Abstract
Objective
Healthcare organizations must de-identify patient records before sharing data. Many organizations rely on the Safe Harbor Standard of the HIPAA Privacy Rule, which enumerates 18 identifiers that must be suppressed (eg, ages over 89). An alternative model in the Privacy Rule, known as the Statistical Standard, can facilitate the sharing of more detailed data, but is rarely applied because of a lack of published methodologies. The authors propose an intuitive approach to de-identifying patient demographics in accordance with the Statistical Standard.
Design
The authors conduct an analysis of the demographics of patient cohorts in five medical centers developed for the NIH-sponsored Electronic Medical Records and Genomics network, with respect to the US census. They report the re-identification risk of patient demographics disclosed according to the Safe Harbor policy and the relative risk rate for sharing such information via alternative policies.
Measurements
The re-identification risk of Safe Harbor demographics ranged from 0.01% to 0.19%. The findings show alternative de-identification models can be created with risks no greater than Safe Harbor. The authors illustrate that the disclosure of patient ages over the age of 89 is possible when other features are reduced in granularity.
Limitations
The de-identification approach described in this paper was evaluated with demographic data only and should be evaluated with other potential identifiers.
Conclusion
Alternative de-identification policies to the Safe Harbor model can be derived for patient demographics to enable the disclosure of values that were previously suppressed. The method is generalizable to any environment in which population statistics are available.
Introduction
The increasing adoption of electronic medical record (EMR) systems has facilitated a rapid escalation in the amount of patient-level data collected and used in primary care settings. Additionally, such systems have become an invaluable resource for a wide range of secondary applications, including comparative effectiveness studies and novel biomedical investigations.1 2 Notably, EMR-derived data have contributed to a number of genome-wide association studies (GWAS), the goal of which is to unearth biomarkers that correlate with various clinical phenotypes.3 These data reuse efforts live in a special sphere for privacy protection. Various regulations, such as those promulgated by the National Institutes of Health (NIH), require that data used in NIH-sponsored research, and GWAS in particular, be shared beyond the initial collecting institution in a de-identified format.4 5 Given that the originating EMR systems are managed by covered entities as defined by the Health Insurance Portability and Accountability Act (HIPAA),6 data sharing is subject specifically to the de-identification standard set forth in the HIPAA Privacy Rule.7
The HIPAA de-identification standard is general and states ‘Standard: de-identification of protected health information. Health information that does not identify an individual and with respect to which there is no reasonable basis to believe that the information can be used to identify an individual is not individually identifiable health information’.7 Technically, to achieve this goal, the Privacy Rule delineates several routes by which data can be rendered de-identified. One route specified in the Privacy Rule has been referred to as the ‘Statistical Standard’. This approach requires an expert to certify that ‘the risk is very small that the information could be used…to identify an individual who is the subject of the information [using] generally accepted…methods for rendering information not individually identifiable’.7 The regulation points to various statistical methods applied by the disclosure limitation community,8 and some candidate methods for supporting this approach were suggested in the comments to the legislation.
An alternative route specified by the Privacy Rule is ‘Safe Harbor’, which enumerates 18 attributes that must be suppressed. The architects of the Privacy Rule designed this route to be ‘an easily followed cookbook approach,’7 for cases where the certification required by the statistical standard was too onerous. The enumerated list includes explicit identifiers, such as personal names and social security numbers, as well as ‘quasi-identifiers’, which, in combination, may lead to the identification of an individual. The latter class of attributes includes (but is not limited to) dates, patient ages over 89, and zip codes that are more specific than the first three digits. Although easy to follow, the Safe Harbor approach has been criticized for being too stringent, partially because it can significantly limit epidemiological and population-based studies, which may depend on some of these suppressed attributes.9 10
Safe Harbor has also been criticized for being an ad hoc approach,11 but many commercial and open-source health information de-identification systems favor this model.12 There are a number of social and technical factors that may have contributed to this preference. Firstly, statistical expertise is not always available and, if it is, there is no clear rule on what constitutes an ‘expert’ under the Statistical Standard. Secondly, many traditional disclosure limitation methods alluded to in the regulation are designed to handle tabular data and are not oriented to handle patient-level records or the semantics of health information. Thirdly, the federal government has not addressed how to apply specific techniques to achieve the Statistical Standard since its passage. In contrast, Safe Harbor is readily interpretable and can be applied without statistical expertise or support.
The goal of this work is to demonstrate how simple computational methods, based in risk analysis, can be designed and applied to support the dissemination of health information in ways that Safe Harbor precludes, and thus to allow data administrators to take advantage of either of the two de-identification standards proposed. We present a de-identification framework that is easy to interpret, straightforward to replicate, and quick to reconfigure across datasets. Additionally, we set up our framework to directly utilize Safe Harbor to parameterize the methods, providing evidence for why these de-identification schemes should be considered. In summary, our approach consists of two core steps. Firstly, we quantify the ‘re-identification’ risk of demographics in patient records inherent in data shared according to the Safe Harbor policy, with an emphasis on patient-specific attributes that have been leveraged to compromise patient privacy in the past.13 Secondly, we alter the granularity of the patient's attributes to generate policy alternatives with risk that is no greater than what Safe Harbor deems acceptable.
To demonstrate its effectives, we assess our approach with real GWAS patient cohorts from five medical centers. While Safe Harbor prohibits the dissemination of the cohorts' patients' ages over 89 years old, we show that such information can be disclosed with minimal impact on the overall scientific utility of the records compared with the original records. Additionally, to assist other institutions in replicating our approach, we offer an open-source version of our software.14
The remainder of this paper is organized as follows. In the following section, we provide background on re-identification risk analysis and existing approaches to measure and mitigate this problem. Next, we provide the setup of our analysis, including the cohorts employed in this study. Afterward, we detail our framework, with a formalization of the risk metrics applied and how risk is estimated for patient-level data. Then, we describe the results of applying the framework to the cohorts, and finally we conclude by discussing the limitations of the methods described and suggestions for future work.
Background and related work
In this section, we review the privacy problem addressed in this work, various methods that have been proposed to mitigate privacy risks, and how such methods relate to the HIPAA de-identification standard.
Re-identification concerns
For the purposes of this project, we function under the premise that the Privacy Rule is designed to prevent the disclosure of individually identifiable records. More specifically, we are concerned with the ‘identity disclosure’ problem, which is the linkage of a shared record to the corresponding patient's identity. Throughout this paper, we refer to a specific identity disclosure as a ‘re-identification’.i We assume that the recipient of health data attempts to re-identify as many records as possible and is not discriminatory toward any particular subset of records. This type of re-identification problem has been called the ‘marketer attack’.15
Various studies have shown that health information, devoid of explicit identifiers (eg, personal name, social security number) can be re-identified through residual features.16–19 For instance, it has been illustrated that the demographics of records that satisfy Safe Harbor overlap with features available in various public resources, such as voter registration databases.20 21 And, empirically, the risk of re-identification being achieved through these residual demographic attributes is small, but not zero.22 Moreover, the risk varies across geographic domains and demographic strata,23 sometimes in non-random ways (eg, college towns versus retirement communities).21 The fact that re-identification is possible after data have been subjected to Safe Harbor is notable because it suggests that risk is inherent in this data-sharing policy. This is why we use the marketer attack scenario as opposed to scenarios that calculate risk according to the highest-risk record.
Privacy protection modeling
Various models have been designed to determine the amount of re-identification risk in a particular record or set of records. In the statistics community, common models have focused on estimating how many records correspond to unique people.24 Other models have taken a broader perspective and model how many individuals a record could have been derived from.25 Such approaches then set a threshold on the probability of identifying a record to a specific individual or set of individuals, considering records at risk if this threshold is exceeded. This is similar to a model studied in the computer science community, known as k-map, which states that each record is protected when it corresponds to k people in the population from which it was derived.26 Under this model, it can be guaranteed that the likelihood of identifying any particular record is at most 1/k. In certain instances, population data are readily available. For instance, in the study described in this paper, we leverage public statistics from the US Census Bureau to estimate the size of the population group for each patient record to be shared. However, there are times when a health data manager may not know the details of the population; for this case, a stricter model called k-anonymity has been proposed. Technically, this model is satisfied when each shared record is equivalent to k−1 other shared records for the values of a quasi-identifier. This model has received increasing attention in the healthcare domain.27 28
Although formally computable, the aforementioned models have several drawbacks if we want to relate the results to Safe Harbor. In particular, they assume that each shared record should be equally risky. In other words, each record must correspond to a group of k or more people. The application of such a k-based model would preclude de-identification solutions where one record is more vulnerable to re-identification while the rest of the records are comparatively less risky—for example, solutions such as Safe Harbor, which leave records in various group sizes, some of which may be unique (ie, k=1). Solutions produced by k-based models may therefore require more information loss. Whether this tradeoff is appropriate is best left to the discretion of administrators or policy makers, but we wish to provide a framework that is amenable to alternative de-identification solutions with properties similar to Safe Harbor.
As an alternative, Truta and colleagues29 proposed a risk model that relates differing levels of risk on an even playing field. In particular, the re-identification risk of each record is proportional to the number of parent records from which it could have been derived. As we explain below, we utilize this risk model, and propose a system for comparing the risks with datasets under Safe Harbor.
Risk-mitigation methods
An array of methods has been designed to modify data to achieve formal privacy models.30 The first class of methods correspond to ‘suppression’ techniques.31 Suppression may be performed at the record level, such as when a particular value in a record is too risky or the entire record is particularly vulnerable, or at the attribute level if a substantial quantity of records are considered too risky. The removal of attributes such as social security numbers is an example of attribute-level suppression. A second class of methods that can be applied for risk mitigation is based on ‘generalization’ (sometimes referred to as ‘abbreviation’) of the information.17 These methods transform data into more abstract representations. For instance, a five-digit zip code may be generalized to a four-digit zip code, which in turn may be generalized to a three-digit zip code, and onward so as to disclose data with lesser degrees of granularity. Similarly, the age of a patient may be generalized from 1-year to 5-year age groups. It is also possible for generalization schemes to deviate from natural hierarchies (eg, patients aged 25 and 55 could be generalized together, with the value for age for each patient being reported as [25, 55] while all other ages are left in their most specific form). A third class of methods that can be applied for risk mitigation is known as ‘randomization’.32 33 In this case, specific values are replaced with equally specific, but different, values, with a certain probability. For example, a patient's age may be reported as a random value within a 5-year window of the actual age. A fourth class of methods corresponds to ‘synthesization’ (sometimes called ‘fabrication’) of the information.34 In this case the original data are never shared. Rather, general aggregate statistics about the data are computed, and new synthetic records are generated from the statistics to create fake, but realistic, data. Notably, health data users are reportedly wary of randomized and synthetic data.15 As such, emerging protection methods in the health domain, including the methods we apply in this study, tend to focus on generalization and suppression.
Methods
Study cohort and data sources
The patient cohorts for this study, summarized in table 1, were constructed for various GWAS with data derived from EMR systems as part of the NIH-sponsored Electronic Medical Records and Genomics (eMERGE) Network.35 As a condition of NIH funding, the patient-level records upon which the studies are based must be shared with the database of Genotype and Phenotype36 for redistribution. The medical centers include Group Health Cooperative (G), Mayo Clinic (Y), Marshfield Clinic (R), Northwestern University (N), and Vanderbilt University (V). This study incorporates cohorts for each medical center's primary clinical phenotype: dementia (GDEM), peripheral arterial disease (YPAD), cataracts (RCAT), type-II diabetes (NT2D), and QRS duration (VQRS). Also, we included two ‘quality control’ cohorts from Northwestern (NQRS) and Vanderbilt (VT2D), which complemented each others' primary phenotypes.ii The number of patients per cohort ranged between 149 and 3616. The patient attributes included birth year, ethnicity (an eight-valued characteristic corresponding to census race categories), and gender. Safe Harbor stipulates that ages over 89 must be top-coded—that is, grouped into a single value the upper bound of which is not known (in this case, usually expressed as ‘90+’)—and all cohorts except one contained between 0% and 1% patients in this age group. In the remaining dementia cohort, 41% of the patients were over this age limit.
Table 1.
Patient cohorts included in the re-identification risk analysis and mitigation study
| Phenotype | Cohort | Institution | US State | State population size (2000 census) | Clinical finding of interest | Cohort size | Patients over 89 years old |
| Primary | GDem | Group Health Cooperative | WA | 5 894 121 | Dementia | 3616 | 1483 |
| RCat | Marshfield Clinic | WI | 5 363 675 | Cataracts | 2646 | 269 | |
| YPAD | Mayo Clinic | MN | 4 919 479 | Peripheral arterial disease | 3412 | 29 | |
| NT2D | Northwestern University | IL | 12 519 293 | Type-II diabetes | 3383 | 6 | |
| VQRS | Vanderbilt University | TN | 5 689 283 | QRS duration | 2983 | 12 | |
| Quality control | NQRS | Northwestern University | IL | 12 519 293 | QRS duration | 149 | 0 |
| VT2D | Vanderbilt University | TN | 5 689 283 | Type-II diabetes | 2015 | 18 |
To estimate the re-identification risk associated with disclosure policies, we compare the patient demographics in each cohort with the population from which they were derived. We use population count tables from the US Census Bureau (tables PCT12 A–G), which report the number of people of each gender, by age and race, in a geographic division.35 Certain portions of the census data are disclosed in an aggregated manner (eg, ages over 100), for which we assumed individuals were distributed at random, a methodology applied in recent medical privacy policy studies20 to approximate the number of individuals in such a region. We use the demographics in the sample cohort as prior knowledge and distribute the remaining individuals in the corresponding aggregated census population uniformly across the range of possible demographics.
Privacy evaluation framework
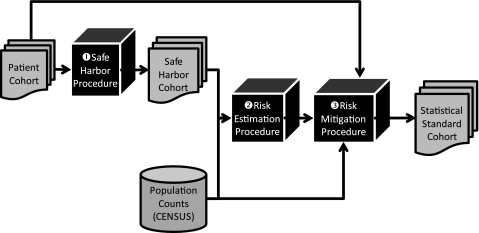
The re-identification risk evaluation framework is outlined in figure 1. Firstly, we transform patient demographics into Safe Harbor-permissible form (Step 1 in figure 1). The set of demographics deemed permissible under this policy correspond to the following:
Age: the age is calculated from birth year. If age is greater than 89, it is reported as a top-coded value of 90+.
Ethnicity: the ethnicity encodings relate to the following eight census race classifications: African-American or Black alone (AA), American Indian and Alaska Native alone, Asian alone, Native Hawaiian and other Pacific Islander alone, some other race alone, two or more races, and white alone (WH).
Gender: the gender is reported as male or female.
State of residence: the state is assumed to be the location of the medical center. Safe Harbor allows geographic information corresponding to a population of 20 000 or greater, a condition satisfied by all US states.
Figure 1.
Workflow of patient privacy risk estimation and mitigation process.
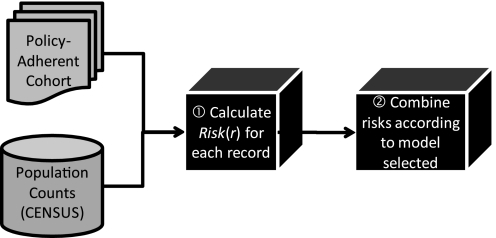
Secondly, given these demographics, we estimate the re-identification risk (step 2 in figure 1). Further details can be seen in figure 2. We utilize the census tables to estimate how many people in the population correspond to each patient in the cohort. We calculate the risk for each patient (step 1 in figure 2) and then combine each patient's risk by summation to calculate a cohort-level risk estimate for the Safe Harbor policy (step 2 in figure 2). Details on the risk calculation and combination are provided in the following section.
Figure 2.
Workflow of privacy risk estimation process.
Thirdly, in the risk-mitigation procedure (step 3 in figure 1), we undertake a process that can be likened to tuning a set of knobs for the fidelity of each demographic attribute. We can dial fidelity down and coarsen an attribute (eg, the age to 5-year age range), or we can dial fidelity up (eg, state to five-digit zip code). If the risk for the altered cohort is no greater than Safe Harbor (risk evaluation for the altered cohort proceeds in the same manner as the evaluation described above, illustrated in figure 2), we certify an acceptable solution in accordance with the Statistical Standard.
Privacy risk measurement
It has long been recognized that perspectives on privacy models vary.37–40 While we focus on a model of privacy that can include the risk of every record, as discussed in the Background section, there is still much flexibility within this model for focusing on different perspectives on privacy. Rather than force health policy makers and medical data administrators to use a universal risk measure, the framework accommodates variations on the definition of risk. Here, we describe several instantiations of the framework that we investigated in this study.
Record-level risk
In general, we base risk on the belief that the greater the number of people in the population to which a patient's attributes (eg, demographics) correspond, the greater the amount of privacy afforded to the patient.32 41 We quantify this notion in a ‘record-level risk’ measure that sets a patient's risk to be inversely proportional to the number of corresponding people in the population. Technically, if gr is the number of corresponding people in the population for a particular patient r demographics, then the patient's risk is:
| (1) |
where a is a scaling factor that enables data managers to weight group size accordingly. The scaling factor provides administrators with the option of tuning the emphasis they place on the most vulnerable individuals. The inclusion of such a parameter is based on the observation that, in certain situations, a data manager may consider the mapping of a patient's record to a group of size x to be much worse than a group of size of x+1. In such a case, setting a to a greater value would capture this belief. Notably, when a is equal to 1, the risk measurement corresponds exactly to an unbiased probability. Specifically, it is the probability that a correct re-identification would be achieved given knowledge of the quasi-identifying values of the dataset and a list of identified individuals. In this case, the best an attacker can do is a random assignment. Thus, a patient who is unique in the population contributes a risk of 1, a patient in a group of two contributes a risk of 0.5, and so on. In the online appendix (see www.jamia.org), we provide an illustration of how varying a influences the risk of a particular cohort in this study.
Dataset-level risk
Given a dataset of records, R, we define the risk for the dataset as the sum of the record-level risks. In this study, we use the record-level risk estimate in two different dataset-level risk estimates: (i) threshold and (ii) total risk. The ‘threshold risk’ is based on the fact that, in certain domains, it is believed that individuals do not contribute risk after a certain group size. Select governmental statistical agencies, for instance, suggest a threshold of five, whereas in other settings only uniques are of sufficient concern.42 Our model permits health policy makers to choose any threshold, which we call T. In this study we investigate thresholds of 1, 10, and 20 000.
We refine the threshold risk into two subclasses called ‘non-graduated risk’ and ‘graduated risk’. The non-graduated risk is based on the perspective that the population group size is, to a certain degree, irrelevant. Rather, it is argued that all patients in a population below T contribute an equal amount of risk. This belief is represented in the k-based privacy models described in the Background section. This notion can be accommodated in our model by setting the scaling factor a in eqn (1) to 0:
| (2) |
The graduated risk, on the other hand, corresponds to the situation in which each record contributes risk proportional to its group size:
| (3) |
Finally, we recognize that many health policy makers find it difficult to settle on any threshold. Thus, we introduce a ‘total risk’ score which includes all patients.
| (4) |
To compare risks across datasets, we compute the percentage of a cohort expected to be re-identified. This is the dataset-level risk normalized by the number of patients in the cohort and multiplied by 100.
Alternative disclosure policies
There are many paths by which we can propose alternative data disclosure policies to mitigate risk. For simplicity and ease of use, we evaluate a predefined set of five alternative policies as outlined in table 2.iii In all policies we fix the geography to the level of US state, and gender to male and female, but consider generalizations of age ranges and ethnicity. For the latter, we use either the eight-valued variable mentioned above, or a three-valued variable of African-American (AA), white (WH), and other (OT).
Table 2.
Patient demographic disclosure policy alternatives
| Disclosure policy | Birth year | Ethnicity | Geography | Sex |
| Safe Harbor | Any year >1920, before 1920 | Any | State | M, F |
| GenEth | Any year | AA, WH, OT | ||
| 5 Year | 5 year range | Any | ||
| 5 Year-GenEth | AA, WH, OT | |||
| 10 Year | 10 year range | Any | ||
| 10 Year-GenEth | AA, WH, OT |
AA, African-American; F, female; M, male; OT, other; WH, white.
The first policy alternative (GenEth) corresponds to the disclosure of any birth year (or age) and generalized ethnicity. The second and third policies (5 Year and 5 Year-GenEth) correspond to age in 5-year ranges, with ethnicity at the original and generalized level, respectively. The fourth and fifth policies (10 Year and 10 Year-GenEth) correspond to 10-year age ranges, with similar ethnicity generalizations.
RESULTS
Re-identification risks of Safe Harbor records
Table 3 reports the estimated re-identification rates for the total, graduated, and non-graduated risks with a threshold of 1, 10, and 20 000 when data are shared according to Safe Harbor. The total risk ranged from 0.01% to 0.19%. More concretely, the 0.01% risk associated with the NQRS cohort implies that less than one person is expected to be re-identified from that dataset. Similarly for the GDEM, the cohort-level risk is 0.12% × 3616, leaving approximately four patients at risk.
Table 3.
Re-identification risk rate of sharing data in accordance with the HIPAA Safe Harbor disclosure policy
| Risk model | Threshold | Percent of cohort at risk | ||||||
| GDEM | RCAT | YPAD | NT2D | VQRS | NQRS | VT2D | ||
| Total risk | N/A | 0.117 | 0.032 | 0.131 | 0.011 | 0.030 | 0.012 | 0.191 |
| Uniques | T=1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Graduated risk | T=10 | 0 | 0.010 | 0.009 | 0 | 0 | 0 | 0 |
| T=20 000 | 0.117 | 0.032 | 0.131 | 0.010 | 0.028 | 0.010 | 0.191 | |
| Non-graduated risk | T=10 | 0 | 0.076 | 0.088 | 0 | 0 | 0 | 0 |
| T=20 000 | 100.000 | 87.302 | 89.918 | 26.369 | 36.943 | 100.000 | 95.533 | |
Notably, none of the cohorts we evaluated harbored a patient that was unique in the corresponding state's population. For a threshold of 10 (ie, patients in a population of greater than 10 do not factor into the risk), we found that five of the seven cohorts had zero risk. The remaining two cohorts had a graduated risk, of 0.076% and 0.088%, which corresponds to the observation that there are two and three patients in the RCAT and YPAD cohorts, respectively, that are in a group of 10 or less. When we consider the non-graduated risk, rates of 0.010% and 0.009% imply that the expected number of re-identifications is 0.28 and 0.30, respectively.
When we shift our threshold to 20 000, the geographic population size threshold designated by HIPAA Safe Harbor, we find that all cohorts carry some level of risk. With respect to non-graduated risk, two of the cohorts, GDEM and NQRS, were completely exposed, with a risk rate of 100%. That is, every patient was in a group of no more than 20 000 in the state's population and suggests that such a measure might not be most useful as a privacy metric, but rather as statistics about the population. The graduated risk at a threshold of 20 000 ranged from 0.01% to 0.19%. Notably, these risk rates are very similar to the total risk rates initially observed. The relative difference between graduated and total risk rates was below 16% for all cohorts and in many cases was less than 2%. This suggests that most patients were in a population group size less than 20 000.
Feasibility of disclosure policy alternatives
With the Safe Harbor risk levels calculated as shown in table 3, we applied these results to determine which of the alternative policies were acceptable. Table 4 provides the re-identification risk for each of the patient cohorts. To place these results in context, we annotated the cells with an asterisk in the table to summarize a ‘thumbs up/thumbs down’ decision. If the application of a policy to a cohort exhibited risk that was worse than Safe Harbor, it received an asterix. If the policy exhibited equal or lesser risk than Safe Harbor, it received no asterisk.
Table 4.
Re-identification risk rate of sharing data via policy alternatives
| Risk model | Threshold | Disclosure policy | Percent of cohort at re-identification risk | ||||||
| GDEM | RCAT | YPAD | NT2D | VQRS | NQRS | VT2D | |||
| Total risk | NA | GenEth | 0.84* | 0.03 | 0.04 | <0.01 | 0.02 | 0.01 | 0.12 |
| 5 year | 0.27* | 0.02 | 0.03 | <0.01 | 0.01 | <0.01 | 0.05 | ||
| 5 year-GenEth | 0.24* | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.02 | ||
| 10 year | 0.12* | 0.01 | 0.01 | <0.01 | <0.01 | <0.01 | 0.03 | ||
| 10 year-GenEth | 0.01 | 0.03 | <0.01 | <0.01 | <0.01 | <0.01 | 0.01 | ||
| Uniques | T=1 | GenEth | 0.08* | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 year | 0.06* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 5 year-GenEth | 0.06* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10 year | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10 year-GenEth | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Graduated Risk | T=10 | GenEth | 0.52* | <0.01 | 0 | 0 | 0 | 0 | 0.01* |
| 5 year | 0.11* | 0.01 | 0 | 0 | 0 | 0 | 0 | ||
| 5 year-GenEth | 0.10* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10 year | 0.03* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10 year-GenEth | 0.02* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| T=20000 | GenEth | 0.84* | 0.03 | 0.04 | <0.01 | 0.01 | 0.01 | 0.12 | |
| 5 year | 0.27* | 0.01 | 0.02 | <0.01 | 0.01 | <0.01 | 0.05 | ||
| 5 year-GenEth | 0.24* | <0.01 | 0.01 | <0.01 | <0.01 | <0.01 | 0.02 | ||
| 10 year | 0.12* | 0.01 | 0.01 | <0.01 | <0.01 | 0 | 0.02 | ||
| 10 year-GenEth | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0 | 0.01 | ||
| Non-graduated risk | T=10 | GenEth | 1.94* | 0.04 | 0 | 0 | 0 | 0 | 0.05* |
| 5 year | 0.39* | 0.04 | 0 | 0 | 0 | 0 | 0 | ||
| 5 year-GenEth | 0.36* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10 year | 0.17* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 10 year-GenEth | 0.11* | 0 | 0 | 0 | 0 | 0 | 0 | ||
| T=20000 | GenEth | 100 | 93.5* | 90.4* | 26.5* | 36.9 | 100 | 95.5 | |
| 5 year | 58.3 | 16.7 | 10.5 | 7.87 | 7.88 | 4.70 | 55.1 | ||
| 5 year-GenEth | 58.3 | 16.7 | 10.5 | 3.98 | 7.88 | 4.70 | 55.1 | ||
| 10 year | 48.2 | 4.61 | 7.74 | 4.83 | 5.06 | 0 | 27.9 | ||
| 10 year-GenEth | 48.2 | 4.61 | 7.68 | 1.58 | 4.96 | 0 | 27.8 | ||
Policies that resulted in more risk than Safe Harbor.
Firstly, we report on the total risk. According to this measure, GDEM can be shared under only one of the alternative policies with birth year generalized to the 10-year range and ethnicity generalized to the three-values. Notably, the remaining six cohorts can be shared under any of the alternative policies.
Turning our attention to unique individuals, we find again that six of the cohorts can be disclosed according to any of the alternative policies. Additionally, the GDEM cohort achieves protection when age is generalized to 10-year ranges, even with ethnicity in its most specific form.
With respect to the graduated risk measure, when the threshold is 10 people, the GDEM cohort is unable to satisfy any of the alternative policies. Five of the cohorts can be shared according to any policy, and the VT2D cohort can be shared when the age range is generalized to a 5-year (or greater) window. When the threshold is 20 000, we find that six of the cohorts can be shared according to any policy. Of the remaining cohorts, GDEM can be shared when age is generalized to a 10-year window and ethnicity is generalized.
The non-graduated risk measure offers a different perspective. At a threshold of 10, although none of the policies suffice to protect the GDEM cohort, only one other cohort failed to satisfy all of the alternative disclosure policies. The VT2D cohort can be shared when the age is generalized to a 5-year window. Finally, with a threshold of 20 000 people, we find that all of the cohorts, GDEM included, satisfy the disclosure policy of 5 year range with most specific ethnicity, but we also find that three of the cohorts are not protected by simply generalizing ethnicity.
Discussion
Our findings indicate there are alternatives to Safe Harbor that hold equal or lesser re-identification risk. The alternatives permit different levels of granularity in patient demographics. Thus, if a healthcare organization determines that age-related features are more important than detailed ethnicity for clinical research, it is possible to shift the level of detail in many cases. We believe that this provides organizations with many options to share data. Although the approach was demonstrated with demographics, it generalizes to any set of attributes for which population-level statistics are known. Additionally, we believe that our approach is interpretable, such that it can be presented before institutional review boards to assist in their decisions about data dissemination in accordance with de-identification requirements and compliance under the Statistical Standard.
Additionally, the results highlight the fact that every dataset is fundamentally different. There is not a one-size-fits-all solution, although alternatives do exist for each of the datasets. The results for GDEM, the cohort with the fewest safe alternatives, indicate that Safe Harbor provides more stringent protection for this dataset. This is also the dataset most affected by the particular suppressions and generalizations required by Safe Harbor, as almost half of the records were affected by age top-coding (ie, 41%), and thus the dataset whose creators and administrators might be most interested in finding alternatives under the Statistical Standard. The fact that only one alternative policy was found (using the total risk measure) does not mean that achieving the Statistical Standard is impossible, only that the alternative policies evaluated in this work were mostly insufficient.
There are several limitations of this work that we wish to highlight. Firstly, we did not work with the geographic or gender variables, which could permit a broader range of alternative policies. The geographic variable was not considered because such details were not available for all cohorts. This is certainly a variable that could be applied within our framework and would be particularly useful to support epidemiological research. Additionally, geographic information may be implied by the location of the provider, even if not explicitly released. Gender was neglected because the study cohorts are tied to genomic sequence data, from which gender can often be inferred. Of course, in a more general setting, removal of gender information would be a viable policy alternative.
Secondly, we adopted a dichotomized view that an alternative disclosure policy was good or poor based on its relationship to Safe Harbor for an entire dataset. Alternatively, we could apply a more fine-grained method based on ‘local recoding’ by generalizing only records with a high risk or suppressing them altogether. We refrained from such an approach because it was our intention to retain some degree of regularity in the demographics for all patient records to include in validation studies.
Thirdly, it should be recognized that the alternative disclosure policies are ‘alternatives’. In particular, biomedical data managers must recognize that, when records from the same underlying cohort are disclosed in two separate releases, it is possible for results to be ‘triangulated’, or overlaid, revealing private information that was concealed by each release independently.
Fourthly, we assume that the harm is re-identification, discovery of the identity corresponding to a record, and that an attacker is equally interested in re-identifying all records. It is possible that an attacker is interested in identifying a subset, or even a single record, in which case, an alternative policy should strive to mitigate the likelihood that any particular record is within a certain risk bound.44 The risk measures proposed in this work can be modified by measuring the maximum risk for the records in the dataset, rather than the sum. This risk metric is essentially the same as the implied risk metric used in k-based models, and alternative policies could be found using algorithms developed in that area.
Finally, although we have focused on patient demographics, there are other patient-specific features that can be exploited for re-identification purposes.19 These attributes should be taken into consideration on a case by case basis. It should be recognized that statistical disclosure control techniques are being designed for aggregate data dissemination for genomic sequence data.45
Conclusions
This work presented a simple, intuitive approach to measure the re-identification risk of sharing de-identified patient demographics in accordance with the HIPAA Safe Harbor Standard. It then illustrated how alternative data disclosure policies can be evaluated with respect to Safe Harbor. We tested our approach with several real world patient cohorts and demonstrated that such risk assessments can be applied to tailor disclosure policies to specific situations. To extend this work, we intend to investigate how to apply such an approach with other potential identifiers and evaluate investigators’ viewpoints on which de-identification alternatives are the most desirable for their studies.
Supplementary Material
Acknowledgments
We thank Gene Hart, MS (Group Health Cooperation), Peggy Peissig, MBA and Luke Rasmussen (Marshfield Clinic), May Law, MPH, MS and Jennifer A Pacheco (Northwestern University), and Deede Wang, MS, MBA (Vanderbilt University) for providing the demographic data associated with the cohort investigated in this study. We thank Ellen Clayton, MD, JD, Aris Gkoulalas-Divanis, PhD (Vanderbilt University), Grigorios Loukides, PhD (Vanderbilt University), Abel Kho, MD, MS (Northwestern University), and Teri Manolio (National Human Genome Research Institute) for insightful discussions and review of this research.
Footnotes
Funding: This was work was supported by grant 1U01HG00460301 from the National Human Genome Research Institute and 1R01LM009989 from the National Library of Medicine. eMERGE network members involved in the review of the manuscript include Eric Larson, MD (Group Health Cooperation), Cathy McCarty, PhD (Marshfield Clinic), Christopher Chute, MD, DrPH (Mayo Clinic), Rex Chisholm, PhD (Northwestern University), and Daniel Roden (Vanderbilt University).
Ethics approval: This study was conducted with the approval of the Vanderbilt University, Group Health Cooperation, Mayo Clinic, Marshfield Clinic, Northwestern University.
Provenance and peer review: Not commissioned; externally peer reviewed.
A second violation voiced in the computer science literature is the ‘attribute disclosure’ problem. This corresponds to the revelation of the value of a sensitive attribute, and can be achieved without identity disclosure. If, for instance, all of the males in the dataset have the same primary diagnosis, and a particular person is known to be represented in the dataset, then their primary diagnosis has been disclosed without the particular record being individually identified. This problem is beyond the scope of this study.
Details of the eMERGE Network phenotypes are online at http://www.gwas.net/ (accessed February 19, 2010).
The selection of these policies was to ensure regularity in the resulting data. One could apply automated algorithms in the manner of those recently proposed.43
References
- 1.Safran C, Bloomrosen M, Hammond W, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association white paper. J Am Med Inform Assoc 2007;14:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roden D, Pulley J, Basford M, et al. Development of a large-scale de-identified DNA biobank to enabled personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio T. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI's office of population genomics. Pharmacogenomics 2009;10:235–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Institutes of Health Final NIH statement on sharing research data. NOT-OD-03–032. 2003 Feb 26. http://grants.nih.gov/grants/guide/notice-files/NOT-OD-03-032.html [Google Scholar]
- 5.National Institutes of Health Policy for sharing of data obtained in NIH supported or conducted genome-wide association studies (GWAS). NOT-OD-07–088. 2007. August 28 http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html [Google Scholar]
- 6.Health Insurance Portability and Accountability Act of 1996. Public L. No. 104-191, 110 Stat. 1936. 1996. http://www.gpo.gov/fdsys/pkg/PLAW-104publ191/pdf/PLAW-104publ191.pdf [PubMed] [Google Scholar]
- 7.Office of Civil Rights, U.S. Dept. of Health and Human Services Standards for privacy of individually identifiable health information: Final Rule. Fed Regist 2000;65:82462–829 [PubMed] [Google Scholar]
- 8.Federal Committee on Statistical Methodology Statistical Policy Working Paper 22, 2005 [Google Scholar]
- 9.Ehringhaus S. Testimony on Behalf of the Association of American Medical Colleges Before the National Committee on Vital and Health Statistics Subcommittee on Privacy. http://www.aamc.org/advocacy/library/research/testimony/2003/111903.pdf/ (accessed 7 Jul 2010).
- 10.Ness R; for the Joint Policy Committee, Societies of Epidemiology Influence of the HIPAA privacy rule on health research. JAMA 2007;298:2164–70 [DOI] [PubMed] [Google Scholar]
- 11.Ohm P. Broken promises: responding to the surprising failure of anonymization. UCLA Law Review 2010;57:1701–77 [Google Scholar]
- 12.DE-ID Data Corp Frequently asked questions. http://www.de-idata.com/index.php?option=com_content&task=view&id=27&Itemid=26 (accessed 1 Jul 2010).
- 13.Gostin L, Nass S. Reforming the HIPAA privacy rule: safeguarding privacy and promoting research. JAMA 2009;301:1373–5 [DOI] [PubMed] [Google Scholar]
- 14.Vanderbilt demographic re-identification assessment tool source code. http://code.google.com/p/vdart (accessed 19 Feb 2010).
- 15.Dankar F, El Emam K. A Method for Evaluating Marketer re-Identification risk. Proceedings of the EDBT/ICDT Workshops. ACM Press, 2010:1–10 [Google Scholar]
- 16.El Emam K, Jabbouri S, et al. Evaluating common de-identification heuristics for personal health information. J Med Internet Res 2006;8:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweeney L. Weaving technology and policy together to maintain confidentiality. J Law Med Ethics 1999;25:98–110 [DOI] [PubMed] [Google Scholar]
- 18.Malin B, Sweeney L. How (not) to protect genomic data privacy in a distributed network: using trail re-identification to evaluate and design anonymity protection systems. J Biomed Inform 2004;37:179–92 [DOI] [PubMed] [Google Scholar]
- 19.Malin B. An Evaluation of the current state of genomic data privacy protection technology and a roadmap for the future. J Am Med Inform Assoc 2005;12:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golle P. Revisiting the uniqueness of the simple demographics in the US population. Proceedings of the ACM Workshop on Privacy in the Electronic Society. ACM Press, 2006:77–80 [Google Scholar]
- 21.Sweeney L. Uniqueness of Simple Demographics in the U.S. Population. Working Paper LIDAP-WP4. Pittsburgh, PA: Data Privacy Lab, Carnegie Mellon University, 2000 [Google Scholar]
- 22.Southern Illinoisian v. IDPH (2006) Docket No. 98712. http://www.state.il.us/court/opinions/supremecourt/2006/february/opinions/html/98712.htm [Google Scholar]
- 23.Benitez K, Malin B. Evaluating re-identification risks with respect to the HIPAA privacy rule. J Am Med Inform Assoc 2010;17:169–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliot M, Skinner C, Dale A. Special uniques, random unique and sticky populations: some counterintuitive effects of geographic detail on disclosure risk. Research in Official Statistics 1998;1:53–67 [Google Scholar]
- 25.Skinner C, Elliot M. A measure of risk for microdata. J R Stat Soc 2002;64:855–67 [Google Scholar]
- 26.Sweeney L. k-Anonymity: A model for protecting privacy. International Journal of Uncertainty, Fuzziness and Knowledge-based Systems 2002;10:557–70 [Google Scholar]
- 27.El Emam K, Dankar F. Protecting privacy using k-anonymity. J Am Med Inform Assoc 2008;15:627–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Emam K, Dankar F, Issa R, et al. A globally optimal k-anonymity method for the de-identification of health data. J Am Med Inform Assoc 2009;16:670–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truta T, Fotouhi F, Barth-Jones D. Disclosure risk measures for microdata. Proc 15th International Conference on Scientific and Statistical Database Management 2003:15–22 [Google Scholar]
- 30.Ohno-Machado L, Silveira P, Vinterbo S. Protecting patient privacy by quantifiable control of disclosures in disseminated databases. Int J Med Inf 2004;73:599–606 [DOI] [PubMed] [Google Scholar]
- 31.Øhrn A, Ohno-Machado L. Using Boolean reasoning to anonymize databases. Artif Intell Med 1999;15:235–54 [DOI] [PubMed] [Google Scholar]
- 32.Cox L. Protecting confidentiality in small population health and environmental statistics. Stat Med 1996;15:1895–905 [DOI] [PubMed] [Google Scholar]
- 33.Evfimievski A. Randomization in privacy preserving data mining. ACM SIGKDD Explorations Newsletter 2002;4:43–8 [Google Scholar]
- 34.Reiter J. Releasing multiply imputed, synthetic public use microdata: an illustration and empirical study. J R Stat Soc 2005;168:185–205 [Google Scholar]
- 35.Electronic Medical Records and Genomics Network http://www.gwas.net (accessed 1 Jul 2010).
- 36.Mailman M, Feolo M, Jin Y, et al. The NCBI database of genotyped and phenotypes. Nat Genet 2007;39:1181–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Census Bureau American fact finder website: http://www.americanfactfinder.gov (accessed 1 Jul 2010).
- 38.Gostin L, Turek-Brezina J, Powers M, et al. Privacy and security of personal information in a new health care system. JAMA 1993;270:2487–93 [PubMed] [Google Scholar]
- 39.Acquisi A, Grosslags J. Privacy and rationality in individual decision making. Proceedings of the 1st ACM International Health Informatics Symposium 2005;3:26–33 [Google Scholar]
- 40.Kohane I, Altman R. Health information altruists—a potentially critical resource. N Engl J Med 2005;353:2074–7 [DOI] [PubMed] [Google Scholar]
- 41.Rudolph P, Shah G, Love D. Small numbers, disclosure risk, security, and reliability issues in web-based data query systems. J Public Health Manag Pract 2006;12:176–83 [DOI] [PubMed] [Google Scholar]
- 42.Elliot M. DIS: A new approach to the measurement of statistical disclosure risk. Risk Management 2000;2:39–48 [Google Scholar]
- 43.Benitez K, Loukides G, Malin B. Beyond Safe Harbor: automatic discovery of health information de-identification policy alternatives. Proc 1st ACM International Health Informatics Symp 2010: forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sparks R, Carter C, Donnelly J, et al. Remote access methods for exploratory data analysis and statistical modeling: privacy-preserving analytics. Comput Methods Programs Biomed 2008;91:208–22 [DOI] [PubMed] [Google Scholar]
- 45.Sankararaman S, Obozinski G, Jordan MI, et al. Genomic privacy and limits of individual detection in a pool. Nat Genet 2009;41:965–7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.