Abstract
The human ATP-binding cassette (ABC) transporter superfamily consists of 48 integral membrane proteins that couple the action of ATP binding and hydrolysis to the transport of diverse substrates across cellular membranes. Defects in 18 transporters have been implicated in human disease. In hundreds of cases, disease phenotypes and defects in function can be traced to nonsynonymous single nucleotide polymorphisms (nsSNPs). The functional impact of the majority of ABC transporter nsSNPs has yet to be experimentally characterized. Here, we combine experimental mutational studies with sequence and structural analysis to describe the impact of nsSNPs in human ABC transporters. First, the disease associations of 39 nsSNPs in 10 transporters were rationalized by identifying two conserved loops and a small α-helical region that may be involved in interdomain communication necessary for transport of substrates. Second, an approach to discriminate between disease-associated and neutral nsSNPs was developed and tailored to this superfamily. Finally, the functional impact of 40 unannotated nsSNPs in seven ABC transporters identified in 247 ethnically diverse individuals studied by the Pharmacogenetics of Membrane Transporters consortium was predicted. Three predictions were experimentally tested using human embryonic kidney epithelial (HEK) 293 cells stably transfected with the reference multidrug resistance transporter 4 and its variants to examine functional differences in transport of the antiviral drug, tenofovir. The experimental results confirmed two predictions. Our analysis provides a structural and evolutionary framework for rationalizing and predicting the functional effects of nsSNPs in this clinically important membrane transporter superfamily.
Keywords: ABC transporters, genetic variation, structural modeling, multidrug resistance
Introduction
ATP-binding cassette (ABC) transporters are found in organisms from archaea to humans.1 In humans, the 48 ABC transporters are divided into seven families, named ABCA–ABCG. Substrates vary widely across the transporters and include chloride ions, lipids, and bile salts. Despite a diversity of substrates, ABC transporters from all organisms share a common overall domain architecture. They are composed of a transmembrane domain (TMD) and a globular nucleotide-binding domain (NBD) that associate to form a complete transporter. These domains can be encoded by separate genes, as in the case of the Escherichia coli vitamin B12 transporter2 and other bacterial ABC transporters, or they can be on the same polypeptide chain, as in human ABC transporters.3 A dimer of paired NBDs and TMDs is thought to be the minimum required unit for transport function, as seen in the recent complete structure of the multidrug resistance transporter (MRP) Sav1866 from the bacteria Staphylococcus aureus.4
Although there are few high-resolution structures of complete, four-domain ABC transporters, there are many structures of isolated NBDs from a variety of organisms. As of January 5, 2010, there were 70 structures clustered at 95% sequence identity in the Protein Data Bank (PDB) with the Structural Classification of Proteins (SCOP) classification “ABC transporter Adenosine Triphosphate (ATPase) domain-like” from 16 different species.5–7 These structures can be used as templates for comparative protein structure modeling of human ABC transporter NBDs without known atomic structures. Comparative models can provide clues to the mechanism of disease association through examination of the structural consequences of mutating particular residue positions as well as by enabling a view of structural conservation across the superfamily. Finally, previous work analyzing the impact of point mutations in breast cancer included structural characterization of SNPs using comparative modeling.7–10
At least two lines of evidence support an important role for ABC transporters in human health. First, they are efflux pumps for xenobiotics such as anticancer agents. For example, some human transporters, such as P-glycoprotein (ABCB1), breast cancer resistance protein (ABCG2), and members of the ABCC family, enable multidrug resistance in cancer therapies because they transport structurally diverse drugs out of cells; a recent review implicated 12 ABC transporters from four families in in vitro multidrug resistance in tissue culture-maintained cell lines.11 Second, defects in ABC transporters cause diseases. Eighteen ABC transporters are associated with a monogenic disease phenotype.12 Examples include ABCD protein family members, likely regulators of long chain fatty acid transport in the peroxisomes of human cells.13 One member of the ABCD family, ABCD1, is implicated in the neurodegenerative disorder adrenoleukodystrophy.13 The cystic fibrosis transmembrane regulator (CFTR) gene (ABCC7) is a chloride ion transporter and genetic aberrations in the protein are responsible for cystic fibrosis.14 Many disease-associated mutations are nonsynonymous single nucleotide polymorphisms (nsSNPs). Here, we focus on naturally occurring nsSNPs to examine their impact on transporter function.
Protein function can be altered by nsSNPs in many ways. Protein expression, folding, and stability can all be affected by single amino acid residue changes. In addition, post-translational modifications, translocation, and interactions with partners can be altered. In ABCC7, the ΔF508 variant is the most common cystic fibrosis-associated variant, seen in about two-thirds of all patients.14 CFTR proteins with this single residue deletion fail to be properly routed to the membrane and are degraded. Mutations in the ATP-binding site also disrupt function, such as G1302R in the Pseudoxanthoma elasticum-associated ABCC6 gene.15 Finally, mutations such as the Dubin-Johnson syndrome-associated Q1382R in ABCC2 are in a mobile region of the NBD (the “Q-loop”) that is hypothesized to transmit a conformational change between the NBDs and TMDs of an ABC transporter.16
While it is preferable to experimentally characterize the effects of each of the thousands of reported variants in ABC transporters, this approach is not feasible given the large number of variants. Of 61,170 variants in the human polymorphism table in Uniprot (January 5, 2010), 1072 are in ABC transporters.17 In addition, the difficulty of working with these large membrane proteins in the laboratory also makes them poor subjects for high-throughput experimental studies, hindering the determination of high-resolution structures for whole eukaryotic ABC transporters. Even the cystic fibrosis-associated mutations are mostly not functionally characterized.14 We propose a combined experimental and computational approach for annotation of both SNPs and putative functional regions in the globular domains of ABC transporters.
Functional impacts of nsSNPs can be rationalized or predicted with methods using protein sequences alone,18 methods using sequence and structure data,8,9,19 and methods using solely structural information.20 Here, a supervised learning approach is taken using the “random forest” method21 to combine a set of sequence and structural features of individual nsSNPs in human ABC transporters.
All nsSNPs from all human ABC transporter NBDs were collected and comprehensively mapped to comparative protein structure models of the NBDs. Multiple sequence alignments and comparative models for NBDs from the 48 human ABC transporters without known atomic structures were generated. Variants previously annotated as “polymorphic” or “disease-associated” (275 in total) were mapped to the alignments and structures, and regions with strong disease association in multiple transporters were identified. The mapped, annotated nsSNPs were used to train multiple classifiers to distinguish between neutral and disease-associated nsSNPs. The classifiers were validated using annotated nsSNPs that were excluded from the training data. Finally, the best performing classifiers were used to predict the effects of 40 unclassified nsSNPs in seven transporters discovered by our Pharmacogenetics of Membrane Transporters (PMTs) project at University of California at San Francisco (http://pharmacogenetics.ucsf.edu/). Two of these predictions for the multidrug resistance-associated transporter MRP4 (ABCC4) were validated experimentally.
Results
Locating structural hot spots associated with disease
Using comparative models, alignments, and mutational data, three previously uncharacterized structural motifs were identified that are associated with disease in multiple ABC transporters (Fig. 1 and Table I).
Figure 1.

Functional hot spots and experimentally characterized nsSNPs mapped to the nucleotide binding domain (NBD) sequences of human MRP4. The previously identified nucleotide-binding loop and the ABC signature motif are indicated in black. C-loop 1, the ARA motif, and C-loop 2 are indicated in blue. Experimentally characterized nsSNPs, G487E and K498E (NBD1), and V1071I (NBD2) are indicated in red.
Table I.
Disease-associated nsSNPs at Three Structural Hotspots in Human ABC Transporter NBDs
| Gene | Disease | Position |
|---|---|---|
| ARA motif | ||
| ABCB11 | BRIC2 | A570T |
| ABCD1 | X-ALD | A616V |
| CFTR | CF | A559T |
| ABCC6 | PXE | R765Q |
| ABCC8 | HHF1 | R841G |
| ABCC8 | HHF1 | R1493Q |
| ABCC8 | HHF1 | R1493W |
| ABCD1 | X-ALD | R617C |
| ABCD1 | X-ALD | R617G |
| ABCD1 | X-ALD | R617H |
| CFTR | CF | R560K |
| CFTR | CF | R560S |
| CFTR | CF | R560T |
| ABCA1 | HDLD1 | A1046D |
| ABCB4 | ICP | A546D |
| C-loop 1 motif | ||
| ABCC8 | HHF1 | D1471H |
| ABCC8 | HHF1 | D1471N |
| CFTR | CBAVD | G544V |
| ABCC8 | HHF1 | G1478R |
| C-loop2 motif | ||
| ABCA4 | STGD1 | H2128R |
| ABCC8 | HHF1 | K889T |
| ABCD1 | X-ALD | R660P |
| ABCD1 | X-ALD | R660W |
| ABCA1 | HDLD2 | M1091T |
| ABCA4 | STGD1 | E2131K |
| ABCA12 | LI2 | E1539K |
| ABCA4 | STGD1 and CORD3 | E1122K |
| CFTR | CF | L610S |
| ABCC8 | HHF1 | L1543P |
| ABCA1 | Colorectal cancer sample; somatic mutation | A2109T |
| ABCC9 | CMD1O | A1513T |
| ABCD1 | X-ALD | H667D |
| CFTR | CF | A613T |
| ABCA1 | HDLD2 | D1099Y |
| ABCD1 | X-ALD | T668I |
| CFTR | CF | D614G |
| ABCA4 | STGD1 | R2139W |
| ABCA4 | STGD1 | R1129C |
| ABCA4 | ARMD2, STGD1, and FFM | R1129L |
Disease abbreviations are as follows: BRIC2, benign recurrent intrahepatic cholestasis type 2; X-ALD, X-linked adrenoleukodystrophy; CF, cystic fibrosis; PXE, Pseudoxanthoma elasticum; HHF1, familial hyperinsulinemic hypoglycemia-1; HDLD1, high density lipoprotein deficiency type 1; ICP, intrahepatic cholestasis of pregnancy; CBAVD, congenital bilateral absence of the vas deferens; STGD1, Stargardt disease type 1; HDLD2, high density lipoprotein deficiency type 2; LI2, ichthyosis lamellar type 2; CORD3, cone-rod dystrophy type 3; CMD1O, cardiomyopathy dilated type 1O; ARMD2, age-related macular degeneration type 2; FFM, fundus flavimaculatus.
All nsSNPs collected from Uniprot.17
Disease-associated variants in the ARA motif, a putative communication interface between the globular and membrane domains
The ARA motif forms a small partially buried α-helix. Mutations in this motif are associated with disease in seven human ABC transporters: CFTR, sulfonylurea receptor 1, ALD, MDR2, bile salt export pump, ABCA1, and MRP6 [Fig. 2(A) and Table I]. There are four total disease-associated positions in the motif. The putative interacting transmembrane region includes the intracellular coupling loops 1 and 2, identified in the structure of the E. coli BtuCD vitamin B12 transporter and the S. aureus transporter Sav18662,4,22 (Fig. 2). The motif is well conserved across bacterial and mammalian orthologs. Thus, the ARA motif may play a role in interactions between the TMDs and NBDs in ABC transporters.
Figure 2.
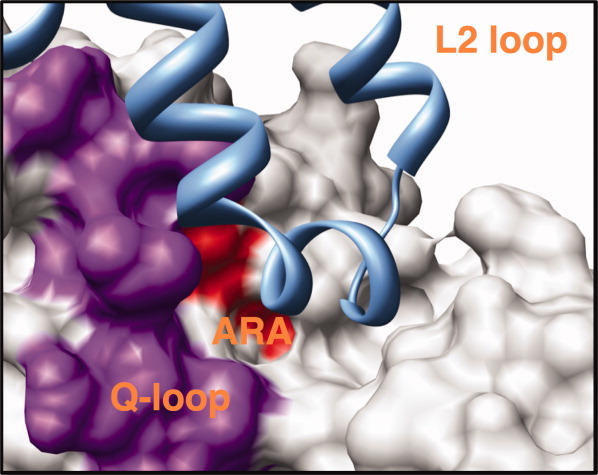
Disease-associated nsSNPs at a putative communication network between the NBD and the transmembrane helices of human ABC transporters. Mutations in the ARA motif, which forms a small, partially buried α-helix that may interact with the transmembrane helices, are associated with disease in seven human ABC transporters (Table I). A surface representation of the human ABCC NBD2 model structure is shown with the ARA motif colored in red, the mobile Q-loop in purple, and the TMD of the structure of the Staphylococcus aureus multidrug ABC transporter Sav1866 in light blue. The transmembrane segment that appears to interact with the Q-loop/ARA region is a portion of the L2 loop.2
C-loop 1: a putative allosteric loop between the membrane and globular domains
Sequences in ABCB and ABCC families include a conserved region that is not conserved in all human ABC transporters; we call this region C-loop 1 for “Conserved-loop 1”. There are four total disease-associated positions in this loop: three are from ABCC8 and associated with familial hyperinsulinemic hypoglycemia and one is from ABCC7 and associated with congenital bilateral aplasia of the vas deferens (Table II). In a bacterial multidrug resistance homolog,22 the loop is oriented toward the TMD.
Table II.
Predictions of the Functional Effects of 40 nsSNPs in ABC Transporters
| Comon name | HUGO name | Mutation | NBD | Prediction |
|---|---|---|---|---|
| BSEP | ABCB11 | E592Q | NBD1 | Neutral |
| BSEP | ABCB11 | N591S | NBD1 | Neutral |
| BSEP | ABCB11 | Q558H | NBD1 | Neutral |
| BSEP | ABCB11 | V444A | NBD1 | Neutral |
| BSEP | ABCB11 | E1186K | NBD2 | Disease |
| MDR1 | ABCB1 | P1051A | NBD2 | Neutral |
| MDR1 | ABCB1 | S1141T | NBD2 | Neutral |
| MDR1 | ABCB1 | T1256K | NBD2 | Disease |
| MDR1 | ABCB1 | V1251I | NBD2 | Neutral |
| MDR1 | ABCB1 | W1108R | NBD2 | Disease |
| MRP2 | ABCC2 | I670T | NBD1 | Disease |
| MRP2 | ABCC2 | L849R | NBD1 | Disease |
| MRP2 | ABCC2 | C1515Y | NBD2 | Disease |
| MRP3 | ABCC3 | D770N | NBD1 | Neutral |
| MRP3 | ABCC3 | K718M | NBD1 | Neutral |
| MRP3 | ABCC3 | T809M | NBD1 | Disease |
| MRP3 | ABCC3 | V765L | NBD1 | Disease |
| MRP3 | ABCC3 | Q1365R | NBD2 | Disease |
| MRP3 | ABCC3 | R1297H | NBD2 | Disease |
| MRP3 | ABCC3 | R1348C | NBD2 | Disease |
| MRP3 | ABCC3 | R1381S | NBD2 | Disease |
| MRP4 | ABCC4 | G487E | NBD1 | Disease |
| MRP4 | ABCC4 | K498E | NBD1 | Neutral |
| MRP4 | ABCC4 | R1220Q | NBD2 | Neutral |
| MRP4 | ABCC4 | T1142M | NBD2 | Neutral |
| MRP4 | ABCC4 | V1071I | NBD2 | Neutral |
| MRP6 | ABCC6 | I1330L | NBD1 | Neutral |
| MRP6 | ABCC6 | I742V | NBD1 | Neutral |
| MRP6 | ABCC6 | P664S | NBD1 | Neutral |
| MRP6 | ABCC6 | R724K | NBD1 | Neutral |
| MRP6 | ABCC6 | R769K | NBD1 | Neutral |
| MRP6 | ABCC6 | A1291T | NBD2 | Neutral |
| MRP6 | ABCC6 | E1369K | NBD2 | Neutral |
| MRP6 | ABCC6 | G1327E | NBD2 | Disease |
| MRP6 | ABCC6 | L1416R | NBD2 | Disease |
| MRP6 | ABCC6 | R1268Q | NBD2 | Disease |
| MRP6 | ABCC6 | R1461H | NBD2 | Disease |
| MXR | ABCG2 | I206L | NBD1 | Neutral |
| MXR | ABCG2 | P269S | NBD1 | Disease |
| MXR | ABCG2 | Q141K | NBD1 | Neutral |
C-loop 2: a putative interaction surface for intracellular partners
The second conserved sequence segment (10–12 residues long) begins with a previously described conserved histidine that plays a role in ATP binding and hydrolysis.23 Twenty disease-associated mutations in seven different transporters were identified. Only one mutation affects the conserved histidine, H2128R in ABCA4. C-loop 2 connects two β-sheets and is exposed to both the cognate NBD and to the intracellular environment (Fig. 1). Interestingly, the sequence of the loop is family-specific, with conserved residues varying from family to family. For example, in the ABCB family, C-loop 2 residues 1–5 [HRLST] are highly conserved, whereas in the ABCA family, the glutamate at position 5 and the arginine at position 12 are the most conserved residues. Mutations with disease association are distributed across the loop (Fig. 3).
Figure 3.

An intracellular surface region with conserved disease associations. C-loop 2 is shown in purple, mapped on the comparative models of NBD1 (dark grey) and NBD2 (light grey) in human P-gp (ABCB1). The previously identified functional histidine is shown in yellow.53 An alignment of sequences containing disease-associated mutations is shown at right with residue coloring indicating sequence conservation; each disease-associated nsSNP is indicated with a red star (Table II).
Predicting the effect of unannotated nsSNPs in ABC transporters
With structural rationalizations in hand, we now turn to predict the functional effects of uncharacterized nsSNPs. We optimized our previously developed classification schemes24,25 for discriminating between disease-associated and neutral nsSNPs in human ABC transporters. The best performer, the clinical classifier, was used to predict the effect of 40 naturally occurring nsSNPs from seven human ABC transporters (Fig. 4, Table II) (http://pharmacogenetics. ucsf.edu/).26 Three variants selected for experimental characterization are discussed below. Additional examples are discussed in Supporting Information.
Figure 4.
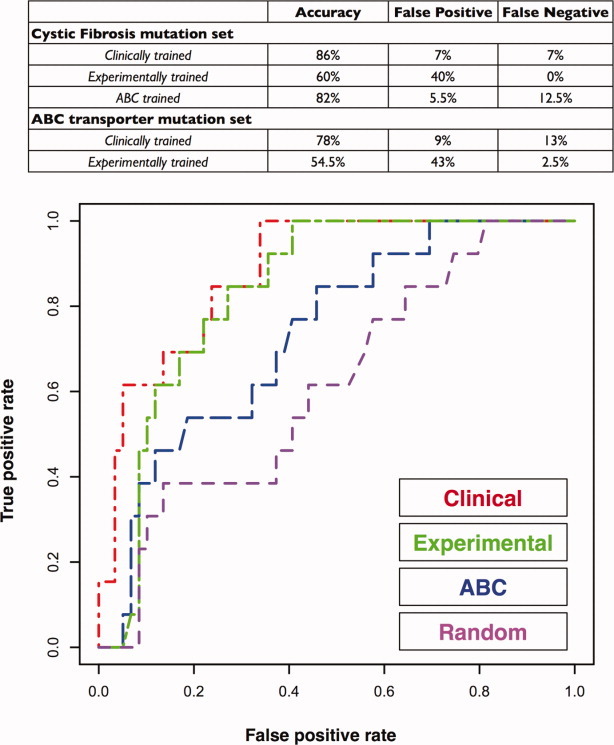
Performance of random forest classifier on a test set of 72 cystic fibrosis mutations. (A) Features used for discriminating between disease-associated and neutral nsSNPs. (B) A receiver-operator curve (ROC) showing the true-positive and false-positive rates for the clinical, experimental, and ABC transporter-trained random forests on the cystic fibrosis test set at left. The clinically trained random forest performs best on both the cystic fibrosis test set and the ABC transporter test set.
A predicted neutral lysine to glutamate variant in MRP4 (ABCC4)
The K498E nsSNP was found with a frequency of 0.025 in the African-American population. The classifier predicted it to be neutral, which can be rationalized as follows. It may be tempting to predict this nsSNP as deleterious because exchanging a glutamate for a lysine residue exchanges a positively charged and large residue for a negatively charged and smaller residue. However, in a comparative model of MRP4 NBD1 [Fig. 5(A)], this residue is on the surface and not near any known functional sites. In the absence of specific interactions with other ABC transporter domains or other proteins, it is likely that a charge change at a solvent-exposed position would not affect the stability or function of the protein.
Figure 5.
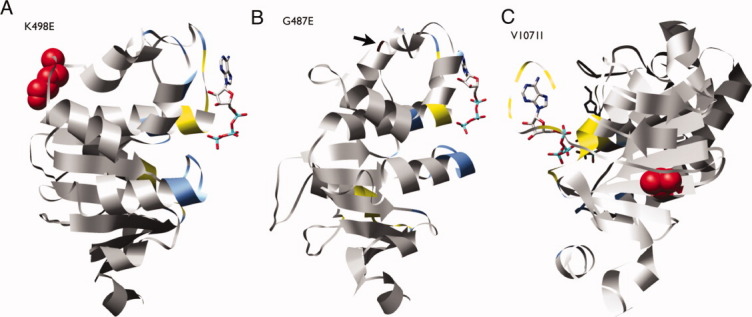
Structural models of MRP4 nsSNPs G487E, K498E, and V1071I. The models for G487E and K498E were based on the crystallographic structure of the human multidrug resistance protein MRP1 (PDB ID: 2CBZ). The model for V1071I was based on the structure of a multidrug resistance protein from Plasmodium yoelii (PDB ID: 2GHI). ATP coordinates are based on a structural alignment of the MRP4 models with the dimeric structure of two ABC NBDs from Methanocaldococcus jannaschii (PDB ID: 1L2T). Residues within 3.5 Å of the cognate NBD are colored light blue. Residues within 3.5 Å of an ATP molecule are colored yellow. An interactive view is available in the electronic version of the article. PRO491 Figure 5
A predicted deleterious glycine to glutamate variant in MRP4 (ABCC4)
The G487E nsSNP was found in only one Asian individual. The classifier predicted it to be deleterious. This residue occurs in a loop near the TMD surface [Fig. 5(B)]. The nonconservative mutation from a small noncharged to a larger negatively charged residue may perturb the loop conformation and thus affect the function of the protein.
A predicted neutral valine to isoleucine variant in MRP4 (ABCC4)
The V1071I nsSNP was found in one African-American. The classifier predicted it to be neutral. The residue is buried, and the substitution from valine to isoleucine is conservative [Fig. 5(C)]. This residue is also outside the range (5 Å) of known functional sites.
Reduced transport of tenofovir by MRP4 variants
Next, we experimentally tested the three MRP4 predictions. Efflux of [3H]-tenofovir (TFV) was compared in human embryonic kidney epithelial (HEK) 293 cells stably expressing MRP4 reference or variants (Fig. 6). TFV efflux was significantly greater in the cells expressing the MRP4 reference transporter compared with the empty pcDNA/FRT vector (mock)-transfected cells. The K498E variant had no effect on MRP4 transport, while the G487E variant showed moderate reduction in function (P < 0.05, Fig. 7). The V1071I variant dramatically decreased TFV transport to ∼40% of the reference (P < 0.001; Fig. 7). Thus, two of the three predictions were validated, while the V1071I nsSNP prediction was invalidated.
Figure 6.
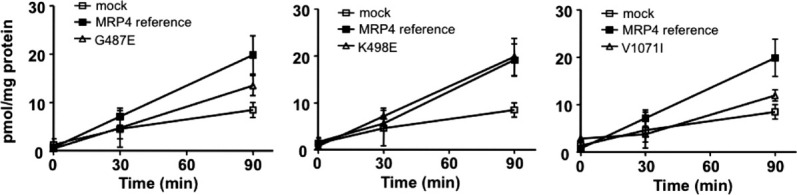
Efflux of [3H]-tenofovir by HEK cells expressing the wild type MRP4 and its variants. HEK293 cells stably expressing empty pcDNA5/FRT vector, the MRP4 wild type (reference), or its variants were preincubated with 1 μM [3H]-tenofovir disoproxil for 2 h under ATP-depleting conditions. After washing, the cells were supplemented with complete DMEM, and extracellular concentrations of tenofovir were determined at 0, 30, and 90 min. Data are mean ± standard deviation of triplicate determinations from one experiment and are representative of multiple experiments.
Figure 7.
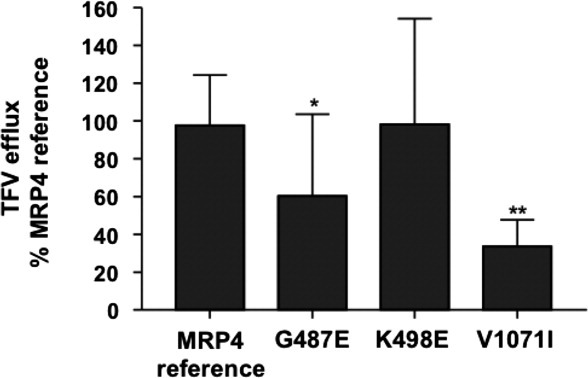
Effect of MRP4 variants on transport of [3H]-tenofovir. The percentage of tenofovir efflux (90 min) for each variant relative to the MRP4 reference was determined. Data are mean ± standard deviation calculated from nine to 22 determinations. Significant differences between the reference and the variants were assessed by one-way ANOVA and Bonferroni's multiple comparison test (*P < 0.05, **P < 0.001).
Expression of MRP4 mRNA, protein levels and localization
To assess whether the decreased TFV transport by MRP4 variants was associated with changes in expression, mRNA and protein levels were measured. No significant differences were noted in the MRP4 mRNA (data not shown) and total cellular protein levels (Supporting Information Fig. 1) between MRP4 and its variants. In contrast, the V1071I variant, which was predicted incorrectly, had significantly less expression on the plasma membrane compared with the reference (Supporting Information Fig. 2).
Discussion
The human ABC transporters are often studied individually rather than as a complete superfamily, due to their diverse functions, cellular locations, and disease associations.14,27–29 However, this approach discounts the clear evolutionary relationships between ABC transporters and misses the opportunity to extrapolate information from one superfamily member to others. Here, evolutionary conservation of sequence and structure in ABC transporters is exploited to identify conserved disease-associated regions in multiple transporters, to predict the impact of nsSNPs, and to apply what was learned from diverse ABC transporters to predict the functional consequences of nsSNPs in MRP4. We consider each point in turn and end with a discussion of the general applicability of our approach.
Clustering of disease-associated mutations reveals three previously unrecognized functional hot spots in ABC transporters
Individual nsSNPs can be thought of as probes of the functional role of the perturbed region in a protein. Here, we tested the hypothesis that clusters of disease-associated nsSNPs at particular positions in the ABC transporter NBDs may reveal functionally important regions. We found three such regions.
ARA motif: a putative communication interface between the NBD and TMD
The arginine in the ARA motif is disease associated when mutated to a number of other residue types, including the relatively conservative mutation to lysine in CFTR [Table I and Fig. 2(A)]. The positively charged arginine could contribute stabilizing contacts with either the mobile Q-loop or the intracellular loops 1 and 2 in the TMDs of ABC transporters [Fig. 2(B)]. Arginine is found in 30 out of 47 NBD1 and 15 out of 31 NBD2 domains of the human ABC transporters (Supporting Information Fig. 3). The importance of this conserved arginine in four transporters from three families combined with its putative role in the NBD–TMD interaction is consistent with previous work indicating that some domain interactions and communication networks are conserved across the ABC transporter superfamily.2,22,30,31
C-loop 1: a putative allosteric loop between the membrane and globular domains
Four nsSNPs in two ABC transporters in C-loop 1 are disease associated (Table I). The most conserved positions in the loop are in a five-residue segment with the consensus motif VG[ED][X]G, corresponding to a short loop closest to the TMD. The final glycine in the motif is absolutely conserved in all human ABCB and ABCC NBDs. We hypothesize that the two highly conserved glycines may be vital for functional loop conformation.
C-loop 2: a putative interaction surface for intracellular partners
The 20 disease-associated nsSNPs in C-loop 2 are from seven different ABC transporters and associated with 17 different human diseases (Table I and Fig. 3). The diversity of disease-associated genes suggests that this loop is functionally relevant across most of the human ABC transporter families; ABCG was the only family with no disease-associated nsSNPs in this motif. We hypothesize that this exposed, conserved loop may be an interaction site for an unidentified binding partner. The sequence diversity in this loop may be explained by the differential expression of ABC transporters in different cell types. For example, transporters such as ABCB1 are expressed in many different cell types,32 whereas ABCA4 is expressed almost exclusively in photoreceptor cells.33 Therefore, interacting partners may depend on cell type, possibly explaining the sequence diversity observed in C-loop 2.
Finally, C-loop 2 is part of an extensive network that stretches across the surface of the intracellular face of the NBDs. The distribution of disease-associated mutations is nonsymmetric in NBD1 and NBD2, rationalizing experimental work indicating that the two NBDs in an individual transporter are not functionally equivalent.34
Experimental characterization of ABCC4 nsSNPs and validation of a prediction model using a cell-based assay
Predictions of functional impact of three ABCC4 nsSNPs (G487E, K498E, and V1071I) were chosen for experimental validation using HEK293 cells stably transfected with the reference transporter and its variants. Previously, we reported that the G487E nsSNP showed significantly reduced transport of two antiviral agents, azidothymidine and adefovir, compared with the reference following transient transfection of HEK293T cells.29 Here, we quantify efflux of TFV directly to further analyze the impact of three nsSNPs in the NBDs of MRP4. The decreased transport of TFV by the G487E variant is in accordance with previous reports using alternate substrates.29,35 Using Xenopus laevis oocytes, it was shown that G487E influenced the transport of 6-mercaptopurine and adefovir.35 As predicted, this effect could be due to the impact of the G487E nsSNP on the conformation of the loop near the membrane, which affects transporter function. It is also consistent with the radical chemical change at this position, as quantified by the relatively higher Grantham value for G487E (D = 98).36
The V1071I variant is buried and lies outside the range of known functional sites. It was predicted to have no effect on MRP4 transport function. The V1071I MRP4 variant shows a significant reduction in transport function (Fig. 7). This functional consequence is likely caused by the observed decreased expression of this variant transporter on the plasma membrane (Supporting Information Fig. 2). The mechanism for the reduced plasma membrane expression of the V1071I variant remains unclear but may be related to changes in stability.
Finally, the lack of an effect of the K498E variant on TFV transport is consistent with our prediction that this surface-exposed site is not affected by a change in charge.
Experimental validation of predictions in other human ABC transporters
An additional three variants in ABCB1 (P-glycoprotein) were experimentally characterized in vitro, two in a yeast system37 and one in a human cell culture system,38 after our predictions were made. These tests provide further support for our approach.
The W1108R variant, predicted correctly by our algorithm to be deleterious (Table II), showed decreased resistance to the anticancer drugs daunorubicin and doxorubicin as well as to valinomycin, when compared with the reference P-glycoprotein in a yeast-based assay, suggesting a functional defect in the transporter. In contrast, the S1141T variant showed either normal or increased transport in the yeast assay, suggesting that the transporter was functioning properly.37 This variant was correctly predicted to be neutral.
Finally, in transfected HEK293T cells, the S1141T variant showed increased function, suggesting that the transporter is functioning normally, but also with substrate dependence as for the V1251I nsSNP.38 This variant was predicted correctly to be neutral.
The V1251I variant of P-glycoprotein showed decreased transport of BODIPY-FL-paclitaxel, but increased transport of calcein-AM in an assay using transfected HEK293T cells. This variant was predicted to be neutral, highlighting the complexities of assessing the impact of genetic variation on ABC transporter function. While substrate binding is currently thought to be limited to the TMDs, our experiments suggest that variation in the NBDs can alter the response of the protein to substrate. Further structural and biochemical characterization is necessary to resolve this issue.
The accuracy of our predictions for P-glycoprotein variants, which is in a different family from that of MRP4 reported here, suggests that our approach is applicable to multiple ABC transporter families.
Combined experimental and computational approaches for nsSNP functional characterization
To evaluate the disease implications of the large number of different nsSNP in ABC transporters that have been found in human populations, we need to know their functional consequences. However, it is impossible to characterize them all experimentally. Therefore, an accurate, efficient, and generally applicable approach is needed. We suggest that our combined experimental and computational approach is a good starting point.
The current variation data available for human transporters is skewed toward disease-associated mutations. There is, however, a wide range of phenotypes associated with these nsSNPs, from severe disease states to more mild impacts on health. Thus, more data on natural variation in human populations must be collected to more accurately determine what features make a nsSNP neutral or disease-associated.
The approach outlined here leverages bioinformatics methods to target nsSNPs of possible clinical interest for experimental validation. We expect that the numbers of nsSNPs discovered in human ABC transporters will increase dramatically39 and that combined computational and experimental approaches represent an efficient and productive framework for future studies on genetic variation in this superfamily.
Materials and Methods
Collection of SNP data
Three datasets of annotated nsSNPs were collected to train classifiers to discriminate between deleterious and neutral variants. The first dataset represents nsSNPs seen in a clinical context, such as those found in the CFTR gene of patients diagnosed with cystic fibrosis (the “clinical” dataset). The clinical dataset consists of 5211 annotated disease nsSNPs and 2910 neutral nsSNPs from Uniprot collected from the SwissProt index of sequence variation in human proteins (http://www.expasy.org/cgi-bin/lists?humsavar.txt). nsSNPs are annotated as “polymorphism,” “disease,” or “unclassified.” Only variants that altered amino acid residue identities were used, and unclassified mutations were discarded. The second dataset contains 275 nsSNPs in human ABC transporters and is a subset of the clinical dataset (the ABC transporter subset). Of these, 215 were disease associated and 60 were neutral. The third dataset represents nsSNPs in an experimental context, such as nsSNPs found to destroy the function of a protein in vitro (the “experimental” dataset). The experimental dataset consists of 1311 nonfunctional nsSNPs and 3644 functional nsSNPs assayed from two comprehensively mutated proteins: E. coli Lac repressor and Bacteriophage T4 lysozyme.
Finally, unannotated nsSNPs from ABC transporters were obtained from the PMT project at UCSF, which is part of the Pharmacogenetics Research Network. PMT investigators screened the exons of 10 ABC transporters in 247 DNA samples from ethnically diverse populations for genetic variation.26,40 The effects of 40 nsSNPs from seven transporters were predicted.
Model building and assessment
Models were built for 76 human ABC transporter NBDs, excluding those with atomic crystallographic structures: MRP1 NBD1 (PDB: 2CBZ),41 CFTR NBD1 (PDB: 2BBS, to be published), and TAP1 (PDB: 1jj7).42 Final models were selected based on sequence coverage and model score; 15 different structures were used as templates for one or more of the final models (Supporting Information Table I). All models are available via the MODBASE database of comparative models (http://salilab.org/modbase) and can be accessed or by searching for any of the human ABC transporter genes.43 We predicted the magnitude of model errors by the DOPE Z-score, a normalized pairwise atomic statistical potential based on globular protein structures in the PDB.44 The mean DOPE Z-score for all NBD1 domains was −0.68, where a Z-score of less than zero indicates a reliable model. The average length of the modeled NBDs was 231 residues. The average target–template sequence identity was 37% (Supporting Information Table I).
Models were used to calculate coarse-grained structural features to assess the likely effect of nsSNPs on the function of the protein. For this purpose, features such as correct surface versus core localization are needed. This level of granularity requires only a correct alignment between the target and template sequences, which is likely given the model assessment scores.44
For each amino acid residue substitution found in a protein sequence, we performed in silico mutation of its comparative model by applying the mutate_model routine in MODELLER, followed by a combination of conjugate gradient minimization and molecular dynamics with simulated annealing of the mutated residue and its neighbors.45
Structural alignments were performed using the SALIGN routine of MODELLER.46 Alignments were visualized using Jalview.47
nsSNP feature generation
A set of features previously demonstrated to perform well on a benchmark set of the comprehensively mutated globular proteins Bacteriophage T4 lysozyme and E. coli lac repressor, as well as for automated annotation of several thousand SNPs collected from dbSNP and OMIM24,25 were generated for each nsSNP. Structural features were generated using the MODPIPE automated comparative structure modeling protocol.43
Sequence features included changes in (1) residue charge, (2) polarity and (3) volume, and (4) Grantham score.36 Structure features included changes in the solvent accessibility for the wild type and the variant (5,6), changes in the relative solvent accessibility for the wild-type and variant residues (7,8), and whether or not a charge change was buried (9). Solvent accessibility features were calculated using DSSP.48 Evolutionary features include two measures of residue conservation in multiple sequence alignments: the relative entropy of a position (10) and the position conservation score (11), as calculated in Ref. 29. Superfamily level alignments for features were automatically calculated by SAMT2K,49via iterative searching of the protein sequences in the nr database from National Center for Biotechnology Information.12
Discriminating between known disease-associated and neutral mutations: random forest training and parameterization
The R package randomForest was used for all random forest training, parameterization, and testing.21 Random forests have been demonstrated to perform well in classification of nsSNPs.50,51 Random forests were trained using the clinical and experimental nsSNP datasets. The random forest with the highest cross-validated accuracy in the parameterization run was selected for testing.
Random forest testing and statistical analysis
The random forests were tested using several different sets of nsSNPs. The first test set was the ABC transporter dataset of 215 disease-associated and 60 neutral nsSNPs in human ABC transporters. This set is an independent test set for the clinical and experimental random forest because none of these nsSNPs were used to train either classifier. The clinical random forest has an accuracy of 78% on this set of nsSNPs (Fig. 4). The accuracy of the experimental classifier is 54.5%.
The second test set was a subset of the ABC transporter set consisting of 72 annotated nsSNPs from CFTR. This set includes 59 nsSNPs annotated as cystic fibrosis associated and 13 annotated as polymorphic. The clinical and experimental random forests, as well as an additional random forest trained on the ABC transporter data minus any CFTR nsSNPs, were tested on this data. Again, the clinical classifier performed the best, with an accuracy of 86% (Fig. 4). The experimental random forest had an accuracy of 60% and the ABC transporter random forest had an accuracy of 82%. A receiver–operator curve demonstrates the relative performances of the clinical, experimental, and ABC-trained random forests (Fig. 4); we generate a “random” random forest by scrambling the labels of the CFTR variants and using the clinical dataset-trained random forest (the best performer) for prediction. These results support previous work suggesting that the clinical set represents the best available information on the effects of nsSNPs in human proteins.52
Construction of MRP4 variants
Human ABCC4 cDNA was subcloned into the pcDNA5/FRT vector (Invitrogen, CA) as described previously.29 MRP4 variant constructs were obtained by site-directed mutagenesis using Phusion high-fidelity DNA polymerase (New England Biolabs, MA). The introduction of the mutations was verified by sequencing.
Construction of MRP4 variant-stable HEK293 cell lines
HEK293 Flp-In cells were seeded at 5 × 105 cells/well in six-well plates in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) without antibiotics. Twenty-four hours later, 3.6 μg pOG44 and 0.4 μg MRP4 reference or its variant plasmids were transfected into HEK293 Flp-in cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Empty pcDNA/FRT vector was transfected as a negative control. At 6 h after transfection, DMEM fresh media was added. The following day, cell selection was started in 75 μg/mL hygromycin (Invitrogen). Media was changed every 2–3 days, and selection of stable transfectants generally took 10–14 days. HEK293 cells stably expressing MRP4 reference or variants were cultured in DMEM with 10% FBS, penicillin (100 Units/mL), and streptomycin (100 μg/mL) at 37°C and 5% CO2.
Functional assays of MRP4 variants
Transport assays for MRP4 function were performed in triplicate in poly-d-lysine-coated 24-well plates (BD Biosciences, CA). Cells were incubated with 1 μM adenine-8-3H-TFV disoproxil (3.8 Ci/mmol, 98.1% purity; Moravek Biochemicals, CA) for 2 h, at 37°C in glucose-free DMEM supplemented with 10 μM NaN3 and 10 μM 2-deoxy-d-glucose. After accumulation, cells were washed with ice-cold phosphate-buffered saline (PBS) and supplemented with complete DMEM. Supernatant fractions were collected at 0, 30, and 90 min, and cells were washed and lysed with 800 μl/well of an aqueous solution of 10% sodium dodecyl sulfate and 1 N NaOH. Supernatants were added to Ecolite scintillation fluid (MP Biomedicals, CA) and extracellular amounts of TFV were counted by scintillation counting. The remaining cell lysate was used to determine the protein concentration with a BCA TM protein assay kit (Pierce Biotechnology, IL) to which TFV levels were normalized.
PCR analysis
Total RNA was extracted from the MRP4 stable cell lines using a RNeasy Mini Kit (Qiagen). RNA was quantitated using a UV spectrophotometer (NanoDrop Technologies, Wilmington, DE), and equal amounts of total RNA were reverse transcribed using SuperScript III First strand (Invitrogen) according to the manufacturer's instructions. The PCR reaction was performed using Phusion DNA polymerase with the following conditions: 1 min at 98°C, followed by 30 cycles of 10 s at 98°C, 20 s at 55°C, and 60 s at 72°C and a final elongation at 72°C for 10 min. The following primer pairs were used: ABCC4, 5′-AGTGTTACTGGAAATCTTA-3′ (forward) and 5′-ACGGACTTGACATTTTGGTTGG-3′ (reverse); glyceraldehyde-3-phosphate-dehydrogenase, 5′-AATCCCATCACCATCTTCCA-3′ (forward) and 5′-TGTGGTCATGAGTCCTTCCA-3′(reverse). PCR products were separated on a 4% agarose gel.
Western blotting
Total protein was isolated from HEK293 cells stably expressing MRP4 by lysing in 150 μM NaCl, 1% Igepal CA-630 (Sigma-Aldrich) with Halt TM protease inhibitor cocktail kit (Pierce). Lysates were centrifuged at 12,000g at 4°C for 10 min, and supernatants were collected. Proteins were separated by gel electrophoresis using 4–12% NuPage Bis-Tris gels (Invitrogen) and transferred to 0.2 mm nitrocellulose membranes (Biorad, CA). Blots were probed with a monoclonal antibody to MRP4 (M4-I10) and IRDye 800 anti-rat IgG (LI-COR, Lincoln, NE). Membranes were scanned and bands were quantified with the Licor Odyssey Infrared Imaging System.
Immunolocalization
HEK293 cells stably expressing MRP4 were seeded on poly-d-lysine-coated coverslips (BD Biosciences) in 24-well plates. After 24 h, cells on the coverslips were washed with PBS and fixed in ice-cold methanol for 5 min. After washing with PBS three times, the cells were permeabilized with 0.3% Triton X-100 in PBS for 5 min and blocked with 5% BSA (15 mg/mL). After staining with M4-I10 for 1 h, cells were incubated with AlexaFluor anti-rat IgG secondary antibody (Invitrogen) for 1 h at room temperature in the dark. Staining was visualized using a Zeiss Axioskop epifluorescence microscope.
Acknowledgments
The authors would like to thank Dr. Adrian S. Ray (Gilead Sciences, Inc.) for expert technical assistance with the MRP4 functional assay. They thank Drs. Kathleen Giacomini and Robert Stroud for helpful discussions on the work. They are also grateful to Hewlett-Packard, NetApps, IBM, Intel, Ron Conway, and Mike Homer for contributions to our computing hardware (AS).
Glossary
Abbreviations:
- ABC
ATP-binding cassette
- ARMD2
age-related macular degeneration type 2
- BRIC2
benign recurrent intrahepatic cholestasis type 2
- CBAVD
congenital bilateral absence of the vas deferens
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane regulator
- CMD1O
cardiomyopathy dilated type 1O
- CORD3
cone-rod dystrophy type 3
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- FFM
fundus flavimaculatus
- HDLD1
high density lipoprotein deficiency type 1
- HDLD2
high density lipoprotein deficiency type 2
- HEK293
human embryonic kidney epithelial 293
- HHF1
familial hyperinsulinemic hypoglycemia-1
- ICP
intrahepatic cholestasis of pregnancy
- LI2
ichthyosis lamellar type 2
- MDR
multidrug resistance
- MRP
multidrug resistance-associated protein
- NBD
nucleotide-binding domain
- nsSNP
nonsynonymous single nucleotide polymorphism
- PXE
Pseudoxanthoma elasticum
- SNP
single nucleotide polymorphism
- STGD1
Stargardt disease type 1
- TMD
transmembrane domain
- TFV
tenofovir
- X-ALD
X-linked adrenoleukodystrophy
References
- 1.Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- 2.Locher K, Lee A, Rees D. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 4.Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 6.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 7.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirkovic N, Marti-Renom MA, Weber BL, Sali A, Monteiro AN. Structure-based assessment of missense mutations in human BRCA1: implications for breast and ovarian cancer predisposition. Cancer Res. 2004;64:3790–3797. doi: 10.1158/0008-5472.CAN-03-3009. [DOI] [PubMed] [Google Scholar]
- 9.Karchin R, Monteiro AN, Tavtigian SV, Carvalho MA, Sali A. Functional impact of missense variants in BRCA1 predicted by supervised learning. PLoS Comput Biol. 2007;3:e26. doi: 10.1371/journal.pcbi.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karchin R, Agarwal M, Sali A, Couch F, Beattie MS. Classifying variants of undetermined significance in BRCA2 with protein likelihood ratios. Cancer Inform. 2008;6:203–216. doi: 10.4137/cin.s618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szakacs G, Ozvegy C, Bakos E, Sarkadi B, Varadi A. Role of glycine-534 and glycine-1179 of human multidrug resistance protein (MDR1) in drug-mediated control of ATP hydrolysis. Biochem J. 2001;356:71–75. doi: 10.1042/0264-6021:3560071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp S, Pujol A, Waterham HR, van Geel BM, Boehm CD, Raymond GV, Cutting GR, Wanders RJ, Moser HW. ABCD1 mutations and the X-linked adrenoleukodystrophy mutation database: role in diagnosis and clinical correlations. Hum Mutat. 2001;18:499–515. doi: 10.1002/humu.1227. [DOI] [PubMed] [Google Scholar]
- 14.Gadsby D, Vergani P, Csanády LS. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilias A, Urban Z, Seidl TL, Le Saux O, Sinko E, Boy CD, Sarkad B, Varadi A. Loss of ATP-dependent transport activity in Pseudoxanthoma elasticum-associated mutants of human ABCC6 (MRP6) J Biol Chem. 2002;277:16860–16867. doi: 10.1074/jbc.M110918200. [DOI] [PubMed] [Google Scholar]
- 16.Dalmas O, Orelle C, Foucher AE, Geourjon C, Crouzy S, Di Pietro A, Jault JM. The Q-loop disengages from the first intracellular loop during the catalytic cycle of the multidrug ABC transporter BmrA. J Biol Chem. 2005;280:36857–36864. doi: 10.1074/jbc.M503266200. [DOI] [PubMed] [Google Scholar]
- 17.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 21.Liaw A, Wiener M. Classification and regression by Randomforest. R News. 2002;2:18–22. [Google Scholar]
- 22.Dawson R, Locher K. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 23.Zaitseva J, Jenewein S, Jumpertz T, Holland B, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 25.Karchin R, Kelly L, Sali A. Improving functional annotation of non-synonymous SNPs with information theory. Pac Symp Biocomput. 2005:397–408. doi: 10.1142/9789812702456_0038. [DOI] [PubMed] [Google Scholar]
- 26.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, Kroetz DL, Ferrin TE, Clark AG, Risch N, Herskowitz I, Giacomini KM. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakos E, Homolya L. Portrait of multifaceted transporter, the multidrug resistance-associated protein 1 (MRP1/ABCC1) Pflugers Arch. 2007;453:621–641. doi: 10.1007/s00424-006-0160-8. [DOI] [PubMed] [Google Scholar]
- 28.Woodahl EL, Yang Z, Bui T, Shen DD, Ho RJ. Multidrug resistance gene G1199A polymorphism alters efflux transport activity of P-glycoprotein. J Pharmacol Exp Ther. 2004;310:1199–1207. doi: 10.1124/jpet.104.065383. [DOI] [PubMed] [Google Scholar]
- 29.Abla N, Chinn LW, Nakamura T, Liu L, Huang CC, Johns SJ, Kawamoto M, Stryke D, Taylor TR, Ferrin TE, Giacomini KM, Kroetz DL. The human multidrug resistance protein 4 (MRP4, ABCC4): functional analysis of a highly polymorphic gene. J Pharmacol Exp Ther. 2008;325:859–868. doi: 10.1124/jpet.108.136523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg MF, Velarde G, Ford RC, Martin C, Berridge G, Kerr ID, Callaghan R, Schmidlin A, Wooding C, Linton KJ, Higgins CF. Repacking of the transmembrane domains of P-glycoprotein during the transport ATPase cycle. EMBO J. 2001;20:5615–5625. doi: 10.1093/emboj/20.20.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newstead S, Fowler PW, Bilton P, Carpenter EP, Sadler PJ, Campopiano DJ, Sansom MS, Iwata S. Insights into how nucleotide-binding domains power ABC transport. Structure. 2009;17:1213–1222. doi: 10.1016/j.str.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clin Pharmacol Ther. 2007;81:265–269. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- 33.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 34.Procko E, Ferrin-O'Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 35.Janke D, Mehralivand S, Strand D, Godtel-Armbrust U, Habermeier A, Gradhand U, Fischer C, Toliat MR, Fritz P, Zanger UM, Schwab M, Fromm MF, Nurnberg P, Wojnowski L, Closs EI, Lang T. 6-Mercaptopurine and 9-(2-phosphonyl-methoxyethyl) adenine (PMEA) transport altered by two missense mutations in the drug transporter gene ABCC4. Hum Mutat. 2008;29:659–669. doi: 10.1002/humu.20694. [DOI] [PubMed] [Google Scholar]
- 36.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 37.Jeong H, Herskowitz I, Kroetz DL, Rine J. Function-altering SNPs in the human multidrug transporter gene ABCB1 identified using a Saccharomyces-based assay. PLoS Genet. 2007;3:e39. doi: 10.1371/journal.pgen.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gow JM, Hodges LM, Chinn LW, Kroetz DL. Substrate-dependent effects of human ABCB1 coding polymorphisms. J Pharmacol Exp Ther. 2008;325:435–442. doi: 10.1124/jpet.107.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang L. Three sequencing companies join the 1000 genomes project. Gastroenterology. 2008;135:336–337. doi: 10.1053/j.gastro.2008.06.053. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen TD, Gow JM, Chinn LW, Kelly L, Jeong H, Huang CC, Stryke D, Kawamoto M, Johns SJ, Carlson E, Taylor T, Ferrin TE, Sali A, Giacomini KM, Kroetz DL. PharmGKB submission update. IV. PMT submissions of genetic variations in ATP-binding cassette transporters to the PharmGKB network. Pharmacol Rev. 2006;58:1–2. doi: 10.1124/pr.58.1.1. [DOI] [PubMed] [Google Scholar]
- 41.Ramaen O, Leulliot N, Sizun C, Ulryck N, Pamlard O, Lallemand JY, Tilbeurgh H, Jacquet E. Structure of the human multidrug resistance protein 1 nucleotide binding domain 1 bound to Mg2+/ATP reveals a non-productive catalytic site. J Mol Biol. 2006;359:940–949. doi: 10.1016/j.jmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Gaudet R, Wiley DC. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP, Sali A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feyfant E, Sali A, Fiser A. Modeling mutations in protein structures. Protein Sci. 2007;16:2030–2041. doi: 10.1110/ps.072855507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, Shen MY, Kelly L, Melo F, Sali A. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 49.Karplus K, Barrett C, Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 50.Bao L, Cui Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21:2185–2190. doi: 10.1093/bioinformatics/bti365. [DOI] [PubMed] [Google Scholar]
- 51.Barenboim M, Masso M, Vaisman II, Jamison DC. Statistical geometry based prediction of nonsynonymous SNP functional effects using random forest and neuro-fuzzy classifiers. Proteins. 2008;71:1930–1939. doi: 10.1002/prot.21838. [DOI] [PubMed] [Google Scholar]
- 52.Care MA, Needham CJ, Bulpitt AJ, Westhead DR. Deleterious SNP prediction: be mindful of your training data! Bioinformatics. 2007;23:664–672. doi: 10.1093/bioinformatics/btl649. [DOI] [PubMed] [Google Scholar]
- 53.Ernst R, Kueppers P, Klein CM, Schwarzmueller T, Kuchler K, Schmitt L. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc Natl Acad Sci USA. 2008;105:5069–5074. doi: 10.1073/pnas.0800191105. [DOI] [PMC free article] [PubMed] [Google Scholar]


