6 μM), which partially stabilizes the precursor dimer from a monomer. However, cleavage at L34/W35 within the TFR, which precedes the TFR1–61/PR1N cleavage at pH ≤ 5, is only partially inhibited. Favorable properties of PR1N relative to PR1M include its suitability for column fractionation by size under native conditions and >10-fold higher dimer dissociation constant (150 nM). Exploiting these properties may facilitate testing of potential dimerization inhibitors that perturb early precursor processing steps.
Keywords: HIV/AIDS, retroviral protease, dimer dissociation, enzyme kinetics, calorimetry, crystal structure, polyprotein processing
Introduction
To date, four genetically distinct groups have been identified for human immunodeficiency virus Type 1 (HIV-1), the predominant virus that causes AIDS. Groups M (Major), N (Non-M non-O), 1 and O (Outlier) 2 derive from SIV strains that infect chimpanzees, whereas a fourth group, first identified in 2009 and designated as Group P, likely arose from a different strain of SIV harbored by gorillas. 3 In distinct contrast to Group M, which is responsible for the vast majority of AIDS infections worldwide, fewer than 20 instances of Group N infections have been identified to date, all in Cameroon. 4 Despite its apparent rarity at present, HIV-1 Group N is transmissible among humans and is capable of causing AIDS. 5
All HIV-1 variants contain a similar organization of the Gag and Gag-Pol polyproteins and code for one subunit of an aspartyl protease. 6 7 The active protease consists of two identical 99-amino acid subunits, each of which contributes one of the two active-site Asp25 residues necessary for catalysis. 7 8 The protease plays an essential role in the viral life cycle by cleaving the Gag and Gag-Pol polyproteins at multiple sites to release the mature structural and functional proteins, including itself, required for viral assembly. 7 8 Thus, the protease is released in its active mature form from the Pol domain of the Gag-Pol polyprotein by an autocatalytic process 8 involving cleavages at its termini (Fig. 1). Cleavage at the N-terminus of the protease, which precedes the C-terminal cleavage, is concomitant with a dramatic lowering of the monomer–dimer equilibrium constant (Kd < 10 nM) giving rise to the fully active dimeric protease with a stable tertiary fold. 8 10 11
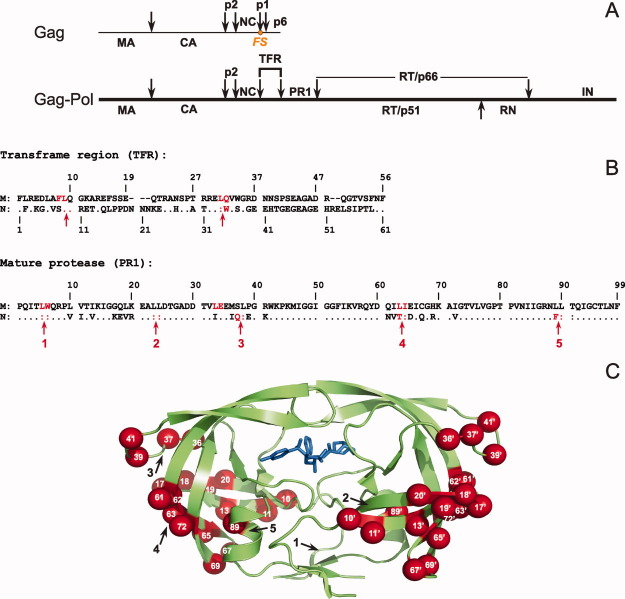
Figure 1.
A: Structural organization of Gag-Pol polyprotein in HIV-1. Arrows indicate the specific sites of cleavage by the viral protease. The protease is flanked at its N- and C-termini by the transframe region (TFR) and the reverse transcriptase, respectively. Nomenclature of HIV-1 proteins is according to Leis et al. 9; MA, matrix; CA, capsid; PR, protease; NC, nucleocapsid; RT, reverse transcriptase; RN, Rnase H; IN, integrase. For clarity, we denote mature proteases corresponding to Groups M and N as PR1M and PR1N. FS denotes the location of the frameshift site required for producing Gag-Pol. 6 B: Sequence alignment of TFR-PR1 domains derived from the Gag-Pol polyprotein of Groups M and N of HIV-1. The numbering system is relative to the N-terminus of TFR and the mature protease. The complete sequence for Group M is shown for reference, with dashes representing residues absent in TFR of Group M but present in the longer TFR of Group N. Dots (and colons at cleavage sites) indicate amino acids that are identical in the Group M and N sequences. Residues shown in red are major cleavage sites for TFR-PR1 autoprocessing (within TFR), and for the autoproteolysis of the mature PR (within PR), observed upon its release from the precursor. C: Sites of polymorphic substitutions mapped on the PR1N dimer described in this study. PR1N is shown as a green cartoon representation bound to inhibitor DRV, shown in blue sticks. The polymorphic substitutions are colored as red spheres on both the monomers and residue positions are labeled in white. Black arrows indicate the locations of the autoproteolytic sites shown in B. An interactive view is available in the electronic version of this article. PRO486 Figure 1
The mature protease of the most prevalent HIV-1 Group M (PR1M) has been exhaustively studied for almost 2 decades and is a major target for structure-based design of clinical protease inhibitors (PIs). 12 However, retaining an effective treatment of patients with PIs has been hampered by the inherent genetic variability of the virus, leading to the selection of variants that propagate in the presence of the drugs. This drug resistance has been a major hurdle even for the regimen involving highly active antiretroviral therapy (HAART). 12 13 PIs in current use are designed with very tight binding (low nM–pM) to the preformed active site of the mature protease. 14 However, to our knowledge, it is not thoroughly understood to what extent these PIs may also interfere with steps preceding release of the mature protease. Alternative methods of targeting the protease, such as by perturbing the transient folding and dimerization of the precursor that permits the crucial intramolecular cleavage at the N-terminus of the protease, may be advantageous for prolonged treatments of HIV infection. 11 15
We envision that studies of the mature protease with polymorphisms arising by natural selection as observed in distinct HIV-1 groups will complement the existing efforts to examine proteases bearing multiple mutations selected during drug pressure in long-term AIDS therapy. At the same time, understanding the precursors and mature proteases of these yet unexplored groups of HIV may have certain advantages for studies targeting protease dimerization. We have undertaken one such study using the mature protease derived from HIV-1 Group N (PR1N) as well as its precursor consisting of the protease fused to the full-length N-terminal transframe region (TFR).
Sequence alignments of the TFR and PR1 regions of the Gag-Pol of Groups M (GenBank accession number K0345516) and N (GenBank accession number AY5326351) are shown in Figure 1. The Group N TFR is five amino acids longer than its Group M counterpart and there is little identity between the M and N sequences. In contrast, in the protease domains of Groups M and N, the N-terminal dimer interface, the active site and flap regions, and the C-terminal residues 70–99 comprising loop and helix motifs as well as the C-terminal dimer interface are highly conserved. 17 Sequence variability is restricted to regions comprising residues 10–20, 36–39, and 61–69 accounting for a total of 20 substitutions (Fig. 1) in PR1N.
We have expressed and purified the mature protease (PR1N) and its precursor TFR-PR1N and characterized PR1N by enzyme kinetics, thermal and chemical denaturation profiles, and interaction with several PIs. PR1N provided insights into the potential role of amino acid polymorphisms in drug resistance from analysis of its atomic resolution crystal structure in complex with the potent PI, darunavir (DRV), and comparison with the corresponding PR1M/DRV complex. In addition, the autocatalytic maturation of PR1N, its inhibition, and the molecular status of the precursor as monomers or dimers were investigated. Desirable properties of PR1N that may aid in developing systems to evaluate inhibition of dimerization of the protease precursor and the mature protease have been highlighted.
Results and Discussion
Expression and purification of mature PR1N and its precursor
PR1N was expressed as a fusion protein, flanked at its N-terminus by its full-length TFR (Fig. 1). However, upon expression in E. coli, a large fraction of the TFR-PR1N precursor undergoes autocatalytic cleavage at the N-terminus of PR1N (TFR/PR1N site) to release the mature PR1N that accumulates in the insoluble fraction (Fig. 2). Thus, accumulation of PR1N and a very small fraction of TFR-PR1N in the insoluble fraction permitted isolation of these two proteins using a combination of three steps, that is, isolation of inclusion bodies followed by fractionation of the protein by size-exclusion chromatography and reverse-phase HPLC. The purity of the proteins is greater than 95% with a yield of ∼10 mg of mature PR1N (Fig. 2, lane P) from a 1-L culture. Because of the miniscule amount of precursor (Fig. 2, lane 3) trapped in the insoluble fraction, it is possible to purify only low microgram quantities of TFR-PR1N, sufficient to assess qualitatively the autoprocessing reaction in vitro.
Figure 2.
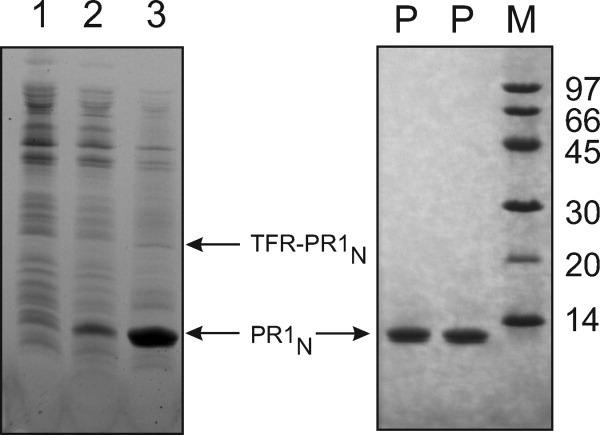
Expression and purification of TFR-PR1N and PR1N. Left: SDS-PAGE to monitor the expression and autoprocessing of TFR-PR1N construct in E. coli. Lanes 1 and 2 represent the total lysates derived from uninduced and induced cells, respectively. Lane 3, insoluble pellet derived from washing the initial insoluble fraction (post cell lysis) with 1M urea and 0.5% Triton X-100. The presence of a major band of PR1N and a faint band of TFR- PR1N in lane 3 indicates that (1) majority of the precursor undergoes autoprocessing upon its expression to release the mature PR1N and (2) both TFR-PR1N and PR1N accumulate in the insoluble fraction and are retained in this fraction even after washing with 1M urea. Right: SDS-PAGE of purified PR1N. Lanes denoted by P represent purified PR1N, used for the various physicochemical and structural analyses described in this article, and M, the molecular weight standards in kDa.
A single Cys95Ala mutation was introduced in PR1N to prevent cysteine-thiol oxidation leading to possible protein aggregation during in vitro studies. The purified proteins were verified by mass spectrometry to match the sequence reported under the GenBank accession number AY532635 with masses of 17,910 (calculated 17,910, includes N-terminal Met and C95A) and 10,869 (calculated 10,869, includes C95A) for the precursor TFR-PR1N and the mature protease PR1N, respectively. PR1N can be maintained stably in 12 mM HCl, presumably as an unfolded protein, for prolonged periods at 4°C. Full enzymatic activity is observed when one part of protein in HCl is diluted in 5.6 parts of buffer between pH values of 4 and 6.5, or as described previously for PR1M (quench protocol of protein folding 18). However, on storage at optimal conditions for activity between pH 4 and 6.5, PR1N exhibits autoproteolysis, consistent with cleavage sites that map between L5/W6, L23/L24, Q37/L38, T63/I64 and F89/L90 [Fig. 1(B), sites indicated in red] as determined by electrospray ionization-mass spectrometry (ESI-MS). Masses of 2122, 2935, and 2735 were easily observed corresponding to fragments 6–23, 38–63, and 64–89, respectively. The locations of sites 1, 3, and 4 [Fig. 1(B)] in PR1N are relatively conserved among PR1M and PR1N. 19 Site 3 is slightly shifted toward the sole major site reported for HIV-2 protease (PR2) at G35/I36 and HTLV-1 at L40/P41. 19 20 This shift could be due to the polymorphic mutation L33I in PR1N, which was shown to retard this cleavage in PR1M, 21 and substitutions in positions 36, 37, and 39 that promote cleavage between Q37/L38. The two sites [2 and 5, Fig. 1(B)] in PR1N that map between L23/L24 and F89/L90 were also identified as cleavage sites in PR2. 19 It is possible that these five sites [Fig. 1(C), black arrows] are accessible for cleavage in the monomeric PR1N rather than the dimer. The higher Kd (see below) observed for PR1N may facilitate an enhanced rate of autproteolysis when compared with PR1M, which has a Kd < 10 nM. 11
The ability to fractionate PR1N by size-exclusion chromatography under native conditions clearly distinguishes it from PR1M and its many variants and mutants (including drug-resistant mutants) that we have studied so far, which interact nonspecifically with most column matrices. In spite of similar isoelectric points exhibited by both proteases, PR1N is a unique example of a PR1 variant which can be readily analyzed by size-exclusion chromatography under native conditions, similar to PR2. 19 This feature of PR1N is an important advantage because it permits the use of physical techniques that are incompatible with PR1M. In the future, redesigning PR1M with substitutions of residues from PR1N in certain positions to enhance its solubility and chromatographic characteristics may be possible.
Dimer dissociation and kinetics
Preliminary experiments (not shown) indicated that PR1N efficiently catalyzes the hydrolysis of substrate IV over a relatively broad range between pH 4 and 6. Michaelis–Menten kinetics [Fig. 3(A)] for hydrolysis of substrate IV catalyzed by 0.36 μM PR1N at pH 5, in the presence of 250 mM NaCl, gave Km = 25 ± 3 μM, kcat = 2.83 ± 0.06 s−1, and kcat/Km = 113 mM−1 s−1. Under these conditions, > 70% of the enzyme is estimated to be dimeric in the presence of a saturating substrate concentration [see below and Fig. 3(B)]. The values of Km and kcat are comparable to those of 48 μM and 2.9 s−1 previously observed with PR1M under the same conditions. 22
Figure 3.
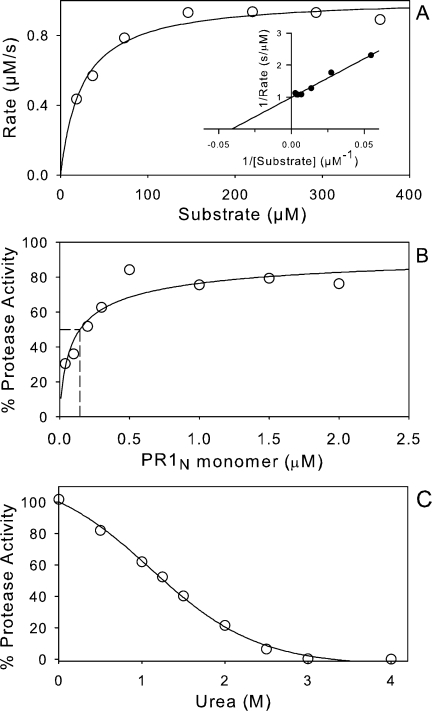
Measurement of kinetic parameters, dimer dissociation, and urea-induced denaturation for mature PR1N in 50 mM sodium acetate buffer, pH 5, containing 250 mM NaCl. A: Michaelis–Menten and Lineweaver–Burk (inset) plots for hydrolysis of substrate IV by 0.36 μM PR1N. Values are Km = 25 ± 3 μM and kcat = 2.83 ± 0.06 s−1. B: Kinetic determination of the dimer dissociation constant, Kd, of PR1N from the dependence of the rate for hydrolysis of substrate IV on enzyme concentration. The Kd of 0.15 ± 0.06 μM is indicated by the dashed line at 50% of the maximal activity. C: A plot of the protease activity as a function of increasing concentration of urea indicates a transition midpoint of ∼1.25M.
In contrast to the very low dimer dissociation constant, Kd, for PR1M, the Kd can be conveniently measured for PR1N [Fig. 3(B)]. Under the above kinetic conditions, the dependence on enzyme concentration of the observed rate for hydrolysis of a saturating (430 μM) concentration of substrate IV gave a Kd of 0.15 ± 0.06 μM that is at least 10-fold larger than that observed for PR1M (< 10 nM). 11 Consistent with the higher value of Kd, denaturation of PR1N at pH 5 occurs at a lower urea concentration than for PR1M [Fig. 3(C)], with a midpoint at ∼1.25M urea in the presence of 250 mM NaCl (which is expected to increase the stability of the dimer fold 23) when compared with ∼1.8M for PR1M10 and 1.8–1.9M for PR2 and its E37K mutant 19 in the absence of added salt. Urea does not affect the binding of substrate to PR1N as shown by a Km of 25.4 μM at 1.25M urea (data not shown). Thus, the loss in catalytic activity with increasing urea concentrations is truly a reflection of the unfolding process rather than an apparent effect of the decreased binding of the substrate as seen for PR2. 19
Calorimetric analysis of inhibitor binding
The increase in thermal denaturation temperature (ΔTm) as measured by differential scanning calorimetry (DSC) on binding of inhibitors to PR1 provides a convenient qualitative assessment of the relative strength of inhibitor binding. 24 25 Unliganded PR1N gave a Tm of 64.3°C at pH 4.6 in 4–5 mM acetate buffer [Fig. 4(A)], which is slightly lower than that of 65.7°C observed for PR1M under similar conditions. 24 In the presence of a twofold molar excess of DRV, the Tm increases to 89.3°C. The ΔTm of 25°C for PR1N in the presence of DRV is comparable to that of 22.4°C for PR1M 24 and is consistent with tight binding of DRV to PR1N. Isothermal titration calorimetry (ITC, see below) confirmed this result, although only an upper limit for the inhibitor dissociation constant could be determined. Similarly, PR1N exhibited large ΔTm values of 20.7, 20.6, and 16.8°C, respectively, on binding to the clinical inhibitors atazanavir (ATV), saquinavir (SQV), and the symmetrical inhibitor DMP323 26 [Fig. 4(B)]. These ΔTm values are very similar to ΔTms of 20.8, 19.3, and 15.3°C observed for PR1M with the same inhibitors. 25 Thus, both these mature proteases bind inhibitors with comparable, high affinities, and consequently PR1N cannot be classified as a drug-resistant variant despite the many residues in common with minor drug-resistance mutations (DRMs) of PR1M.
Figure 4.
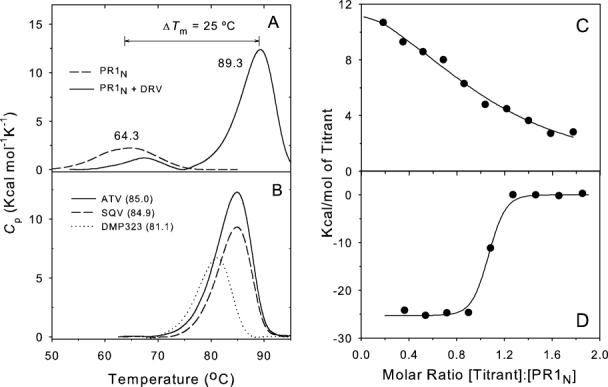
Calorimetric characterization of interaction of PR1N with inhibitors. A: DSC thermograms of mature PR1N in the absence of inhibitor and in the presence of approximately twofold molar excess of DRV. The weak thermal transition (small apparent ΔH) in the absence of inhibitor was explained by autoproteolysis during the course of the DSC experiment. B: DSC thermograms of mature PR1N in the presence of approximately twofold molar excess of the clinical inhibitors ATV (solid line) and SQV (dashed line) and the symmetrical inhibitor DMP323 (dotted line). Tm values (°C) are in parentheses. C: Isothermal titration calorimetry of PR1N (4.4 μM) in 25 mM Tris buffer, pH 7.0, containing 50 mM NaCl, at 28°C with 40 μM acetyl pepstatin. The KA determined for acetyl pepstatin is (5.6 ± 1.8) × 105M−1. D: Displacement titration of the PR1N-acetyl pepstatin complex with 40 μM DRV in the presence of 150 μM acetyl pepstatin gives an apparent binding constant ≥ 1.0 × 108M−1 for DRV, from which the KA for DRV in the absence of acetyl pepstatin is estimated to be ≥ 8 × 109M−1 (see text).
The binding constant for DRV was too large for ITC measurement by displacement titration 27 of the weakly binding inhibitor acetyl pepstatin; a lower limit for the binding constant was estimated to be ∼8 × 109M−1 [Fig. 4(B,C)]. This corresponds to a ligand dissociation constant (KL) ≤ 120 pM; thus, for PR1N this dissociation constant may be within an order of magnitude or less of the 5–10 pM 14 28 reported for the PR1M/DRV complex. The inability to obtain sufficient data for accurate estimation of KL for DRV by displacement titration results in part from the lower affinity of PR1N for acetyl pepstatin under the present conditions at pH 7 (KL = 1.8 μM), relative to PR1M (KL = 0.25 μM at pH 527), such that at 100–500 μM concentrations it does not adequately compete with the strongly binding DRV.
PR1N sequence analysis
The polymorphic sites of PR1N are mapped onto the structure of the protease dimer shown in Figure 1, and positions of major and minor DRMs and natural variations observed in PR1M are indicated (Fig. 5). The polymorphisms do not alter residues in the active-site cavity or flexible flaps. Most of them are located at distal regions, except for M36I, S37Q, P39E, and R41K at the hinge of the flaps, whereas H69R is the only substitution at the intersubunit interface. None of the polymorphic substitutions in PR1N are selected as major DRMs as defined by the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu/) and the International AIDS Society-US panel for antiretroviral resistance. 13
Figure 5.
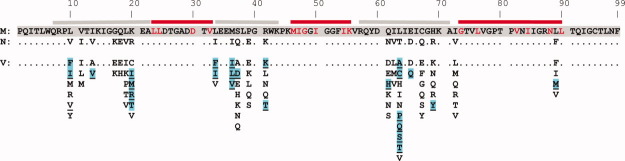
The wild-type PR1M sequence is aligned with PR1N. Only the amino acid differences relative to PR1M are indicated in PR1N. The red and gray lines on top indicate Stanford database DRM hot spots (major DRMs shown in red letters) and areas of greatest variability among the sequences, respectively. Note there is very little overlap between the natural variability and major DRM sites. Natural variations are suggested to coselect with major DRMs upon drug pressure to offset the detrimental effect of major DRMs alone. 12 13 V lists all the natural variants and DRMs selected under drug pressure in PR1M (extracted from Ref. 11) for the 20 positions which vary in PR1N. Natural variations in PR1M are listed alphabetically, selected DRMs are indicated in cyan, and residues common to both are underlined.
All of the substitutions in PR1N occur in the regions where natural variations occur among the proteases (Fig. 5). 11 However, these polymorphisms include changes at sites of minor resistance mutations, which likely arise as a result of natural selection together with the major DRMs rather than direct selection by drug pressure. 12 13 These minor DRMs persist in the drug-resistant strains because they provide a survival advantage by compensating for the reduced fitness of the enzyme (catalytic competence to undergo autoprocessing and process the polyproteins essential for proper virus assembly) arising from the selection of major DRMs. L10V appears as a minor mutation in PR1M for the drug ATV. K20R is observed as a minor mutation in PR1M for ritonavir-boosted ATV, indinavir, lopinavir, and tipranavir. Similarly, the M36I mutation is observed in isolates from patients treated with ATV, indinavir, nelfinavir, and tipranavir. The K20R mutation in protease occurs rarely in the absence of the M36I mutation, whereas M36I is found in the absence of K20R; this implies that the M36I mutation generally precedes K20R, 29 and the two substitutions may act synergistically during the selection of major DRMs. Also, the I62V mutation is implicated in resistance to ATV and SQV. Mutations at 13 and 69 are suggested to be associated with resistance to tipranavir, whereas mutations at residues 63 and 89 are observed when using lopinavir and DRV, respectively. 12 13
Overall structural comparison of DRV-bound PR1N and PR1M
PR1N was crystallized in complex with the PI DRV in the orthorhombic space group P212121 with one protease dimer in the asymmetric unit. The crystallographic data and refinement statistics are summarized in Table I. The atomic resolution (1.09 Å) structure showed excellent electron density for all the protease residues, which enabled unambiguous modeling of all the polymorphic substitutions in PR1N. A representative electron density map for L89′F in monomer B is shown in Figure 6(A). The mean B-factor for the main-chain atoms of the protein is 12.1 Å2. The average B-factors for the bound DRV inhibitor and the solvent molecules are 10.5 and 27.7 Å2, respectively. In total, 35 alternate conformations were modeled for residues in the crystal structure with monomer A containing 21 alternate conformations in comparison to 14 in monomer B. The main-chain atoms of Ile50, Gly51, Ile50′, and Gly 51′ in the two subunits exhibited two alternate conformations with relative occupancies of 0.6 and 0.4. The carbonyl groups of Ile50 and Ile50′ can flip 180° and retain a hydrogen bond with the amide groups of Gly51′ and Gly51 linking the tips of the two flaps.
Table I.
Crystallographic Data Collection and Refinement Statistics
| PR1N | |
|---|---|
| Space group | P212121 |
| a (Å) | 45.94 |
| b (Å) | 53.37 |
| c (Å) | 66.09 |
| Resolution range | 50–1.09 |
| Total observations | 233,527 |
| Unique reflections | 64,819 |
| Completeness | 93.0 (57.6)a |
| <I/σ(I)> | 14.1 (2.2) |
| Rsym (%)b | 9.6 (33.6) |
| Refinement resolution range | 10–1.09 |
| R (%)c | 16.48 |
| Rfree (%)d | 21.68 |
| Number of solvent | 146.3 |
| Mean B-factor (Å2) | |
| Main chain | 12.2 |
| Side chain | 19.2 |
| Inhibitor | 10.5 |
| Solvent | 27.3 |
| RMS deviations from ideality | |
| Bond length (Å) | 0.013 |
| Angle distance (Å) | 0.035 |
Values in parentheses are given for the highest resolution shell (1.11–1.09).
Rsym = Σhkl|Ihkl − 〈Ihkl〉|/ΣhklIhkl.
R = Σ||Fobs| − |Fcal||/Σ|Fobs|.
Rfree = Σtest||Fobs| − |Fcal||/Σtest|Fobs|.
Figure 6.
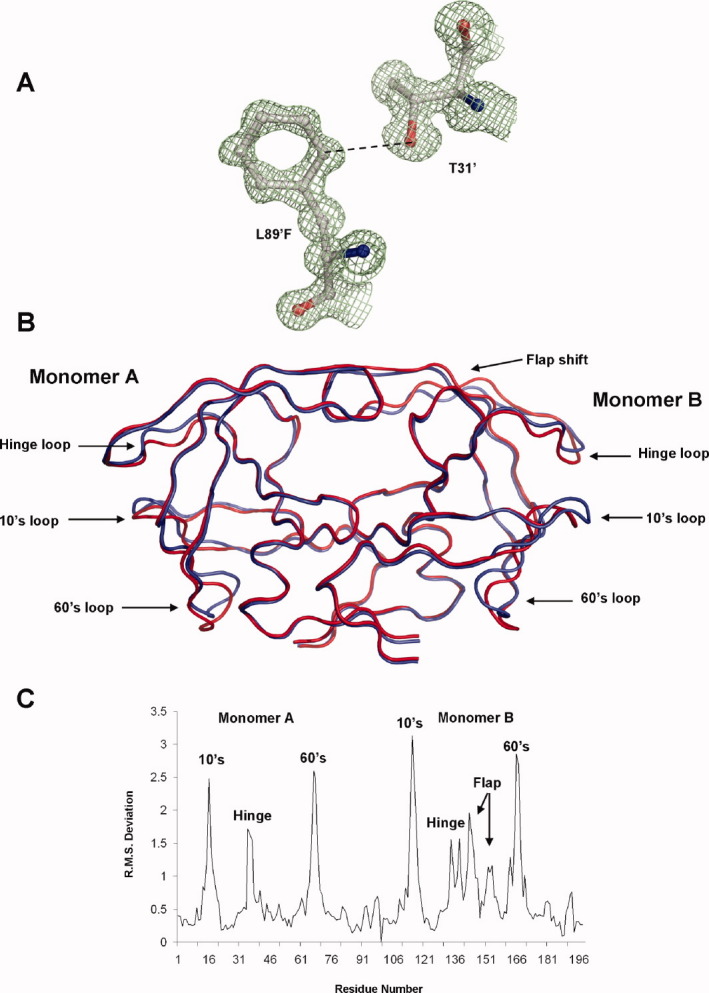
A: 2Fo − Fc electron density map contoured at 1.5σ for L89′F polymorphic substitution in monomer B of PR1N/DRV complex. The shortened CH…O interaction with Thr31′ is indicated by the broken line. B: Superposition of PR1N dimer in red against PR1M dimer in blue. The core of the PR1N structure including the active site shows excellent agreement with the PR1M structure. The two structures deviate in the 10s loop, 60s loop, and the flap hinge regions where the majority of polymorphic substitutions map. The flap of monomer A agrees well with that of PR1M structure, but the flap of monomer B deviates by ∼1 Å. C: The RMS deviations (Å) per residue when compared with those of PR1M are plotted for the Cα atoms of PR1N. The two monomers are numbered 1–99 and 101–199. The deviations corresponding to 10s, 60s, and hinge loops for both the monomers and the flap of monomer B are indicated. An interactive view is available in the electronic version of the article. PRO486 Figure 6
PR1N and PR1M share essentially the same overall structure, although the crystal structures were determined in different unit cells. The catalytic dimer of the PR1N/DRV complex can be superimposed with that of PR1M/DRV (2IEN) with RMSD of 0.86 Å for 198 topologically equivalent Cα atoms. This RMSD is larger than the average value of 0.56 Å reported for 73 PR1M dimers complexed with different inhibitors in a variety of lattices, 30 and the RMSD of < 0.6 Å observed in comparisons of PR1M from subtypes B, C, and F, 31 32 consistent with a significant effect arising from the 20 amino acid substitutions in PR1N. Within the dimer, monomer A of PR1N/DRV is more similar to the monomer A of PR1M/DRV with RMSD of 0.67 Å, whereas the B monomers show a larger RMSD of 0.84 Å for 99 equivalent Cα atoms. The largest deviations of more than 2.5 Å occur in the 10s and 60s loops [Fig. 6(B,C)], whereas another large deviation of more than 1.5 Å is seen for the flap hinge region (residues 36–41). Interestingly, the 10s loop is sandwiched between the 60s loop and the flap hinge permitting correlated structural perturbations due to the polymorphic substitutions that are all located in these neighboring loops. These deviations in the loops, flap hinge, and the flap itself are more pronounced in monomer B than in monomer A [Fig. 6(C)]. We cannot, however, rule out the possibility that these deviations may result from crystal packing effects.
The effect of M36I and K20R substitutions
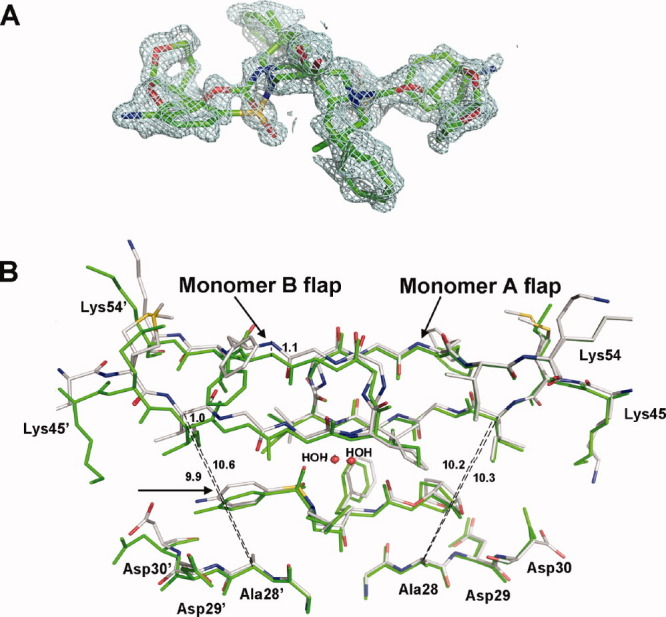
Changes in the 10s loop and the flap hinge propagate to shift the flap in monomer B away from the inhibitor. Polymorphisms in residue 36 in the hinge region of the flaps are associated with minor drug resistance, and M36I is the most frequently observed substitution in PR1M non subtype B. 33 34 Molecular dynamics studies indicate that mutation at residue 36 can alter the volume of the inhibitor-binding cavity. 35 Substitution of the shorter Ile36 in the PR1N/DRV structure causes part of the hinge loop to buckle inward to maintain all the interactions formed by Met36 in the PR1M/DRV complex [Fig. 7(A,B)]. This buckled loop is anchored by the K20R substitution in PR1N, which forms two hydrogen-bond interactions with the main-chain carbonyls of Glu34 and Glu35 and new van der Waals interactions with Ile36. The side chains of Arg20 and Glu35 exhibit alternate conformations in monomer A [Fig. 7(A)]. In monomer B, these residues show a single conformation, and the side chain of Gln37′ is flipped out in comparison with its conformation in monomer A, whereas Glu18′ forms an ion pair with Arg20′, a water-mediated hydrogen bond with the main-chain amide of Gln37′ [Fig. 7(B)], and van der Waals interactions with Ile36′ and the main-chain carbonyl of Gln37′. The hinge shows similar shifts when M36I is present in the crystal structures of PR1M subtypes F and B with different inhibitors, suggesting that the changes are independent of the type of inhibitor or space group. 31 36 In the case of monomer A, the hinge shift tapers off at residue Lys41 leaving the flap in an equivalent position in PR1N/DRV and PR1M/DRV. However, in monomer B, the shift propagates through the entire flap excluding the residues 49′–52′ at the tip, such that the flap residues are shifted more than 1 Å away from the bound inhibitor; however, only minor changes in the affinity for substrate and inhibitor were observed for PR1N relative to PR1M.
Figure 7.
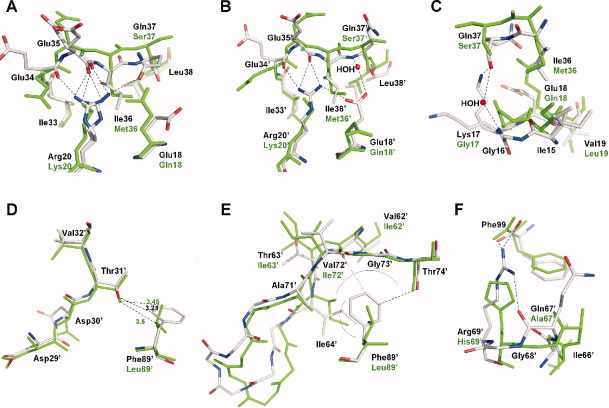
Atomic descriptions of regions in the PR1N/DRV complex showing significant changes from the PR1M/DRV complex. The PR1N residues are colored by element type, whereas PR1M residues are colored green. The hydrogen bond and ion pair interactions are shown as broken lines. The PR1N residue positions are labeled in black, and the polymorphic differences in PR1M are labeled in green. A: Key deviations at the site of M36I substitution in the monomer A of PR1N/DRV complex. The M36I substitution causes part of the hinge loop to buckle inward, which is anchored by another polymorphic substitution K20R. Flexibility of monomer A is indicated by the alternate conformations of Arg20 and Glu35. B: The effect of M36I substitution in monomer B of PR1N/DRV. All the residues show single conformations, and the hinge loop is more tightly held in place by polymorphic substitution Glu18′, which forms an ion pair with Arg20′ and water-mediated hydrogen bond with the main-chain amide of Gln37′. Glu18′ also forms a total of six van der Waals interactions with Ile36′ and the main-chain carbonyl of Gln37′. C: The buckling of the hinge loop shifts Ile15 resulting in downward movement of the 10s loop. The carbonyl of Gly16 at the tip of the loop is flipped by 180°. D: Superposition of PR1N/DRV and PR1M/DRV structures at the site of L89′F substitution in monomer B. The L89′F substitution results in a shortened CH…O interaction with Thr31′ that induces a shift of ∼0.5 Å in the position of inhibitor-interacting residues Asp29′, Asp30′, and Val32′. E: Superposition of the 60s loops of PR1N/DRV and PR1M/DRV structures in monomer B. L89′F substitution in PR1N makes van der Waals interactions with Ile64′. The residues C-terminal to Ile 64′ in the 60s loop exhibit large deviations when compared with those of the PR1M/DRV structure. The other end of the loop is fixed by the extensive new interactions between Phe89′ and the main-chain atoms of Val72′ and Gly73′ and the carbonyl of Thr74′. The side chains of residues 65′–70′ are omitted for clarity. The van der Waals interactions are indicated by an arc. F: In addition to L89′F, the H69′R substitution is also responsible for the large deviations seen in the 60s loop. The H69′R is the only substitution with intersubunit interactions in PR1N and sits close to the tip of the loop. H69′R in monomer B forms ionic and van der Waals interactions with the C-terminal residue Phe99 in monomer A of PR1N/DRV complex.
Changes in the 10s and 30s loops are interconnected and result in deviations of residues 15–20 in comparison with PR1M. The buckled M36I in the hinge of the flaps pushes down Ile15 and the rest of the 10s loop of both the monomers in PR1N/DRV, and thus Ile15 exhibits two alternate conformations in monomer A of PR1N/DRV, while the carbonyl of Gly16 at the tip of the loop is flipped by 180° [Fig. 7(C)]. The amide nitrogen of the G17K polymorphic residue in PR1N forms a water-mediated hydrogen bond with the carbonyl of the S37Q substitution in the flap hinge. The Lys17 side chain forms a hydrogen bond with the carbonyl of Gln37 in PR1N. In monomer B, the interaction of Glu18′ with Arg20′ and Ile36′ described in the previous paragraph prevents the contact between Gln37′ and the 10s loop.
Significance of L89F and H69R substitutions
Substitutions in the 60s loop and L89F propagate to perturb residues 29–32 that interact with inhibitor. Accessory mutations at the internal residue Leu89 are associated with DRV treatment. In the analysis of the crystal structure of HIV-1 protease subtype F, it was proposed that the substitution L89M of the longer methionine side chain pushes the Leu90 side chain toward the catalytic Asp25 as observed in the PR1M structure with major DRM L90M. 31 37–39 However, in the PR1N/DRV structure, the Leu90 side chain adjacent to L89F is in an identical conformation to that seen in PR1M/DRV. Instead, the bulkier Phe89 side chain results in a shortened CH…O interaction with the side-chain hydroxyl of Thr31, which lies between residues Asp30 and Val32 in the inhibitor-binding cavity [shown for monomer B in Fig. 7(D)]. The substitution of L89F shifts the main-chain atoms of residues 29–32 by ∼0.4–0.5 Å, thereby perturbing the inhibitor-binding site.
Additionally, the Phe89 side chain in PR1N makes extensive new interactions with residues 62, 64, and 71–74 resulting in dramatic shifts of the 60s loop by ∼0.7 and ∼1.3 Å at Ile64 in monomers A and B, respectively [Fig. 7(E)]. Phe89 forms 17 van der Waals interactions with the two conformations of Ile64 in monomer A of PR1N/DRV compared with only one in the PR1M/DRV structure, and Phe89′ has seven van der Waals interactions with the single conformation of Ile64′ in monomer B. Also, Phe89′ forms new CH…O interactions with the main-chain carbonyl of Val62′ [Fig. 7(E)]. The other end of the 60s loop is fixed by multiple interactions of Phe89′ with the main-chain atoms of Val72′ and Gly73′ as well as new CH…O interactions with the carbonyl of Ala71′ and Thr74′. Six of the polymorphic substitutions are present in this loop (residues 62–72). H69R, which lies at the tip of the 60s loop, is the only substitution forming intersubunit interactions in the PR1N dimer. In the PR1M/DRV complex, His69 forms an ionic interaction with the C-terminal carboxylate and five van der Waals interactions with the side chain of the C-terminal residue Phe99′. In the PR1N/DRV complex, Arg69 forms ionic interactions and eight van der Waals interactions with the C-terminal Phe99 of the other subunit [Fig. 7(F)]. Rearrangements at the tip of the 60s loop in PR1N/DRV allow the long Arg69 side chain to form a hydrogen bond with the main-chain carbonyl of Gln67.
PR1N interactions with DRV
The DRV molecule binds PR1N in two alternative conformations related by 180° with 50% occupancy each [Fig. 8(A)]. The conformation in which the bis-THF group of DRV interacts with Asp29′ and Asp30′ is designated as orientation A, and the other is defined as orientation B for the sake of description. The conserved water molecule that normally anchors the inhibitor and the two flaps is well resolved into two positions separated by ∼1 Å with each position interacting with one inhibitor orientation. Several residues contributing to the inhibitor-binding site exhibit alternate conformations as reported for the PR1M/DRV complex. 40 DRV was designed to form hydrogen bonds with the conserved region of PR1M to overcome drug resistance. 40 41 All the protease-inhibitor hydrogen-bond interactions observed in the PR1M/DRV structure are conserved in orientation A of the PR1N/DRV complex. However, in orientation B, the P2′ aminophenyl moiety of DRV is rotated as well as shifted from the corresponding position in the PR1M/DRV complex [Fig. 8(B)], which weakens its hydrogen bonds with the main-chain amide and carbonyl oxygen group of Asp30, although the interaction with the side chain of Asp30 is maintained. A similar rotation of the aminophenyl moiety that weakens DRV binding to the main chain of Asp30 has been reported for PR1M structures with single mutations of I50V, I54M, and V82A. 38 40 42 The size of the S2′ binding pocket of orientation B in the PR1N/DRV structure is increased over that in PR1M/DRV as shown by an ∼0.7 Å increase in the distance between Cαs of Ala28′ and Ile47′ [Fig. 8(B)] because of the shift of the monomer B flap, whereas no significant difference is seen in monomer A.
Figure 8.
A: Electron density map of DRV in PR1N/DRV complex. The 2Fo − Fc map was contoured at 1σ level. The DRV molecule binds PR1N in two alternative orientations related by 180° with 50% occupancy each. B: Superposition of DRV binding in orientation B at the active site of PR1N/DRV and PR1M/DRV complexes. The PR1N residues are colored by element type, whereas PR1M residues are colored green. An arrow indicates the aminophenyl moiety in DRV. The flap of monomer B in PR1N/DRV is shifted by 1 Å, which increases the size of the S2′ pocket as measured by the distance between Cαs of Ala28′ and Ile47′. The shift in the position of aminophenyl moiety is in agreement with the increased size of the S2′ pocket.
In vitro precursor processing
Extensive studies of the maturation of PR1M using various precursor analogs 8 11 have shown that the mature protease is cleaved from the Gag-Pol polyprotein by an ordered, stepwise, autocatalytic process likely requiring the initial, transient formation of a dimeric intermediate containing the two Asp residues essential for catalytic activity. 8 11 43 Before cleavage, the TFR-PR1M precursor is mainly a monomer with very low catalytic activity. 11 Because of the important role of the TFR in shifting the monomer–dimer equilibrium toward the dimer upon its cleavage from the PR domain, 10 11 43 the present studies focus on the N-terminal processing of the TFR-PR1N precursor, shown schematically in Figure 9(B). Near neutral pH, the predominant reaction pathway involves cleavage between the C-terminus of the TFR and the N-terminus of PR1N to give mature PR1N (band C) and full-length TFR (band D) with no significant build-up of intermediates [Fig. 9(A)]. The TFR is subsequently cleaved by the mature protease at the L34/W35 site (cf. Fig. 1). In contrast, at pH 4 [Fig. 9(D)], initial cleavage occurs preferentially at L34/W35 (bands B and F), and mature catalytic activity does not appear until a second cleavage occurs to release PR1N and the fragment TFR 35–61. Thus, a lag in the development of mature protease activity is observed when activity is measured as a function of time [Fig. 9(C)].
Figure 9.
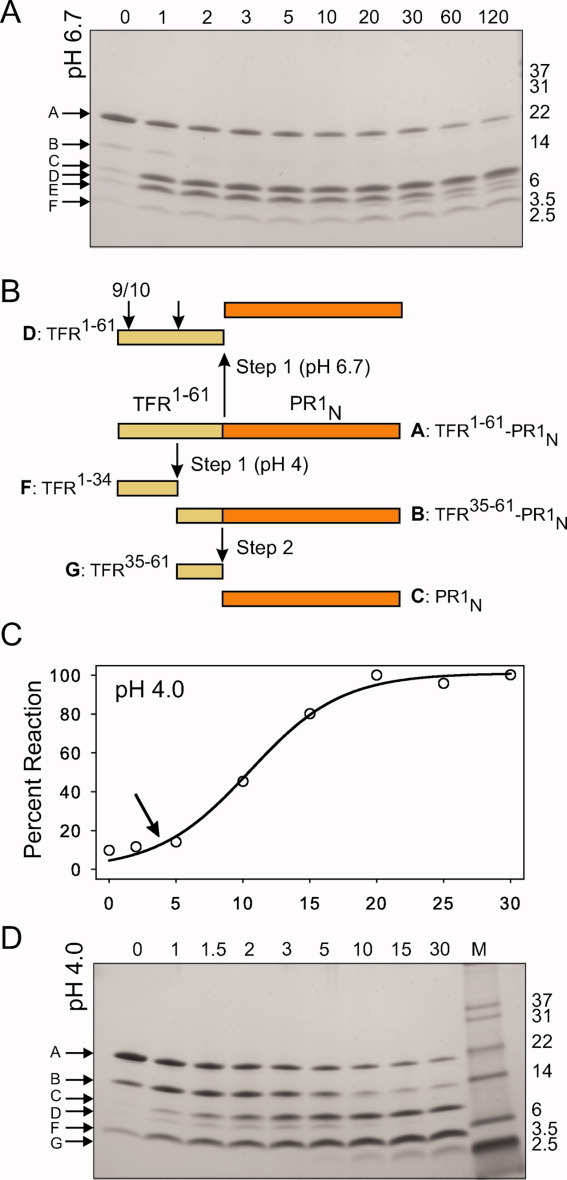
Pathways for the autocatalytic maturation of TFR-PR1N at pH 4 and 6.7. For experimental details, see text. A: Time course of the autoprocessing reaction at pH 6.7 showing a single major cleavage between the TFR 1–61 (band D) and the N-terminus of the protease domain (band C, PR1N). Subsequent cleavage of the TFR 1–61 occurs between residues 9/10 and 34–35 (band E, TFR 10–61 and band F, TFR 1–34). B: Schematic representation of the sites of sequential cleavages at pH 4 (downward arrows) and the predominant cleavage at pH 6.7 (upward arrow). Letter designations shown for the intermediates and products correspond to the band labels in Figure 9(A,D). C: Time course of appearance of mature protease activity at pH 4.0 measured by kinetic assays of samples taken at the times indicated in minutes. The lag (arrow) is due to the formation of intermediate B, which exhibits the same low activity as that of the full-length TFR-PR1N. Cleavage at the N-terminus of the protease is concomitant with the appearance of the mature protease (band C) and mature-like catalytic activity. D: Time course of processing at pH 4 showing initial cleavages at L34/W35 of the TFR (to give B and F) followed by cleavage of the resultant intermediate B to release the TFR 35–61 (G) and the mature protease (C).
These pH-dependent maturation pathways resemble those previously observed for PR1M with some distinct differences. 8 At pH values near neutrality, the predominant process is cleavage between the TFR and PR1 domains with no lag in the appearance of activity. At low pH, the initial cleavage of TFR-PR1M occurs at F8/L9 of the TFR releasing the N-terminal transframe octapeptide (TFP) of the TFR and an intermediate that is subsequently converted to the mature PR1M. Although these cleavages occur at different positions in TFR-PR1M and TFR-PR1N, it is of interest that both cleavage sites have neighboring acidic residues (Asp and Glu at P4 and P5 for TFR-PR1M and Glu at P2 for TFR-PR1N). A requirement for protonation of acidic residues has been suggested to explain the preference at low pH for cleavage at these positions. 44 It is most likely that processing at the N-terminus of PR1N also leads to a drastic decrease in the Kd, consistent with the appearance of mature-like protease activity as shown for both PR1M and PR1N autoprocessing, and the appearance of a stable tertiary fold of the mature dimer, as shown by NMR for PR1M autoprocessing. 10
Molecular mass analysis of PR1N and its precursor
Unlike PR1M, both PR1N and its precursor can be analyzed by size-exclusion chromatography under native conditions. Thus, it was possible to take advantage of this property to determine the molecular masses of these proteins by size-exclusion chromatography coupled with multiangle light scattering and refractive index measurements (SMR). Although it is not possible to conduct these measurements with TFR-PR1N in the absence of an inhibitor because of the intrinsic rapid autoprocessing of TFR-PR1N, addition of excess DRV to inhibit autoprocessing during folding enabled its mass analysis and comparison with the mature protein [Fig. 10(A)]. A retention volume of ∼15 mL for PR1N on a Superdex-75 column (1 cm × 30 cm) is similar to that observed for PR2 dimer under similar conditions, whereas the larger TFR-PR1N is less retained as expected. Both folded proteins form dimers in the presence of the inhibitor. This represents the first direct demonstration that a PI shifts the equilibrium of the PR precursor from a monomer to a dimer. This result appears inconsistent with the suggestion based on FRET data using chimeric constructs that DRV may act as a dimerization inhibitor at the precursor stage. 45
Figure 10.
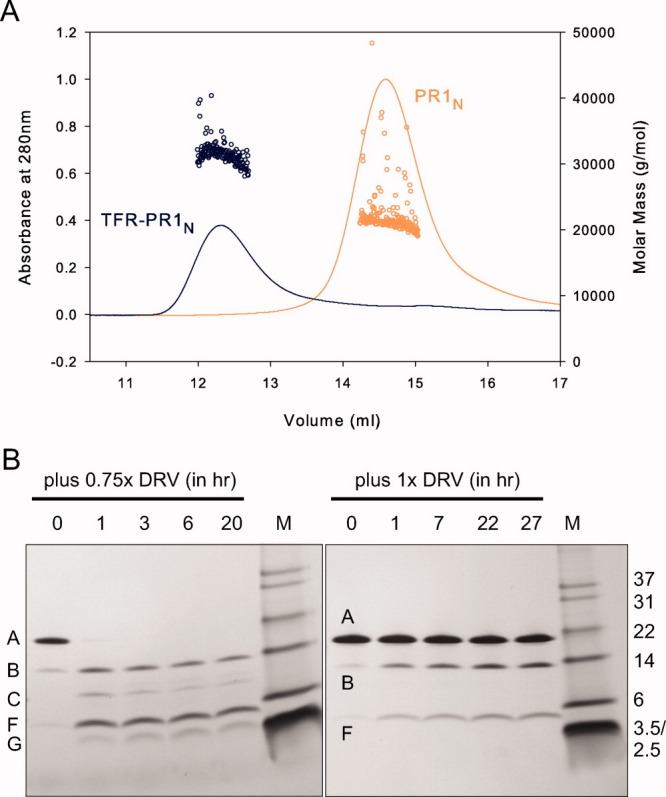
A: Molecular mass analysis of PR1N (orange) and TFR-PR1N (blue) by SMR. Calculated molecular masses are indicated by circles. PR1N and TFR-PR1N were folded in the presence of a two- and threefold molar excess of DRV, respectively, before loading onto the column. For chromatographic conditions, see Materials and Methods. Measured average masses (circles) are 22,450 ± 584 for PR1N (calcd. dimer 21,738) and 35,760 ± 1073 for TFR-PR1N (calcd. dimer 35,820, this includes an N-terminal methonine residue that is not excised during expression in E. coli). B: Effect of inhibitor DRV on the autocatalytic maturation of TFR-PR1N. Processing of TFR-PR1N at pH 4 in the presence of 0.75 (1.6 μM) and 1.0 (6.1 μM) molar equivalents of DRV relative to precursor (as dimer). Band labels are shown as in Figure 9. Note the much longer time scale relative to Figure 9(D) in the absence of inhibitor.
Inhibition of precursor processing by DRV
Because DRV can bind to the precursor to induce dimerization, it was of interest to assess if DRV, which exhibits a sub-nM affinity to the active site of mature PR1N, completely blocks the processing of TFR-PR1N. At pH 4, processing of TFR-PR1N is nearly complete within less than an hour in the absence of inhibitors [Fig. 9(C,D)]. In the presence of 0.75 molar equivalent of DRV, TFR-PR1N undergoes initial cleavage between Leu34 and Trp35 of the TFR, and the full-length precursor almost completely disappears within the first hour [Fig. 10(B), left, bands B and F]. However, further processing to mature PR1N is significantly slowed, although some formation of the mature enzyme can be inferred by the diminished intensity of the TFR 35–61-PR1N band over time as well as the appearance of a weak band corresponding to the mature protease (band C, most apparent at 1 h). At a 1:1 molar ratio of inhibitor to precursor dimers, the full-length precursor is still present even after 27 h [Fig. 10(B), right]. Cleavage proceeds at the L34/W35 site, but no mature PR1N is observed. Thus, cleavage at the N-terminus of the protease domain is inhibited much more strongly than the pH-dependent intermediate cleavage at L34/W35, resulting in accumulation of a minimally active TFR 35–61-PR1N intermediate. Cleavage at an analogous site (TFP/p6pol) in the processing of the Gag-Pol precursor from Group M is also poorly inhibited, even in the presence of 1 μM inhibitor in cell culture. 44 46 Structural studies of the TFR-PR1 precursor may provide insights into the differential inhibition of cleavage within the TFR when compared with the cleavage at the N-terminus of the protease.
Concluding Remarks
From the time of its discovery in the late 1980s, pioneering studies of the protease isolated from Group M HIV-1 and its subtypes, responsible for nearly 90% of the infections globally, have led to the development of potent PIs used in the treatment of AIDS patients. Resistance to PIs also develops quickly because of the natural polymorphisms that occur frequently in HIV, and this presents an ongoing challenge in treating patients even using the HAART regimen.
The appearance of mature-like catalytic activity of PR1N is concomitant with the cleavage at the N-terminus of the TFR-PR1N precursor, consistent with observations for TFR-PR1M autoprocessing. PR1N, in spite of having 20 substitutions in naturally variable residues and many small but cumulative structural changes in the 10s and 60s loops and flap hinge region that propagate to slightly enlarge the inhibitor-binding cavity, is comparable in its catalytic properties with PR1M. Although a number of these substitutions correspond to minor DRMs in PR1M, this study indicates that several inhibitors designed for PR1M bind to PR1N with similar affinities. This result is consistent with the suggestion 12 13 that these polymorphic mutations, or so-called minor DRMs, do not confer resistance to PIs on their own but may contribute to resistance in conjunction with major DRMs. Thus, it is possible that high levels of viral drug resistance may develop quickly in patients with Group N HIV-1 infection by selection for one or more of the major DRMs.
Apart from these similarities, PR1N bears some striking differences from PR1M. Unlike PR1M or its variants, PR1N has enhanced solubility at pH values above 5.8 and can be fractionated in its native form by size-exclusion column chromatography in simple buffers as reported recently for PR2. 19 This property permits easy assessment of the molecular masses of the species eluting from the column by SMR. Furthermore, the dimer dissociation constant (Kd) of PR1N is > 15-fold higher than for PR1M facilitating measurement of its Kd by a straightforward kinetic assay. The step-wise cleavage (L34/W35 site in TFR) occurring before the cleavage at the N-terminus of PR1N to produce a native-like active protease also differs in its location from that of the PR1M precursor.
Numerous rational approaches to inhibition of mature PR dimerization for the past decade or longer 11 have not reached fruition partly because (1) the mature PR1M forms a very tight dimer (Kd < 10 nM), requiring the putative inhibitors to have a higher affinity for effective dissociation and (2) it exhibits poor solubility and chromatographic properties, precluding use of a whole complement of biophysical and structural studies to investigate dimerization inhibition. PR1N demonstrates specific properties that may help to circumvent these difficulties in the screening and validation of potential dimerization inhibitors, 47 48 namely the higher Kd in conjunction with mass analysis by SMR under native conditions. Furthermore, it will be advantageous to develop reliable and simple approaches toward the inhibition of the protease at the level of its folding and dimerization before its autoprocessing. The observation that DRV, a sub-nM inhibitor of the mature PR, only partially inhibits the autoprocessing of TFR-PR1N at the L34/W35 site in TFR even at 6 μM DRV [Fig. 10(B)] suggests that the interactions of PIs with the active site of the protease precursor may differ from those with the mature protease. It is essential to develop methods to isolate sufficient quantities of the active precursor to validate the inhibition of its autoprocessing by PIs other than DRV. In addition, future studies possibly describing the structure of the protease precursor bound to a PI may reveal structural differences aiding the design of specific inhibitors of autoprocessing. Such inhibitors, by targeting an early event in polyprotein processing, may have the advantage of curtailing the occurrence of drug resistance with HAART.
Materials and Methods
Expression and purification
A synthetic gene encoding 160 amino acids of the TFR and the protease derived from the genomic sequence of HIV-1 Group N (GenBank accession number AY5326351) and optimized for expression in E. coli, termed TFR-PR1N, was cloned between the Nde1 and BamH1 sites of pET11a vector (Novagen, San Diego, CA) and transformed into E. coli BL-21 (DE3; Agilent Technologies, Santa Clara, CA). Cells were grown in Luria-Bertani medium, and expression was induced at 0.7 OD600nm with a final concentration of 2 mM IPTG for a period of 2 h. Cells derived from 1 L of culture were suspended in 80 mL of buffer A [50 mM Tris-HCl, pH 8, 10 mM ethylenediaminetetraacetic acid (EDTA) and 10 mM dithiothreitol (DTT)], followed by the addition of lysozyme (100 μg mL−1) and sonicated at 4°C. The insoluble recombinant protein was washed by resuspension in 70 mL of buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, 10 mM DTT, 2M urea, and 1% Triton X-100 and subsequently in buffer A. In all cases, the insoluble fraction was pelleted by centrifugation at 20,000g for 30 min at 4°C. The final pellet was solubilized in 50 mM Tris-HCl, pH 8.0, 7.5M guanidine hydrochloride (GnHCl), 5 mM EDTA, and 10 mM DTT to yield a concentration of ∼20 mg mL−1. A maximum of 30 mg of protein was applied on a Superdex-75 column (HiLoad 2.6 cm × 60 cm, GE HealthCare, Piscataway, NJ) equilibrated in 50 mM Tris-HCl, pH 8, 4M GnHCl, 5 mM EDTA, and 1 mM DTT at a flow rate of 3 mL min−1 at ambient temperature. Peak fractions were pooled and subjected to reverse-phase HPLC on POROS 20 R2 resin (PerSeptive Biosystems, Framington, MA) and eluted using a linear gradient from 99.95% water (v/v) and 0.05% TFA to 60% acetonitrile (v/v), 0.05% TFA (v/v), and 39.95% water (v/v) over a period of 16 min at a flow rate of 4 mL min−1. Peak fractions, eluting in an unusually broad peak, were combined, estimated for protein content at 280 nm, and stored at −70°C. A fraction of the protein was dialyzed against 12 mM HCl, concentrated to ∼2 mg mL−1, and stored at 4°C. The protein was folded according to the protocols described before. 18
Microgram quantities of the full-length TFR-PR1N precursor were purified from residual precursor that is trapped in the insoluble fraction without undergoing processing during expression using the same methodology as described above for PR1N. TFR-PR1N eluted as a distinct peak earlier than PR1N in the S-75 column under denaturing conditions, as described above, and was then subjected to reverse-phase HPLC. This allowed monitoring the autocatalytic maturation reaction of TFR-PR1N and its inhibition in vitro by enzyme assays and SDS-PAGE.
Enzyme assays
For determination of kinetic parameters, initial rates of enzymatic hydrolysis of the chromogenic peptide substrate IV [Lys-Ala-Arg-Val-Nle-(4-nitrophenylalanine)-Glu-Ala-Nle-NH2, California Peptide Research, Napa, CA] were measured by following the absorbance change at 310 nm in 50 mM sodium acetate buffer, pH 5, containing 250 mM sodium chloride, at 28°C. For each assay, 5 μL of a 0.19 mg mL−1 (8.74 μM) stock solution of the enzyme in 12 mM HCl was folded 18 by addition of 45 μL of 5 mM sodium acetate buffer, pH 6 (buffer A), immediately followed by 60 μL of 100 mM sodium acetate buffer, pH 5, containing 500 mM sodium chloride (buffer B). Reactions were initiated by adding varying amounts of water and substrate to give final substrate concentrations of 18–366 μM and a final enzyme concentration of 0.36 μM in a final volume of 120 μL. Observed initial rates (vi) of absorbance change were converted to M−1 s−1 by use of Δɛ310 = 1797 M−1 cm−1 for hydrolysis of the substrate, and the data were analyzed by use of the enzyme kinetics module of SigmaPlot 10 (Systat Software, Richmond, CA).
The effect of urea on enzymatic activity was determined by analogous kinetic measurements in the presence of urea at concentrations ranging from 0 to 4M. Dilutions of an 8M urea solution in buffer B with the same buffer containing no urea were used as the second buffer in the quench protocol described above. After protein folding, each enzyme solution was allowed to stand for 10 min at room temperature before assaying for activity by addition of substrate IV.
Evaluation of the dissociation constant
The above kinetic method was also used for determination of the dimer dissociation constant, Kd, from the dependence of the rate of hydrolysis of 430 μM substrate IV on enzyme concentrations from 40 to 250 nM in buffer solutions with the same composition and pH as above. The enzyme activity measured at each concentration was expressed as vi/[PR], where [PR] is the enzyme concentration as monomers. Even at the highest enzyme concentration used, a small fraction of monomers remains in equilibrium with the predominant dimers, and hence the maximum value of vi/[PR] (100% activity) was extrapolated and Kd was calculated as follows. The fraction (fu) of PR present as unfolded monomer 49 at a given total PR concentration (expressed as monomer units) is given by
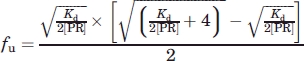 |
The fraction as dimer at a given PR concentration, fd = (1 − fu), is equal to Vx/Vmax, where Vx and Vmax are the values of vi/[PR] at each experimental PR concentration and at the maximum value of vi/[PR] where all the PR is dimeric. Thus,
and combining these relationships gives Eq. (1):
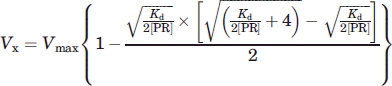 |
(1) |
The parameters Kd and Vmax were estimated from nonlinear curve fitting of Eq. (1) to the experimental data; observed rates were converted to percentages of the extrapolated Vmax as shown in Figure 3(B).
Differential scanning calorimetry
Measurements were performed using a MicroCal VP-DSC microcalorimeter. Mature PR1N was folded in the presence or absence of inhibitors by addition of 5.66 vol of 5 mM sodium acetate, pH 5.3, to 1 vol of PR1N (67 μM) in 12 mM HCl or by addition of 2.33 vol of 5 mM sodium acetate, pH 6 (with or without inhibitor), followed by 3.33 vol of 100 mM sodium acetate buffer, pH 5, to give a final protein concentration of ∼10 μM and a final pH of 4.5–5.0. For experiments in the presence of the PIs DRV, ATV, SQV, and the symmetrical inhibitor DMP323, 26 the final concentration of inhibitor was approximately twice that of PR1N (as dimer) after mixing. Thermal denaturation scans were begun at 20°C and run at a rate of 90°C h−1. The final temperature ranged from 85 to 100°C, depending on the observed Tm. Data were processed as described previously. 24 Values of the Tm were determined from the maxima of the transitions. In the absence of inhibitor, the apparent ΔH (integrated area under the transition peak) for PR1N was low [cf. Fig. 4(C)]. SDS-PAGE and ESI-MS of the sample after DSC (data not shown) showed that extensive autoproteolysis had occurred during the course of the DSC experiment giving rise to a major fragment (W38-F99) with a mass of 6711 Da, along with several smaller fragments.
Isothermal titration calorimetry
Measurements were performed at 28°C using a MicroCal high-precision iTC200 titration calorimeter. PR1N at a concentration of 4.4 μM (estimated from the titration endpoint with DRV) in 25 mM Tris buffer, pH 7, containing 50 mM NaCl, was titrated with 11 3.85-μL injections of 40 μM acetyl pepstatin in the same buffer, and the binding constant for acetyl pepstatin was determined using the Origin software provided with the instrument. Acetyl pepstatin (8 mM in 8 mM NaOH) was added to the solution recovered after titration to give a final concentration of 150 μM and a final DMSO concentration of 0.07%. The resultant solution was titrated as above with 40 μM DRV in the same buffer, containing 0.07% DMSO. The titration curve obtained on displacement of the weakly binding inhibitor, acetyl pepstatin, by DRV exhibited too steep a transition to permit accurate determination of the association constant 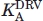 [see Fig. 4(B)]. A lower limit for
[see Fig. 4(B)]. A lower limit for 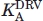 was estimated from the apparent binding constant (
was estimated from the apparent binding constant (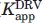 ) in the presence of acetyl pepstatin by use of the equation
) in the presence of acetyl pepstatin by use of the equation 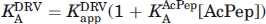 , where
, where 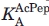 is the KA for acetyl pepstatin as determined in the first titration. 27
is the KA for acetyl pepstatin as determined in the first titration. 27
Crystallization, X-ray data collection, and structure determination
PR1N was prepared according to the dialysis protocol of protein folding 18 in the presence of DRV and concentrated. Crystals of the PR1N/DRV complex were grown in hanging drops at room temperature by mixing 1 μL of protein solution (6 mg mL−1 of protein) and 1 μL of reservoir solution (1.7M sodium chloride and 0.1M sodium acetate buffer at pH 4.4). The crystals were cryoprotected with 1.7M sodium chloride and 0.1M sodium acetate buffer at pH 4.4 and 30% glycerol. Diffraction data were collected at 100 K on beamline 22-ID (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory. The data were integrated and scaled with HKL2000. 50
The crystal structure was solved by molecular replacement with the published structure of PR1M in complex with DRV (2IEN 40) as the initial model using PHASER. 51 52 The inhibitor was fitted into unambiguous electron density. The model was subjected to several rounds of refinement in Shelx 53 and model building with Coot. 54 Solvent molecules were inserted at stereochemically reasonable positions using 2Fo − Fc and Fo − Fc maps at 1 and 3 sigma levels, respectively. The final refined model has good protein geometry with no disallowed ϕ/ψ values on the Ramachandran plot. Molecular figures were prepared with Molscript, Raster3D, and PyMOL (http://www.pymol.org).
In vitro precursor processing
TFR-PR1N in 12 mM HCl was folded by addition of 4 vol of buffer A (5 mM sodium acetate, pH 6), followed immediately by 5 vol of buffer B (100 mM Tris, pH 7, containing 100 mM NaCl, final pH 6.7, or 50–100 mM sodium formate, pH 4, containing 50–100 mM sodium chloride, final pH 4.1) to give a final concentration of 0.18 mg mL−1 TFR-PR1N (5 μM as dimer) and final buffer concentrations of 25–50 mM buffer, 25–50 mM sodium chloride. Processing in the presence of varying amounts of the inhibitor DRV used a similar protocol with a final concentration of 0.075–0.22 mg mL−1 TFR-PR1N (2.05–6 μM as dimer) at pH 4. Samples (15 μL) were removed at selected times, mixed with 6 μL SDS-PAGE sample buffer, and frozen immediately. Samples were analyzed on 10–20% gradient Tris-Tricine gels (Invitrogen, Carlsbad, CA) and Coomassie stained. Identity of intermediates and products from TFR-PR1N was confirmed by ESI-MS of samples that were removed at specific time points, diluted in 5% acetic acid, and frozen. The appearance of mature protease activity with time was also followed by kinetic assays. For each time point, the TFR-PR1N (2 μL of a 0.5 mg mL−1 solution in 12 mM HCl) was folded as above by addition of 48 μL of buffer A and 60 μL of 100 mM sodium formate buffer, pH 4, containing 100 mM NaCl. Processing was allowed to proceed for the desired times, and assays (see above) were initiated by addition of 10 μL of substrate IV to give a final concentration of ∼360 μM substrate and ∼0.25 μM TFR-PR1N (as dimer).
Assessment of the monomer–dimer status of PR1N and its precursor by SMR
Size-exclusion chromatography with detection by multiangle light scattering (DAWN EOS, Wyatt Technology, Santa Barbara, CA) and refractive index (OPTILAB DSP, Wyatt Technology) (SMR) was used for mass analysis of PR1N and TFR-PR1N in the presence of excess DRV. A solution (75 μL) containing 150 μg of PR1N or TFR-PR1N was folded by addition of 2.3 vol of 5 mM sodium acetate buffer, pH 6, containing a two- or threefold molar excess of DRV, followed by 3.3 vol of 0.5M ammonium acetate, pH 5.4. The sample (500 μL) was injected onto a Superdex-75 column (1.0 cm × 30 cm) pre-equilibrated with the same ammonium acetate buffer and eluted at a flow rate of 0.5 mL min−1 at room temperature. Molecular masses were calculated using the Astra software provided with the instrument.
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3MWS.
Acknowledgments
This research was authored, in whole or in part, by National Institutes of Health staff. The authors thank Annie Aniana for technical assistance and the staff at the Southeast Regional-Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory, for assistance during X-ray data collection. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. DRV, ATV, and SQV were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH and DMP323 from DuPont Pharmaceuticals.
Glossary
Abbreviations:
- ATV
atazanavir
- DRMs
drug-resistance mutations
- DRV
darunavir
- DSC
differential scanning calorimetry
- ESI-MS
electrospray ionization-mass spectrometry
- HAART
highly active antiretroviral therapy
- HIV-1
human immunodeficiency virus type 1
- ITC
isothermal titration calorimetry
- PI
clinical inhibitor of PR1M
- PR1
mature HIV-1 protease
- PR1M
mature HIV-1 protease from Group M
- PR1N
mature HIV-1 protease from Group N
- PR2
mature HIV-2 protease
- SMR
size exclusion chromatography coupled with multiangle light scattering and refractive index measurements
- SQV
saquinavir
- TFR
transframe region.
References
- 1.Bodelle P, Vallari A, Coffey R, McArthur CP, Beyeme M, Devare SG, Schochetman G, Brennan CA. Identification and genomic sequence of an HIV type 1 group N isolate from Cameroon. AIDS Res Hum Retroviruses. 2004;20:902–908. doi: 10.1089/0889222041725262. [DOI] [PubMed] [Google Scholar]
- 2.Kane CT, Montavon C, Toure MA, Faye MA, Ndiaye AG, Diallo AG, Ndoye I, Liegeois F, Delaporte E, Mboup S, Peeters M. Full-length genome sequencing of HIV type 1 group O viruses isolated from a heterosexual transmission cluster in Senegal. AIDS Res Hum Retroviruses. 2001;17:1211–1216. doi: 10.1089/088922201316912817. [DOI] [PubMed] [Google Scholar]
- 3.Plantier JC, Leoz M, Dickerson JE, De OF, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 4.Vallari A, Bodelle P, Ngansop C, Makamche F, Ndembi N, Mbanya D, Kaptue L, Gurtler LG, McArthur CP, Devare SG, Brennan CA. Four new HIV-1 group N isolates from Cameroon: prevalence continues to be low. AIDS Res Hum Retroviruses. 2010;26:109–115. doi: 10.1089/aid.2009.0178. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi J, Coffey R, Vallari A, Ngansop C, Mbanya D, Ndembi N, Kaptue L, Gurtler LG, Bodelle P, Schochetman G, Devare SG, Brennan CA. Identification of HIV type 1 group N infections in a husband and wife in Cameroon: viral genome sequences provide evidence for horizontal transmission. AIDS Res Hum Retroviruses. 2006;22:83–92. doi: 10.1089/aid.2006.22.83. [DOI] [PubMed] [Google Scholar]
- 6.Hatfield DL, Levin JG, Rein A, Oroszlan S. Translational suppression in retroviral gene expression. Adv Virus Res. 1992;41:193–239. doi: 10.1016/S0065-3527(08)60037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oroszlan S, Luftig RB. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 8.Louis JM, Weber IT, Tozser J, Clore GM, Gronenborn AM. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv Pharmacol. 2000;49:111–146. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 9.Leis J, Baltimore D, Bishop JM, Coffin J, Fleissner E, Goff SP, Oroszlan S, Robinson H, Skalka AM, Temin HM. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat Struct Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 11.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 protease: structure, dynamics, and inhibition. Adv Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 12.Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antiviral Res. 2010;85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1. Top HIV Med. 2008;16:138–145. [PubMed] [Google Scholar]
- 14.Brower ET, Bacha UM, Kawasaki Y, Freire E. Inhibition of HIV-2 protease by HIV-1 protease inhibitors in clinical use. Chem Biol Drug Des. 2008;71:298–305. doi: 10.1111/j.1747-0285.2008.00647.x. [DOI] [PubMed] [Google Scholar]
- 15.Levy Y, Caflisch A, Onuchic JN, Wolynes PG. The folding and dimerization of HIV-1 protease: evidence for a stable monomer from simulations. J Mol Biol. 2004;340:67–79. doi: 10.1016/j.jmb.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 16.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 17.Ceccherini-Silberstein F, Erba F, Gago F, Bertoli A, Forbici F, Bellocchi MC, Gori C, D'Arrigo R, Marcon L, Balotta C, Antinori A, Monforte AD, Perno CF. Identification of the minimal conserved structure of HIV-1 protease in the presence and absence of drug pressure. AIDS. 2004;18:F11–F19. doi: 10.1097/01.aids.0000131394.76221.02. [DOI] [PubMed] [Google Scholar]
- 18.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J Biol Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 19.Louis JM, Ishima R, Aniana A, Sayer JM. Revealing the dimer dissociation and existence of a folded monomer of the mature HIV-2 protease. Protein Sci. 2009;18:2442–2453. doi: 10.1002/pro.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis JM, Oroszlan S, Tozser J. Stabilization from autoproteolysis and kinetic characterization of the human T-cell leukemia virus type 1 proteinase. J Biol Chem. 1999;274:6660–6666. doi: 10.1074/jbc.274.10.6660. [DOI] [PubMed] [Google Scholar]
- 21.Mildner AM, Rothrock DJ, Leone JW, Bannow CA, Lull JM, Reardon IM, Sarcich JL, Howe WJ, Tomich CS, Smith CW. The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33:9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- 22.Ishima R, Torchia DA, Lynch SM, Gronenborn AM, Louis JM. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J Biol Chem. 2003;278:43311–43319. doi: 10.1074/jbc.M307549200. [DOI] [PubMed] [Google Scholar]
- 23.Szeltner Z, Polgar L. Conformational stability and catalytic activity of HIV-1 protease are both enhanced at high salt concentration. J Biol Chem. 1996;271:5458–5463. doi: 10.1074/jbc.271.10.5458. [DOI] [PubMed] [Google Scholar]
- 24.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active-site D25N mutation on the structure, stability and ligand binding of the mature HIV-1 protease. J Biol Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayer JM, Louis JM. Interactions of different inhibitors with active-site aspartyl residues of HIV-1 protease and possible relevance to pepsin. Proteins. 2008;75:556–568. doi: 10.1002/prot.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam PY, Jadhav PK, Eyermann CJ, Hodge CN, Ru Y, Bacheler LT, Meek JL, Otto MJ, Rayner MM, Wong YN. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science. 1994;263:380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]
- 27.Velazquez-Campoy A, Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat Protoc. 2006;1:186–191. doi: 10.1038/nprot.2006.28. [DOI] [PubMed] [Google Scholar]
- 28.King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Bethune MP, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman NG, Schiffer CA, Swanstrom R. Covariation of amino acid positions in HIV-1 protease. Virology. 2003;314:536–548. doi: 10.1016/s0042-6822(03)00484-7. [DOI] [PubMed] [Google Scholar]
- 30.Zoete V, Michielin O, Karplus M. Relation between sequence and structure of HIV-1 protease inhibitor complexes: a model system for the analysis of protein flexibility. J Mol Biol. 2002;315:21–52. doi: 10.1006/jmbi.2001.5173. [DOI] [PubMed] [Google Scholar]
- 31.Sanches M, Krauchenco S, Martins NH, Gustchina A, Wlodawer A, Polikarpov I. Structural characterization of B and non-B subtypes of HIV-protease: insights into the natural susceptibility to drug resistance development. J Mol Biol. 2007;369:1029–1040. doi: 10.1016/j.jmb.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 32.Coman RM, Robbins AH, Fernandez MA, Gilliland CT, Sochet AA, Goodenow MM, McKenna R, Dunn BM. The contribution of naturally occurring polymorphisms in altering the biochemical and structural characteristics of HIV-1 subtype C protease. Biochemistry. 2008;47:731–743. doi: 10.1021/bi7018332. [DOI] [PubMed] [Google Scholar]
- 33.Kantor R, Katzenstein DA, Efron B, Carvalho AP, Wynhoven B, Cane P, Clarke J, Sirivichayakul S, Soares MA, Snoeck J, Pillay C, Rudich H, Rodrigues R, Holguin A, Ariyoshi K, Bouzas MB, Cahn P, Sugiura W, Soriano V, Brigido LF, Grossman Z, Morris L, Vandamme AM, Tanuri A, Phanuphak P, Weber JN, Pillay D, Harrigan PR, Camacho R, Schapiro JM, Shafer RW. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:325–337. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kantor R, Katzenstein D. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 2003;5:25–35. [PubMed] [Google Scholar]
- 35.Ode H, Matsuyama S, Hata M, Neya S, Kakizawa J, Sugiura W, Hoshino T. Computational characterization of structural role of the non-active site mutation M36I of human immunodeficiency virus type 1 protease. J Mol Biol. 2007;370:598–607. doi: 10.1016/j.jmb.2007.04.081. [DOI] [PubMed] [Google Scholar]
- 36.Clemente JC, Moose RE, Hemrajani R, Whitford LR, Govindasamy L, Reutzel R, McKenna R, Agbandje-McKenna M, Goodenow MM, Dunn BM. Comparing the accumulation of active- and nonactive-site mutations in the HIV-1 protease. Biochemistry. 2004;43:12141–12151. doi: 10.1021/bi049459m. [DOI] [PubMed] [Google Scholar]
- 37.Hong L, Zhang XC, Hartsuck JA, Tang J. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance. Protein Sci. 2000;9:1898–1904. doi: 10.1110/ps.9.10.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovalevsky AY, Tie Y, Liu F, Boross PI, Wang YF, Leshchenko S, Ghosh AK, Harrison RW, Weber IT. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J Med Chem. 2006;49:1379–1387. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agniswamy J, Weber IT. HIV-1 protease: structural perspectives on drug resistance. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J Mol Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 41.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother. 2003;47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F, Kovalevsky AY, Tie Y, Ghosh AK, Harrison RW, Weber IT. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J Mol Biol. 2008;381:102–115. doi: 10.1016/j.jmb.2008.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature. 2008;455:693–696. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis JM, Dyda F, Nashed NT, Kimmel AR, Davies DR. Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease. Biochemistry. 1998;37:2105–2110. doi: 10.1021/bi972059x. [DOI] [PubMed] [Google Scholar]
- 45.Koh Y, Matsumi S, Das D, Amano M, Davis DA, Li J, Leschenko S, Baldridge A, Shioda T, Yarchoan R, Ghosh AK, Mitsuya H. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J Biol Chem. 2007;282:28709–28720. doi: 10.1074/jbc.M703938200. [DOI] [PubMed] [Google Scholar]
- 46.Lindhofer H, von der Helm K, Nitschko H. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology. 1995;214:624–627. doi: 10.1006/viro.1995.0074. [DOI] [PubMed] [Google Scholar]
- 47.Sluis-Cremer N, Tachedjian G. Modulation of the oligomeric structures of HIV-1 retroviral enzymes by synthetic peptides and small molecules. Eur J Biochem. 2002;269:5103–5111. doi: 10.1046/j.1432-1033.2002.03216.x. [DOI] [PubMed] [Google Scholar]
- 48.Uhlikova T, Konvalinka J, Pichova I, Soucek M, Krausslich HG, Vondrasek J. A modular approach to HIV-1 proteinase inhibitor design. Biochem Biophys Res Commun. 1996;222:38–43. doi: 10.1006/bbrc.1996.0694. [DOI] [PubMed] [Google Scholar]
- 49.Todd MJ, Semo N, Freire E. The structural stability of the HIV-1 protease. J Mol Biol. 1998;283:475–488. doi: 10.1006/jmbi.1998.2090. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 52.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 53.Sheldrick GM, Schneider TR. High resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]