Abstract
Amyloid fibrils are filamentous protein aggregates implicated in several common diseases like Alzheimer’s disease and type II diabetes. Similar structures are also the molecular principle of the infectious spongiform encephalopathies like Creutzfeldt-Jakob disease in humans, scrapie in sheep, and of the so-called yeast prions, inherited non-chromosomal elements found in yeast and fungi. Scanning transmission electron microscopy (STEM) is often used to delineate the assembly mechanism and structural properties of amyloid aggregates. In this review we consider specifically contributions and limitations of STEM for the investigation of amyloid assembly pathways, fibril polymorphisms and structural models of amyloid fibrils. This type of microscopy provides the only method to directly measure the mass-per-length (MPL) of individual filaments. Made on both in vitro assembled and ex vivo samples, STEM mass measurements have illuminated the hierarchical relationships between amyloid fibrils and revealed that polymorphic fibrils and various globular oligomers can assemble simultaneously from a single polypeptide. The MPLs also impose strong constraints on possible packing schemes, assisting in molecular model building when combined with high-resolution methods like solid-state nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR).
Keywords: Amyloid, mass measurement, fibril, filament, oligomer, protein aggregation, electron microscopy, scanning transmission electron microscopy, STEM, Alzheimer Disease, Type 2 Diabetes Mellitus, prion, scrapie, yeast, fungi
1. Introduction
The term amyloid was originally introduced with reference to proteinaceous fibrillar structures that accumulate in the extracellular space in various diverse diseases - prominent examples include type 2 diabetes mellitus (pancreatic amyloid deposits), dialysis-related amyloidosis (amyloid deposits in vasculature), and Alzheimer’s disease (amyloid deposits in brain parenchyma and vessels). There are also examples of amyloid fibrils that accumulate intracellularly in certain neurodegenerative diseases - like Parkinson’s disease, Huntington’s disease and other so-called poly-glutamine extension diseases as well as Alzheimer’s disease and frontotemporal dementias (Ross and Poirier, 2004; Mandelkow et al., 2007). Both plasma-derived and cellular proteins form systemic (in many different organs) or organ-specific amyloid deposits that characterize the different forms of amyloidosis. Amyloid fibrils also afford the molecular mechanism that underlies infectious self-propagating protein polymers called prions in mammals (eg. leading to Creutzfeldt-Jakob disease in humans and scrapie in sheep) and fungal systems (Baxa et al., 2006, 2008).
Correlation with particular disease states affords a clinical basis for distinguishing various types of amyloid. Interestingly, the proteins that form the major components of these amyloids are relatively few in number (about 20). More generally, many different polypeptides are capable of forming structurally related amyloid fibrils or amyloid-like structures in vitro (Kodali & Wetzel, 2007; Fändrich, 2007; Baxa, 2008). The hallmark of amyloid fibrils is a cross-β structure in which the β-strands run approximately perpendicular to the fibril axis. Further defining characteristics are that they are unbranched, smooth-surfaced, physically and chemically robust (e.g. protease-resistant) and that they bind the dyes Congo Red and thioflavin T.
Cytotoxicity and organ failure in amyloid-associated diseases is attributed to misfolding of the normally soluble polypeptides into aberrant β-sheet conformations (Ross and Poirier, 2004, Novitskaya et al., 2006; Bitan et al., 2005; Konarkowska et al., 2006, Mocanu et al., 2008). Resultant fibril assemblies, metastable intermediates and off-pathway oligomeric aggregates have all been proposed to be cytotoxic (Wogolis et al., 2005; Lesné et al., 2006; Lambert et al., 2007; Novitskaya et al., 2006; Konarkowska et al., 2006; Lansbury and Lashuel, 2006). This has initiated many investigations into fibrillar and oligomer assembly, which have revealed remarkably complex assembly pathways and polymorphic structures (Kodali & Wetzel, 2007; Fändrich, 2007; Baxa, 2008). Due to the complexity of nomenclature in the literature for various amyloid-related aggregates, a glossary of terminology is provided in Box 1.
Box 1: Glossary of terms.
Amyloid
Protein deposits in the form of filamentous aggregates that are implicated in a broad spectrum of diseases. Although the deposits share conserved tertiary cross β-sheet structural motifs, the fibril/filament (terms used interchangeably) forming protein and organ involved is disease specific; for example, deposits of Aβ form amyloid plaques in Alzheimer’s disease brains and deposits of amylin (Islet Amyloid Polypeptide: IAPP) form in the islets of Langerhans in the pancreas in the course of type 2 diabetes mellitus. Fibrils can be straight or twisted and sometimes contain multiple protofilaments that align parallel to the fibril axis or coil around each other. The term “amyloid” has been extended by structural biologists to additionally encompass the fibrillization of synthetic and recombinant polypeptides from various sources that also assemble cross β-sheet structure (Kajava et al., 2009).
Protofibril
Intertwining strands that make up mature amyloid fibrils. This term is used interchangeably with the term “protofilament”. However, in the case of Aβ, “protofibril” has additionally been used to describe metastable fibrils that appear early in solutions of this polypeptide in vitro before mature amyloid fibrils appear (Walsh et al., 1997; Harper et al, 1997). These are short flexible fibrils, rods or globular structures with no clear axial periodicities. Although the term implies it, these transient “protofibrils” are not necessarily on-pathway intermediate structures and may represent semi-stable off-pathway assemblies (Kodali and Wetzel, 2007).
Protofilament
Intertwining strands that make up mature amyloid fibrils.
Oligomer
Macromolecular complexes of self-assembled amyloid polypeptides but not in fibril/filament form. Oligomers can be off-pathway stable structures (Gellermann et al., 2008). Other oligomers represent unstable transient nuclei that initiate fibril assembly. When visible in electron micrographs or atomic force microscopy images, stable oligomers resemble globules or ring-like assemblies (Lambert et al., 1998; Nybo et al., 1999; Goldsbury et al., 2000; Lashuel et al., 2003). A complex set of terminologies exists in the literature describing oligomers - for example, for Aβ: ADDLs (Aβ-derived-diffusible-ligands; Lambert et al., 1998), globulomers (n≈12 oligomers; Gellermann et al., 2008) and Aβ* (n≈12 oligomers; Lesné et al., 2006).
Paired Helical Filaments (PHFs)
Name given to amyloid fibrils formed by tau protein. PHFs have only relatively recently been widely recognized structurally as amyloid fibrils. This is due to the atypical nature of tau as the molecular subunit in the fibrils. Only a relatively small segment of this large protein contributes the β-sheet core structure that forms these filaments (Mandelkow et al., 2007). The rest of the tau protein lies peripherally and does not have β-structure. This made detection of defining cross-β diffraction patterns more challenging. PHFs also form intracellularly, in contrast to the classical extracellular amyloid deposits.
Electron microscopy (EM) of negatively stained or vitrified specimens has been applied successfully to determine the structures of many kinds of protein filaments, but has been relatively unproductive in the field of amyloid. The polymorphism of amyloid fibrils and their tendency to aggregate poses problems. In addition their smooth surface tends to generate little contrast in micrographs recorded by cryo-EM. In this situation, scanning transmission electron microscopy (STEM) has proved a powerful tool helping to delineate the assembly mechanism and structural properties of amyloids. Importantly, STEM is the only method that can directly measure the mass-per-length (MPL) of individual filaments. Working from an image, it also links the MPL to the sample appearance allowing a direct correlation to negative stain or cryo-EM images that reveal structure. If the monomer mass of the subunits that make up a fibril is known, the MPL determined by STEM imposes strong constraints on possible packing schemes within fibrils, assisting in molecular model building (Kajava et al., 2009; Petkova et al., 2005; Paravastu et al., 2008; Sen et al., 2007) and illuminating hierarchical relationships between fibrils of different kinds (Bauer et al., 1995; Goldsbury et al., 2005).
Many different filamentous protein assemblies have been examined by STEM, among them bacterial pili (Hahn et al., 2002; Köhler et al., 2004), viral filaments (Kendall et al., 2008) and neurofilaments (Leapman et al., 1997). The method can also be used to measure the masses of individual particles and, thus, define their stoichiometry. In the field of amyloid, data of this kind document the compositional heterogeneity of oligomeric particles assembled from amyloidogenic polypeptides and proteins (Lashuel et al., 2003; Goldsbury et al., 2000). Further, STEM mass measurements have helped to show that polymorphic fibrils and oligomers can assemble simultaneously from a single polypeptide. Such morphological or molecular-level polymorphisms are believed to underlie the strain phenomena and species barrier in prion transmission, as well as differences in cytotoxicity in amyloid diseases.
2. STEM
STEM is used for diverse applications in various fields but few instruments are tuned to make quantitative measurements on biological specimens (Müller et al, 1992): namely the dedicated instruments at the Brookhaven National Laboratory (BNL), (Upton, New York, USA), at the Biozentrum, University of Basel (Basel, Switzerland), and at the National Institutes of Health (Bethesda, USA). In addition, there is a TEM with STEM attachment at the Wellcome Trust Centre for Cell-Matrix Research, University of Manchester (Manchester, UK) (Holmes et al., 2006; Sherratt et al., 2009). In particular, wide-angle detector systems capable of single electron counting are a prerequisite for all quantitative measurements (reviewed in Engel, 2009). Quantitative biological dark-field STEM exploits the linear relationship between the number of electrons elastically scattered by the specimen and the mass of the irradiated region (Figure 1A; Müller and Engel, 2001; Ksiezak-Reding and Wall, 2005). Visualization is thereby combined with the ability to measure the masses, for example, of individual protein complexes. Calibration is assisted by measuring the MPL of tobacco mosaic virus (TMV) in the same session. The RNA containing TMV rods are highly uniform, ~300 nm long and 18 nm wide, and have a known MPL of 131 kDa/nm. Note, however, that a fully calibrated STEM measures absolute mass so a calibration standard such as TMV is not strictly required to achieve mass measurements but is very useful for detecting any drift in the microscope calibration or, when adsorbed to the same grid as the sample, problems with specimen preparation (Wall and Hainfield, 1986). Once images have been collected, mass analysis of sample and standard is carried out offline using custom software. Two packages are freely available; MASDET (Basel; Krzyžáneka et al, 2009) and PC-Mass (Brookhaven; Wall et al., 2008; http://www.bnl.gov/biology/stem/files/PCMASS30.pdf).
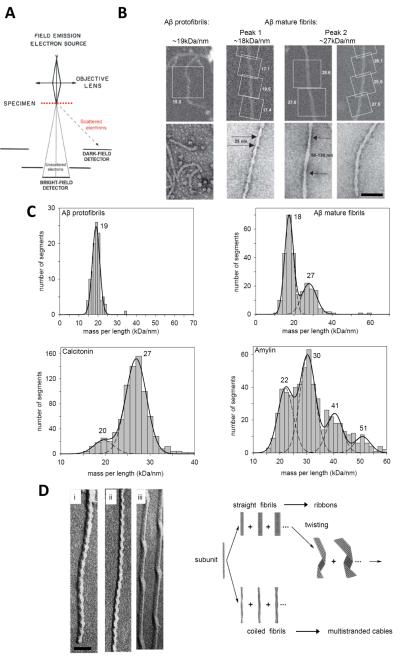
Figure 1. Collection of STEM data.
(A) Schematic representation of STEM optics. Electrons elastically scattered from the specimen are detected by an annular dark-field detector system. For thicknesses t < Λ where Λ is the average mean free path between two subsequent scattering events, multiple scattering may be neglected and this signal is proportional to the mass of the irradiated region. (B) ROI selection on fibrils formed in vitro by synthetic Aβ and prepared for mass measurement (upper panels, square boxes). The calculated MPLs are noted; units kDa/nm. Data from hundreds of such segments are pooled to generate MPL histograms (shown in upper row C). The images illustrate the spectrum of different fibril polymorphs including metastable protofibrils (left), and mature fibrils (right) that form sequentially on incubating the solutions. These images from unstained samples can be linked by dimensions and morphology to fibrils imaged by negative stain microscopy (lower panels). (C) STEM MPL histograms fitted with Gaussian distributions, from independent measurements made on synthetic calcitonin, amylin and Aβ fibrils. The peak MPLs are indicated (kDa/nm). It is useful to express MPLs as the number of monomeric subunits per 0.47 nm step (the inter-strand spacing in a β-sheet, an axial spacing in cross-β structures). The corresponding numbers are 2.1 for Aβ protofibrils (monomer mass, 4.3 kDa); 2.0 and 2.9 for Aβ mature fibrils (monomer mass, 4.3 kDa); 2.6, 3.6, 4.9 and 6.1 for amylin fibrils (monomer mass, 3.9 kDa) and 2.7 and 3.7 for calcitonin fibrils (monomer mass, 3.4 kDa). (D) TEM images of unidirectional metal shadowed samples. Aβ (i) and amylin (ii) both show left handed axial cross-over spacings of ~25 nm. Twisted ribbons, shown here for A (iii) are also left-handed. Combined with information from STEM, these TEM data were used to generate a model illustrating the hierarchical protofilament substructure common to polymorphic amyloid fibril assembly. Adapted from Ksiezak-Reding et al., 2005;, with permission from Wiley & Sons; Bauer et al., 1995; Goldsbury et al., 2000a,b; Goldsbury et al., 2005, all with the permission of Elsevier. Scale bars, 50 nm.
The overall accuracy of a STEM mass measurement is typically 5-10% depending on the sample. Instrument-related uncertainty predominantly involves the counting statistics of the scattered electrons: the quantum efficiency and the collection efficiency of the annular detector (Wall and Hainfield, 1986; Müller et al., 1992, Müller & Engel, 2006). Precise knowledge of the incident beam current and microscope magnification are also prerequisites (for a detailed discussion of instrumental accuracy and calibration see: Wall and Hainfield, 1986 and Müller et al., 1992). Mass measurements have been achieved with samples at room temperature (Müller et al., 1992), and with samples at −160°C (Wall and Hainfield, 1986) the latter condition reducing beam-induced mass loss. Since mass loss kinetics can be measured for each sample and appropriately corrected for (Müller et al., 2001), the results of room temperature measurements are equivalent to those obtained at −160°C. Specimen-related factors are also a large contributor to the experimental error (Müller et al., 1992). These include the thinness and uniformity of the carbon support film and the purity of the sample (Wall and Hainfield, 1986; Müller and Engel, 2006). To routinely achieve accuracies of 5-10% for mass measurements, it is essential to minimize statistical variation in the support scattering by preparing highly reproducible thin, clean carbon films, to avoid residual salts and small polypeptide contaminants in the sample and to carry out low dose measurements. In the case of amyloids, the presence of unassembled protein monomers and small aggregates (below 100 kDa) in the sample is often unavoidable. This can decrease the accuracy of the measurements in the same way as other polypeptide contaminants.
Images are analyzed by selecting regions-of-interest (ROIs; Figure 1B) and using the software to calculate their total mass, average MPL (Supplementary Material Figure S1) or average mass-per-area (MPA; Kržyzáneka,et al.,2009; Wall et al., 2008). Correction is made for electron beam-induced mass-loss and the data is pooled and binned into histograms (Figure 1C). The histograms immediately reveal the extent of heterogeneity of a specimen, and in the case of amyloids formed from short polypeptides (~3-5 kDa), can often be described by multiple Gaussian distributions demonstrating hierarchical assemblages of protofilament strands in mature fibrils (Figure 1C). As each pixel of a STEM scan is a quantitative measurement it is also possible to obtain detailed mass maps (Wall and Hainfeld, 1986; Wall et al., 2008; Reichelt et al, 1990). In particular, this allows MPL variations along the length of filaments that are not uniform to be more finely monitored (Herrmann et al., 1996; Steinmetz et al., 2008).
It should be noted that recently, an attempt was made to use dark-field images obtained by tilted-beam TEM to estimate the MPL of amyloid fibrils (Chen et al., 2009). Unlike the dedicated STEMs, this technique utilizes a relatively ill-defined electron signal making the measurement entirely dependent on a mass standard; absolute mass values cannot be obtained. Accordingly, the standard must be of high-quality, a criterion that may not have been optimal in the work in question; the TMV particles show clear signs of retained salt (high electron scattering at the edges of the virus rods).
3. Polymorphisms in amyloid fibrils revealed by STEM
Amyloid fibrils are visualized by electron microscopy as unbranched, ~6-20 nm wide, rope-like polymers of up to several μm in length that, as stated above, have cross-β core structures (Eanes and Glenner, 1968). Variations in the packing of the β-strands in their cores are now thought to distinguish different amyloids from each other and to distinguish different polymorphs assembled from the same amyloidogenic building blocks (Baxa et al., 2006; White et al., 2009; Petkova et al., 2005, Kajava et al., 2009). Broadly speaking, there are two types of polymorphism: (1) Variability in fibril morphology and the number of filamentous strands termed protofilaments. This form of polymorphism is clearly discernable in images (Figure 1D and Goldsbury et al., 1997) and can be further dissected by STEM MPL measurements (Figure 1, B and C). (2) Molecular level polymorphism arising from slight differences in the conformation of polypeptide molecules within fibrils (Petkova et al., 2005). This second type of polymorphism usually cannot be dissected by STEM MPL measurements and will only be visible by electron microscopy if different long-range repeats result.
STEM MPL measurements have been instrumental in determining the relationship between polymorphic variants that co-exist in some fibril populations (Bauer et al., 1995; Goldsbury et al., 1997, 2000a,b; Sen et al., 2007). Aβ, calcitonin, amylin, and other relatively short amyloid-forming polypeptides have in common that they form hierarchical assemblages of fibrils with different numbers of protofilament strands (Figure 1, C and D). The resulting fibrils typically have from two to six protofilaments that are either coiled in cable-like structures or packed side-by-side in ribbon-like structures (Figure 1D) (Bauer et al., 1995). The various fibril polymorphs can often be distinguished in STEM MPL histograms as Gaussian components whose peaks are spaced at equal increments corresponding to the MPL of one protofilament (Figure 1C). Their coiling gives rise to axially periodic crossover points that are visible in side-views of the fibril. The coiled fibrils and ribbons of some amyloids, e.g., those formed by amylin and Aβ, have a distinct left-handed twist. The periodicity is distinctive for a given amyloid, although some fibrils formed by different polypeptides have the same axial periodicities, e.g., both amylin and Aβ form 2-protofilament coiled fibrils that have an ~25 nm cross-over spacing (Figure 1D) (Goldsbury et al., 2000a,b). Although, there is virtually no amino acid sequence homology, the amylin and Aβ polypeptides are of similar length and have corresponding short internal sequences with high β-sheet forming propensity. This, in turn, may explain a common tertiary fold resulting in similar fibril morphologies (Goldsbury et al., 2000a,b) and structural models (Wiltzius et al., 2008, Petkova et al., 2002). Conversely, polymorphic fibrils assembled from the same polypeptide subunit can have different discrete periodicities (Baxa et al., 2002, Goldsbury et al., 2000a,b).
STEM is able to link the packing density indicated by the MPL to the axial periodicity and width of fibrils, imposing strong constraints for model building (Kajava et al., 2005; Petkova et al., 2002, Luca et al., 2007, Wiltzius et al., 2008). In this context, it is often helpful to express fibril MPLs in terms of the number of monomeric subunits (Nsub) per 0.47 nm axial step, which corresponds to the inter-strand spacing in a -sheet. To within experimental error, the MPLs of many amyloid fibrils come out as simple integral numbers in these units (e.g. Aβ (Figures 1 and 2), some tau-related fibrils (Figure 3), several kinds of fibrils of Ure2p (Figure 4) and Sup35p fusions (Diaz-Avalos et al, 2005) (Table 1). The non-unitary cases (Nsub > 1) generally represent fibrils consisting of multiple protofilaments. Along the same lines, a fibril may entail more than one 0.47 nm step per monomeric subunit, as in the case of HET-s, which has two steps (Nsub ~0.5 monomers per 0.47 nm step; Figures 2 and 4). According to current data, there are some apparent exceptions to this trend, e.g., amylin fibrils (Goldsbury et al., 2000a; Figure 1), some tau fibrils (von Bergen et al., 2006; Figure 3), and one type of Sup35p fusion fibril (Diaz-Avalos et al, 2005) (Table 1).
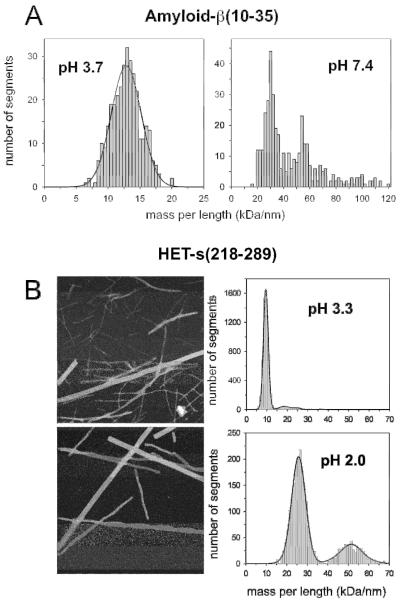
Figure 2. Fibril mass-per-length distributions depend on the solution conditions.
Shown here for (A) Aβ10-35 and (B) HET-s-(218 –289); MPL of 9 kDa/nm at pH 3.3 indicating 0.5 monomers per 0.47 nm (monomer mass, 8.5 kDa) and the MPLs of 26 and 52 kDa/nm at pH 2.0 indicating 1.4 and 2.9 monomers per 0.47 nm, respectively. Adapted from Antzutkin et al., 2002 and Sen et al., 2007. Scale bar, 100 nm.
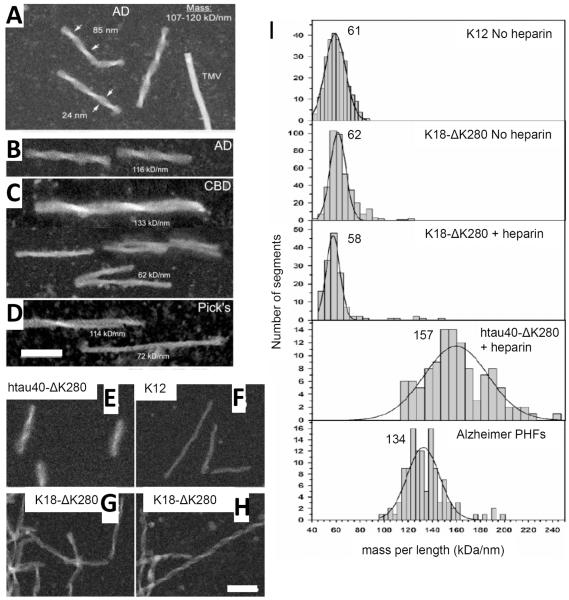
Figure 3. STEM measurements of tau fibrils isolated from diseased brain or assembled in vitro.
Fibrils isolated from (A,B) Alzheimer disease (C) Corticobasal Degeneration and (D) Picks disease brains. (E-H) Synthetic fibrils assembled from recombinant tau constructs as indicated. The full-length tau protein construct shown here is htau40-ΔK280, while K18- ΔK280 and K12 represent shorter C-terminal fragments of tau (von Bergen et al., 2006). (I) STEM MPL histograms of Alzheimer brain PHFs and recombinant tau fibrils as indicated. The Gauss peak positions are noted; units kDa/nm. The corresponding number of monomers per 0.47 nm step are 2.2 for K12 (subunit mass, 13.1 kDa), 2.0 or 2.1 for K18ΔK280 (13.7 kDa) and 1.6 for htau40ΔK280 (45.7 kDa – full-length tau). Note that the full-length tau Nsub per 0.47 nm value is ~30% less than the values obtained for constructs of repeat domain fragments. This indicates that full-length tau packs less densely in filaments than the repeat domains alone. Adapted from von Bergen et al., 2006 (Copyright 2006 American Chemical Society) and Ksiezak-Reding et al., 2005, with permission from Wiley & Sons. Scale bars, 100 nm.
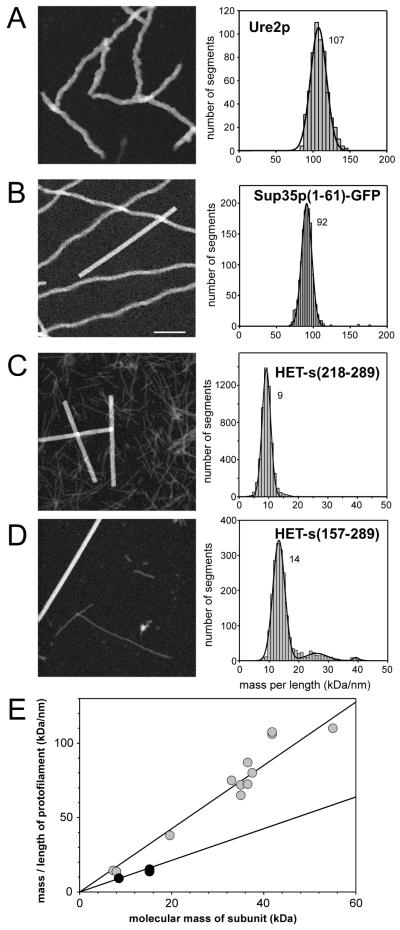
Figure 4. STEM measurements of prion protein fibrils.
(A) Ure2p. (B) Sup35p(1-61)-GFP, VK variant (Diaz-Avalos et al., 2005). (C) HET-s(218-289). (D) HET-s(157-289). Left dark-field STEM images of the unstained samples, right MPL histograms. The Gauss peak positions are noted; units kDa/nm. The corresponding numbers of monomeric subunits per 0.47 nm step are ~ 1 for Ure2p (monomer mass, 41.8 kDa), ~1 for Sup35p(1-61)-GFP (37.2 kDa), 0.5 for HET-s(218-289) (8.5 kDa), and ~ 0.5 for HET-s(157-289) (15.5 kDa). Note that Ure2p and Sup35p filaments both have ~1 monomers per 0.47 nm step, whereas the HET-s filaments have only ~ 0.5 monomers per 0.47 nm step, indicating a different packing arrangement. (E) The graph shows the average MPL of several Ure2p constructs (fusions of prion domain with different C-terminal moieties) in gray, HET-s(218-289) and HET-s(157-289) in black against the respective subunit molecular mass of each construct. The linear regressions of Ure2p constructs has a slope of ~1 monomeric subunit per 0.47 nm whereas the regression for HET-s constructs has a slope of ~0.5 monomers per 0.47nm. Adapted from Baxa et al., 2003; Diaz-Avalos et al., 2005; and Sen et al., 2007. Scale bar for A to D, 100 nm.
Table 1.
Summary of MPLs and corresponding monomeric units per 0.47 nm step along the fibril axis for different amyloid fibrils investigated by STEM
|
Monomer MW |
Fibril MPL kDa/nm |
Subunits /0.47nm |
Reference | ||
|---|---|---|---|---|---|
| Prion constructs | Ure2p | 41.8 | 107 | ~1 | Baxa et al., 2003 |
| Sup35p1-61-GFP | 37.2 | 92 | ~1 | Diaz-Avaloz et al., 2005 | |
| HET -s(218-289) | 8.5 | 9 | ~0.5 | Sen et al., 2007 | |
| HET -s(157-289) | 15.5 | 14 | ~0.5 | Sen et al., 2007 | |
|
Tau, in vivo isolates |
Alzheimer | ~41*# | 134 | ~1.5# | von Bergen et al., 2006 |
| Alzheimer | ~41*# | 107-120 | # | Ksiezak-Reding and Wall, 2005 | |
|
Alzheimer;
pronase treated |
# | 65 | # | Wischik et al., 1988 | |
| Picks | ~41*# | 71-113 | # | Ksiezak-Reding and Wall, 2005 | |
| Tau constructs | K12 | 13.1 | 61 | ~2 | von Bergen et al., 2006 |
| K18deltaK280 | 13.7 | 58 | ~2 | ||
|
K18deltaK280 +
heparin |
13.7 | 62 | ~2 | ||
| htau40deltaK280 | 45.7 | 157 | ~1.6 | ||
| Aβ1-40 | Protofibrils | 4.3 | 19 | ~2 | Goldsbury et al., 2005 |
| Mature fibrils | 18 | ~2 | |||
| 27 | ~3 | ||||
| Aβ1-42 | Mature fibrils | 4.5 | 24 | ~2.5 | Schmidt et al., 2009 |
| 46 | ~5 | ||||
| Amylin | 3.9 | 22 | ~2.6 | Goldsbury et al., 2000 | |
| 30 | ~3.6 | ||||
| 41 | ~5 | ||||
| 51 | ~6 | ||||
| Calcitonin | 3.4 | 20 | ~3 | Bauer et al., 1996 | |
| 27 | ~4 |
Average tau subunit MW.
Probably include lower molecular weight species; degradation can occur after death and during filament isolation.
Interestingly, for many of the shorter polypeptides (up to ~5 kDa), the minimum number of protofilaments in a mature fibril is two, with single protofilaments rarely being observed (Figure 1C). As mentioned above, many of these polypeptides have a propensity to form even higher-order fibrils. For example, the predominant amylin fibrils contain 3 protofilaments. Although the most prevalent Aβ1-40 fibrils contain 2 protofilaments, higher MPL species are also present (Figure 1C). These higher MPL peaks are spaced at equal 9 kDa/nm increments corresponding to the MPL of one protofilament (Figure 1C) (Table 1). However, it should be noted that another study envisaged a higher MPL Aβ protofilament of 23 kDa/nm rather than 9 kDa/nm indicated by the data in Figure 1C (Schmidt et al., 2009).
Further to this, the prevalence of different fibril polymorphs can be influenced by solution conditions (Fraser et al., 1991, Gosal et al., 2005). For example, Aβ fibrils assembled under shaking or quiescent conditions also showed differences in the most prevalent fibril polymorph (Goldsbury et al., 2000b; Petkova et al., 2005). Similarly, Aβ10-35 fibrils prepared at pH 3.7 revealed thinner fibrils of lower MPL than at pH 7.4 where heavier fibrils predominated (Figure 2A) (Antzutkin et al., 2002). HET-s prion domain fibril interactions were also found to be dependent on pH with triplets predominating at pH 2 and singlets at pH 7.4 (Figure 2B) (Sen et al., 2007). STEM measurements showed that the MPL of singlet fibrils did not change. Rather, interactions between charged side-chains influenced by pH caused the aggregation of singlet fibrils into higher order assemblies (Sen et al., 2007; Figure 2B). By contrast, deleting the first 8 amino acids of the 37-amino acid residue amylin polypeptide did not change the overall fibril morphology, although it altered the relative frequency of 2- versus 3-protofilament fibril polymorphs present as shown by STEM MPL analysis, increasing the relative number of the 2-protofilament polymorph (Goldsbury et al., 2000a). This indicates an influence of the N-terminus on protofilament-protofilament interactions in this case. On the other hand, fibrils formed from larger proteins are often more homogenous and as such can give rise to single Gaussian distributions in STEM MPL histograms. Examples may be found in Figures 3 and 4. One explanation for the absence of hierarchal assembly in many cases may be that flanking domains outside the β-sheet core of the protein sterically inhibit filament-filament association, as for example in prion fibrils (Figure 4). The situation for some tau filaments (Figures 3) is discussed further below.
As described above, Aβ fibrils formed under different growth conditions – quiescent vs. agitated or low vs. high pH – exhibit different morphologies and differentiable STEM MPL distributions (Fraser et al., 1991; Goldsbury et al., 2000a,b; Antzutkin et al., 2002; Petkova et al., 2005) (Figure 2A). Petkova et al., (2005) made the additional observation that quiescent vs. agitated fibril preparations are distinct, not only at the morphological level, but also at the molecular level by their solid state NMR spectra. From seeding experiments, they proposed a mechanism of self-propagating molecular polymorphism whereby the underlying molecular structure of “parent” fibrils defined by solid-state NMR spectra dictates the ultimate structure of “daughter” fibrils. In this particular case, the molecular-level polymorphism also gave rise to discernable differences in the numbers of protofilaments per fibril detectable by STEM MPL measurements and in TEM micrographs (Petkova et al., 2005). However, the effect of molecular-level polymorphism on the overall morphology of fibrils may in other cases be more subtle and not clearly discernable by STEM, for instance as discussed below for yeast prions.
4. Structural polymorphism underlies variant phenotypes in yeast prions
In infectious amyloids (prions), filament structures are thought to correlate with phenotypical behavior in the infected organisms (e.g. neurotoxicity in humans). In yeast, for example, the prions [URE3] and [PSI+] are directly linked to loss of function of the respective prion proteins, Ure2p and Sup35p (Wickner 1994) (Figure 4, A and B). In wild-type (uninfected) yeast strains, these proteins are soluble and exercise biological functions - for Sup35p in translation termination and for Ure2p in nitrogen catabolism. In the prion form (in infected cells), they form filaments with no (Ure2p) or greatly reduced (Sup35p) activity. The direct connection to a phenotype makes these systems uniquely advantageous for studying the so-called prion strain or variant phenomenon. Prion variants were first observed for the mammalian prion protein (Collinge et al.,2007, Fraser and Dickinson 1973) and refer to slight phenotypical differences that are inheritable. Recently, cell-cell transmission of amyloid aggregates has also been recognized as a potential mechanism for the spread of non-prion amyloids within organs – for example the progression of α-synuclein aggregates in the Parkinson’s disease brain (Desplats et al., 2009). In yeast, different prion variants can be distinguished by differences in the residual biological activity of the prion protein (strength of prion) and the mitotic stability (stability of prion) or some other phenotypic features.
It has been shown that in vitro assembled yeast prion fibrils give rise to a range of variants when transformed into yeast cells (Brachmann et al., 2005, Tanaka 2004) suggesting that there is a corresponding structural polymorphism. Interestingly, only one fibril type is isolated from any selected yeast clone despite the original recombinant prion inoculum containing a polymorphic mixture of fibrils (Baxa et al., 2006, Brachmann et al., 2005, Tanaka et al., 2004). This outcome indicates a mechanism of selection whereby, although there could be more than one fibril polymorph in a single cell, only the “dominant” one prevails (possibly because it grows faster and depletes the soluble protein from the other fibril variants). The in vitro assembled fibrils all give rise to MPL histograms with a single peak (Figure 4A) (Baxa et al., 2003; Diaz-Avalos et al., 2005; Sen et al., 2007). Since by and large they also have indistinguishable MPLs indicating the presence of ~1 monomer per 0.47 nm (Table 1), their polymorphism exists at the level of subtle differences in conformation that are currently beyond the resolution of STEM (see above). Other features, e.g. repeat length and growth rate, may reflect these subtle differences in structure (Baxa et al., 2006; DePace et al., 2002, Diaz-Avalos et al., 2005). However, in one study of a Sup35p1-61-GFP construct, apparent small deviations in MPL were reported, indicating the presence of slightly more than 1 monomer per 0.47 nm for some fibril polymorphs (Table 1; Diaz-Avalos et al., 2005).
Nevertheless, the general architecture of the fibril variants is very similar and the subunits have been shown to be organized in an in-register, parallel, stacked arrangement of β-sheets (Shewmaker et al., 2006, Baxa et al., 2007, Wickner et al., 2008). In summary, the structural polymorphism underlying yeast variants is likely to reflect subtle conformational differences in the parts of the polypeptide chain involved in the amyloid structure (Baxa et al., 2006). The “parallel superpleated β-structure” model (Kajava et al, 2004) is in compliance with these constraints and affords a framework wherein such variants could be constructed. The distinction between infectious, propagating amyloids and non-infectious amyloids is very likely not based on general morphological differences but due to slight differences in growth and breakage rates of fibrils and their interactions with the environment (Baxa 2008; Tanaka et al., 2006).
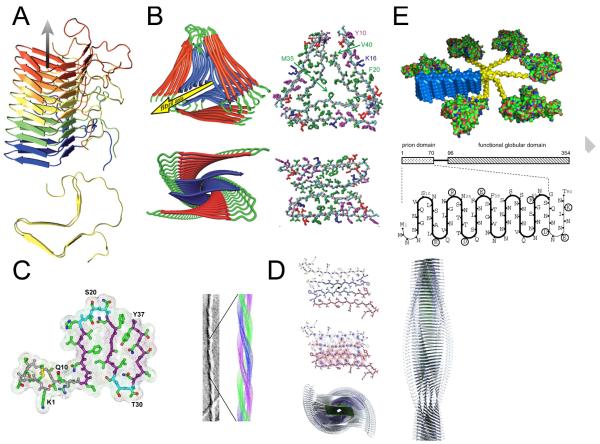
The MPL and the in-register parallel architecture of yeast prions is in marked contrast to the HET-s prion protein found in the filamentous fungus Podospora anserina. Fibrils of HET-s have a MPL of ~0.5 monomeric subunits per 0.47nm (Figure 4E; Table 1; Sen et al., 2007) and their structure has recently been elucidated by solid state NMR (Figure 5A; Wasmer et al., 2008; Ritter et al., 2005). Appending an extra 50 amino acid residues to the 72-residue HET-s prion domain did not alter the axial packing density of subunits per 0.47nm in the fibrils, consistent with the proposition that the prion domains form a β-sheet backbone that is decorated peripherally with other domains (Figure 4, C and D). HET-s also differs from the other yeast prions in that it has a biological function in the prion form, which therefore might be seen as the “native” structure of the protein.
Figure 5. Molecular structure models of amyloid fibrils aided by packing constraints derived from STEM measurements.
(A) HET-s prion structure (Wasmer et al., 2008; adapted with permission from AAAS). (B) Aβ fibril structures (Paravastu et al., 2008). (C) Amylin fibril structure (Kajava et al., 2005). (D) Amylin fibril structure (Wiltzius et al., 2008; adapted with permission from The Protein Society). (E) Ure2p (Kajava et al., 2004).
5. Differentiation of Aβ oligomers, metastable-protofibrils and mature fibrils using STEM
Solutions of amyloid polypeptides often form metastable structures at early time points before mature amyloid fibrils appear. This has been especially well investigated in the case of the Alzheimer’s disease-associated Aβ polypeptide whereby these various assemblies have been given a complex set of terminology in the literature – labels include “oligomers”, “globulamers”, “protofibrils” and “annular pores” (see glossary of terms: Box 1) (Kodali and Wetzel, 2007). Since in the disease context, smaller oligomeric structures formed by Aβ have been linked to cytotoxicity (Glabe, 2008), it is of considerable interest to determine the relationship of these transient assemblies to stable amyloid fibrils, which are the predominant structures that accumulate in brain tissue. Despite the characterization of oligomers of various sizes and metastable protofibrils in solutions of Aβ in vitro, the actual molecular pathways of assembly are poorly understood. Indeed, there are many examples of “off-pathway” stable and metastable oligomeric and fibrillar amyloid species (Gellermann et al., 2008; Necula et al., 2007; Ehrnhoefer et al., 2008; Gosal et al., 2005). The dynamic and heterogeneous nature of the aggregates makes kinetic studies challenging (Yun et al., 2007, Bitan et al., 2005). At the same time, given the proposed role of oligomers in affecting cytotoxicity in diverse diseases, further understanding of their placement within or separate from mature amyloid fibril assembly pathways is needed.
In the case of Aβ, STEM was used to compare the MPLs of metastable protofibrils with mature fibrils (Figure 1, B and C). The homogenous appearance of the protofibrils in both STEM and TEM images suggested that at the morphological level, they comprised a single polymorph. This inference was supported by the MPL histogram, which could be described by a single Gaussian distribution (Figure 1C; Table 1). In particular, STEM revealed that the MPL of the metastable protofibrils was too large for them to correspond to protofilament strands of the polymorphic mature fibrils (Figure 1, B and C). STEM was also used to measure the mass distribution of the globular oligomers that appear in solutions of Aβ (Table 1). Aβ oligomerization was thus found to be highly heterogeneous, realizing small oligomers (e.g., up to hexamers) as well as larger spheroidal structures containing from ~12 to 200 Aβ monomers (Glabe, 2008; Bitan, 2006; Goldsbury et al., 2000b). As the lower limit for STEM detection of unstained particles is approximately 100 kDa, this method is restricted to the analysis of oligomers of n >~20 monomers in the case of Aβ. Subject to this constraint, STEM mass measurements of oligomers formed from Aβ1-40 recorded a continuum of particle masses corresponding to up to ~200 monomers. Most of the masses measured fell into a broad peak at corresponding to 72±21 monomers, with a shoulder at 128±21 monomers (Goldsbury et al., 2000b). Further, STEM mass measurements of oligomers formed by Aβ polypeptide sequences containing the familial Alzheimer’s disease “Arctic mutation” revealed populations containing 18-93 monomers or 81-186 monomers, in reasonable agreement with assemblies of the wild-type Aβ1-40 (Lashuel et al., 2003). Other techniques including electrophoretic and cross-linking assays are needed to identify the smaller oligomers formed by Aβ (Bitan, 2006; Barghorn et al., 2005). Cross-linking assays have documented transiently stable Aβ pentamers and hexamers that are particularly abundant in solutions of Aβ1-42 (Bitan, 2006). Another in vitro study characterized stable dodecameric globular oligomers also formed by Aβ1-42 that are proposed to assemble through an aggregation process distinct from the fibril-forming pathway (Barghorn et al., 2005; Gellermann et al., 2008).
Coupled to TEM imaging and STEM mass measurements, time-lapse atomic force microscopy has given a detailed picture of how the various Aβ fibril polymorphs form and elongate in vitro, helping to clarify the inter-relation of the various assembly pathways (Goldsbury et al., 1999, 2005). Importantly, seeding with the species of interest considerably increased assembly rates. The propensity of specific amyloid structures to propagate themselves in seeded growth was further exploited in a recent study employing MPL and NMR measurements to characterize the species present in Alzheimer’s disease brains (Paravastu et al., 2009). Here fibrils extracted from the brains were used to seed fibril formation in synthetic Aβ1-40 solutions.
6. STEM analysis of tau filaments isolated from brains of Alzheimer’s disease and related dementias
It is desirable to be able to relate structural information gained from in vitro assembled amyloid to what occurs in organs of the body as amyloid-associated diseases progress. STEM provides a way to compare amyloid fibrils purified from tissue to fibrils assembled in vitro. However, the only ex vivo fibril preparations so far extensively analyzed by STEM are the paired helical filaments (PHFs) made of the microtubule-associated protein tau that are isolated from intracellular neurofibrillary tangles, a hallmark in Alzheimer’s and other neurodegenerative diseases (note: a recent study has investigated ex vivo Alzheimer’s disease-associated Aβ fibrils, using energy filtering transmission electron microscopy (EFTEM) (Feja et al., 1997) to make MPL measurements (Paravastu et al., 2009)). Due to characteristic intraneuronal accumulations of tau, all of these neurodegenerative diseases are classified as “tauopathies”. Understanding how the normally soluble and microtubule-bound axonal tau protein forms aberrant amyloid aggregates in the somatodendritic compartment of neurons is of great interest. A number of studies have been carried out by the Brookhaven STEM group (reviewed in Ksiezak-Reding and Wall, 2005), including extensive MPL measurements on tau fibrils (PHFs) isolated from different Alzheimer’s disease brains as well as from brains of other tauopathies including corticobasal degeneration and Pick’s disease (Figure 3, A to D; Ksiezak-Reding and Wall, 1994; Ksiezak-Reding et al., 1996, Ksiezak-Reding et al., 1998, King et al., 2001). Following this work, the Basel STEM group independently analyzed PHFs isolated from Alzheimer’s disease brains and compared their packing density to in vitro assembled fibrils made from various tau constructs (Figure 3, E to I; von Bergen et al., 2006).
The results obtained by the two STEM groups for tau PHFs isolated from various Alzheimer’s disease cases are in good agreement (Figure 3I; Table 1). Variations between the individual Alzheimer’s disease cases and in purification procedures likely explain the differences. Synthetic fibrils made from the full-length tau protein construct htau40-ΔK280 assemble into filaments with MPLs that span a large range indicating sample heterogeneity, although there appears to be no hierarchal filament association detectable by STEM (Figure 3I). The MPLs measured for PHFs isolated from Alzheimer’s brains may be interpreted in the same way (Figure 3I). This reflects the more homogenous morphological appearance of fibril preparations from synthetic full-length tau constructs, which generally comprise filaments of consistent width and periodicity in electron micrographs (Figure 3E-H). By comparison, synthetic fibril preparations formed from ~3-5 kDa amyloid polypeptides look relatively heterogeneous whereby the MPL histograms can clearly be described by multiple Gaussian distributions (Figure 1B-D).
Short fragments of the tau C-terminus corresponding to its microtubule-binding repeats, form fibrils that, when examined by negative stain TEM, appear morphologically indistinguishable from full-length tau fibrils except that they are narrower and lack the surrounding characteristic “fuzzy coat” (Figure 3, E to H) (von Bergen et al., 2006). The MPLs of these fibrils fall into single histogram peaks (Figure 3I). Synthetic fibrils assembled from the full-length construct of the largest tau isoform yielded MPLs corresponding to an axial packing of monomers per nm that was in very good agreement with that of preparations isolated from Alzheimer’s brains, particularly, when keeping in mind that the ex vivo Alzheimer’s disease fibrils represent a mixture of isoforms including lower MW species (Figure 3I). Notably, tau fragments formed denser fibrils corresponding to slightly more monomers per nm (Figure 3I). Hence, it was proposed that this difference arises because the peripheral N-terminal domain of full-length tau partially interferes with the fibril assembly process, thereby causing a slightly looser, suboptimal, packing (von Bergen et al., 2006).
A minor subset of the Alzheimer’s disease tau fibrils analyzed by the Brookhaven group had distinctly lower MPLs and could be isolated from the heavier fraction by sucrose density gradients (Ksiezak-Reding and Wall, 2005). As according to immunogold labeling experiments these lighter \fibrils lacked the N-terminal antibody epitopes of tau, it was concluded that they most likely arose from endogenous degradation. Indeed, both recombinant and ex vivo tau PHFs contain a central β-sheet core surrounded by a fuzzy coat that gives TEM images of these assemblies a blurred appearance (Figure 3E). The peripheral coat can be proteolytically digested in vitro, leaving behind a protease-resistant C-terminal core domain made up of tau’s microtubule binding repeats plus flanking sequences thereby yielding sharper TEM images (Wischik et al., 1988; Ksiezak-Reding and Wall, 2005; von Bergen et al., 2006). Interestingly, early MPL measurements of tau fibrils isolated from Alzheimer’s disease brains after pronase treatment (Wischik et al., 1988) demonstrate that the MPL of the protease resistant fibril core is in very good agreement with that of synthetic fibrils assembled from short tau fragments comprising the assembly domains alone (Figure 3I; Table 1; von Bergen et al., 2006). Degradation also occurs to varying extents in vivo, during postmortem delay or during purification procedures, resulting in fibrils with lower MPLs than expected for fibrils made of full-length tau. This can be seen in Figure 3C and 3D for cases of corticobasal degeneration and Picks disease; the degraded filaments also sometimes appear to be laterally associated (Ksiezak-Reding and Wall, 2005). In contrast to samples derived from Alzheimer’s disease, preparations from Picks disease and corticobasal degeneration exhibited a higher abundance of the lighter fibrils, indicating a difference in fibril stability depending on the form of tauopathy (Figure 3, A to D; Ksiezak-Reding and Wall, 2005). A trend to lower MPL values from fibrils isolated from older compared to younger brains has also been observed, possibly indicating more fibril degradation at later disease stages (Ksiezak-Reding and Wall, 2005, von Bergen et al., 2006).
Importantly, STEM data has helped to prove that cofactors such as heparin act catalytically, i.e. to promote assembly without being incorporated into the fibril structure itself. Most remarkably, the subunit packing in synthetic fibrils assembled in vitro is similar to that in authentic fibrils, i.e. those isolated from Alzheimer’s brains (Table 1; Ksiezak-Reding and Wall, 2005, von Bergen et al., 2006). This finding is very encouraging suggesting that in vitro models of amyloid fibrils are physiologically relevant and, hence, will aid the rational design of inhibitors of tau filament formation.
7. STEM provides constraints for molecular structure model building of amyloid fibrils
High-resolution structures of amyloid fibrils (see Kajava et al., 2009 for recent review) have been (with rare exceptions (Wiltzius et al., 2008) notoriously difficult to obtain due to the insolubility, lack of crystallographic order and inaccessibility of filamentous specimens to X-ray crystallography or solution NMR. By contrast, solid-state NMR and electron paramagnetic resonance (EPR) have in some instances enabled secondary structure and tertiary conformations of peptide sequences within amyloid fibrils to be resolved (Wasmer et al., 2008; Ritter et al., 2005; Cobb et al., 2007; Petkova et al., 2002; Chen et al., 2007). Merging as many constraints from different methods as possible is therefore an important way to obtain high-resolution information about amyloids (Baxa 2008). To this end, STEM MPL measurements provide strong constraints and complement the information obtained from other methods including solid-state NMR, EPR and fluorescence labeling. Whereas NMR constraints give information about distances at the atomic level, STEM MPL data provide an indication of how the molecules are stacked or aligned at the macromolecular level, for example, within protofilaments.
To date, the most detailed experimentally determined structure for an amyloid fibril is the solid-state NMR-derived model of the C-terminal domain of the fungal prion protein HET-s (Figure 5A; Wasmer et al., 2008; Ritter et al., 2005). This model, which is based on >100 solid-state NMR constraints, envisages a fibril made of a single protofilament that is formed by stacking subunits in a β-solenoid conformation with each subunit contributing two turns of the solenoid. As such, this model predicted an MPL of Nsub = 0.5 monomers per 0.47nm, which was confirmed by STEM data (Table 1; Sen et al., 2007). MPL measurements have also played a crucial role in developing models for yeast prion fibrils Ure2p and Sup35p (Figure 5E; Kajava et al., 2004; Shewmaker et al., 2006). Another well-defined amyloid model is of an Aβ fibril polymorph (Petkova et al., 2002) and recently, a model for a different Aβ fibril polymorph was reported by the same group (Figure 5B; Paravastu et al., 2008). The conformation of the subunit within the protofilament in this second model is close to that in the first, although there are small but significant differences. The solid-state NMR signals were used to define secondary and tertiary structural elements of the polypeptides and interactions in the main-chain hydrogen-bonding direction for both fibril polymorphs. STEM MPL measurements (Antzutkin et al., 2002, Goldsbury et al., 2000, Paravatsu et al., 2008) were essential to ascertain that each protofilament consists of two continuous β-sheets stacked perpendicular to the fibril axis and that the Aβ fibril is built from two or three such protofilaments (Figure 5B; Petkova et al., 2002, Paravatsu et al., 2008). For another Aβ fibril polymorph, which has been studied by cryo-electron microscopy, STEM MPL measurements have reported Nsub = 5 monomers per 0.47nm (Table 1; Schmidt et al., 2009). It is not clear how this corresponds to the apparent two-fold symmetry of the fibrils however.
Several models of single amylin fibril polymorphs have likewise been proposed by using, in part, restrictions imposed by STEM MPL data (Figure 5, C and D; Kajava et al., 2005; Luca et al., 2007; Wiltzius et al., 2008). The latter models relied heavily on constraints from solid-state NMR (Luca et al., 2007) or X-ray crystallography (Figure 5D; Wiltzius et al., 2008) as well as information from biochemical and fluorescence resonance energy transfer (FRET) data (Padrick and Miranker 2001). Both propose a fibril polymorph, consistent with STEM MPL data, containing four parallel β-sheets differing only slightly in the details of side-chain packing. The structure of the fibrils formed by a 2.2 kDa model peptide called ccβ-Met has also been determined using a combination of several methods (Steinmetz et al., 2008). This peptide forms an α-helical coiled-coil structure at ambient temperature but converts to amyloid when the temperature is raised (Kammerer et al., 2004). Detailed analysis of MPL variations along individual filaments indicated that heterogeneity can even exist along the same fibril, an observation that might reflect the dynamic nature of amyloid fibrils described by Carulla et al., (2005).
A model recently derived from cryo-EM data for beta-2 microglobulin (β2m) fibrils assembled in vitro at pH 2.5 also used STEM MPL data as a major constraint (White et al., 2009). This model challenges the generic idea of amyloid as a continuous β-ribbon. Instead the β2m protofilaments, with an MPL of 53 kDa/nm resemble a string of beads and the tetrameric building blocks form at least three different subunit interfaces within the homopolymeric assembly. Importantly, the 3D EM maps reveal that the globular repeat forms an integral part of the fibril structure rather than being a folded “passenger” domain located externally to the fibril backbone, as for example seen in fibrils of Sup35p (Glover et al., 1997) and Ure2p (Kajava et al., 2004).
8. Concluding Remarks
The first contributions of STEM to the amyloid field were published in the 1990s (Ksiezak-Reding and Wall 1994; Bauer et al., 1995). Since then this technique has been widely employed to characterize amyloid fibril structure and polymorphism. In combination with constraints imposed by other experimental techniques, STEM-derived data on fibril MPL and on their morphological features and dimensions have made a major contribution to the current generation of structural models. Given the putative role of amyloid in a number of diseases, such models or their successors may ultimately bear fruit in the design of new diagnostic amyloid-binding probes and ultimately, rationally designed therapeutic compounds that suppress oligomer formation or fibril growth in vivo.
Supplementary Material
Acknowledgements
We thank all current and past operators of the Basel STEM for their contributions to the cited work and Robert Häring, Robert Wyss and Roland Burki for maintenance of the instrument and its computer system. We thank Robert Tycko, Beat Meier and David Eisenberg for data and images adapted in figures 2 and 4. The Basel STEM has been continuously supported by Swiss National Foundation grants (present Grant: 3100A0-108299 to A.E.) and by the Maurice E Müller Foundation of Switzerland. This work was supported in part by the Intramural Research Program of NIAMS, National Institutes of Health. The BNL STEM is supported by USDOE-OHER (Department of Energy, Office of Health and Environmental Research).
Abbreviations
- EM
electron microscopy
- EPR
electron paramagnetic resonance
- MPA
mass-per-area
- MPL
mass-per-length
- PHF
paired helical filament
- ROI
region of interest
- STEM
scanning transmission electron microscopy
- TEM
transmission electron microscopy
- TMV
tobacco mosaic virus
- NMR
nuclear magnetic resonance
- EFTEM
energy-filtering transmission electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- Antzutkin ON. Amyloidosis of Alzheimer’s Aβ peptides: solid-state nuclear magnetic resonance, electron paramagnetic resonance, transmission electron microscopy, scanning transmission electron microscopy and atomic force microscopy studies. Magn. Reson. Chem. 2004;42:231–246. doi: 10.1002/mrc.1341. [DOI] [PubMed] [Google Scholar]
- Barghorn S, Mandelkow E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry. 2002;41:14885–14896. doi: 10.1021/bi026469j. [DOI] [PubMed] [Google Scholar]
- Barghorn S, Nimmrich V, Striebinger A, Krantz C, Keller P, Janson B, Bahr M, Schmidt M, Bitner RS, Harlan J, Barlow E, Ebert U, Hillen H. Globular amyloid β-peptide oligomer - a homogenous and stable neuropathological protein in Alzheimer’s disease. J. Neurochem. 2005;95:834–847. doi: 10.1111/j.1471-4159.2005.03407.x. [DOI] [PubMed] [Google Scholar]
- Bauer HH, Aebi U, Haner M, Hermann R, Müller M, Arvinte T, Merkle HP. Architecture and polymorphism of fibrillar supramolecular assemblies produced by in vitro aggregation of human calcitonin. J. Struct. Biol. 1995;115:1–15. doi: 10.1006/jsbi.1995.1024. [DOI] [PubMed] [Google Scholar]
- Baxa U, Cheng N, Winkler DC, Chiu TK, Davies DR, Sharma D, Inouye H, Kirschner DA, Wickner RB, Steven AC. Filaments of the Ure2p prion protein have a cross-β core structure. J. Struct. Biol. 2005;150:170–179. doi: 10.1016/j.jsb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB, Steven AC. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J. Biol. Chem. 2003;278:43717–43727. doi: 10.1074/jbc.M306004200. [DOI] [PubMed] [Google Scholar]
- Baxa U, Cassese T, Kajava AV, Steven AC. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv. Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- Baxa U. Structural basis of infectious and non-infectious amyloids. Curr. Alz. Res. 2008;5:308–318. doi: 10.2174/156720508784533367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, Tycko R. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- Bitan G. Structural study of metastable amyloidogenic protein oligomers by photo-induced cross-linking of unmodified proteins. Methods Enzymol. 2006;413:217–36. doi: 10.1016/S0076-6879(06)13012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G, Fradinger EA, Spring SM, Teplow DB. Neurotoxic protein oligomers - what you see is not always what you get. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO. J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso I, Goldsbury CS, Müller SA, Olivieri V, Wirtz S, Damas AM, Aebi U, Saraiva MJ. Transthyretin fibrillogenesis entails the assembly of monomers: a molecular model for in vitro assembled transthyretin amyloid-like fibrils. J. Mol. Biol. 2002;317:683–695. doi: 10.1006/jmbi.2002.5441. [DOI] [PubMed] [Google Scholar]
- Carulla N, Caddy GL, Hall DR, Zurdo J, Gairi M, Feliz M, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558.23. 327–341. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. U S A. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register β-structure. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- DePace AH, Weissman JS. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat. Struct. Biol. 2002;9:389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Avalos R, King CY, Wall J, Simon M, Caspar DL. Strain-specific morphologies of yeast prion amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- Engel A. Scanning transmission electron microscopy: biological applications. In: Hawkes Peter., editor. Advances in Imaging and Electron Physics. Vol. 159. Academic Press; Burlington: 2009. pp. 357–386. [Google Scholar]
- Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- Feja B, Durrenberger M, Müller SA, Reichelt R, Aebi U. Mass determination by inelastic electron scattering in an energy-filtering transmission electron microscope with slow-scan CCD camera. J. Struct. Biol. 1997;119:72–82. [Google Scholar]
- Fraser H, Dickinson AG. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J. Comp. Pathol. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- Fraser PE, Nguyen JT, Surewicz WK, Kirschner DA. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys. J. 1991;60:1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellermann GP, Byrnes H, Striebinger A, Ullrich K, Mueller R, Hillen H, Barghorn S. A β-globulomers are formed independently of the fibril pathway. Neurobiol. Dis. 2008;30:212–220. doi: 10.1016/j.nbd.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Glabe CG. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Goldsbury CS, Cooper GJ, Goldie KN, Müller SA, Saafi EL, Gruijters WT, Misur MP, Engel A, Aebi U, Kistler J. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- Goldsbury C, Goldie K, Pellaud J, Seelig J, Frey P, Müller SA, Kistler J, Cooper GJS, Aebi U. Amyloid fibril formation from full-length and fragments of amylin. J. Struct. Biol. 2000a;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- Goldsbury CS, Wirtz S, Müller SA, Sunderji S, Wicki P, Aebi U, Frey P. Studies on the in vitro assembly of Aβ1-40: implications for the search for Aβ fibril formation inhibitors. J. Struct. Biol. 2000b;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- Goldsbury C, Frey P, Olivieri V, Aebi U, Müller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J. Mol. Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Gosal WS, Morten IJ, Hewitt EW, Smith DA, Thomson NH, Radford SE. Competing pathways determine fibril morphology in the self-assembly of β2-microglobulin into amyloid. J. Mol. Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Goux WJ. The conformations of filamentous and soluble tau associated with Alzheimer paired helical filaments. Biochemistry. 2002;41:13798–13806. doi: 10.1021/bi016079h. [DOI] [PubMed] [Google Scholar]
- Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Häner M, Taschner N, Burkhard P, Aebi U, Müller SA. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- Harper JD, Wong SS, Lieber CM, Lansbury PT. Observation of metastable Abeta amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Haener M, Brettel M, Müller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J. Mol. Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Holmes DF, Kadler KE. The 10+4 microfibril structure of thin cartilage fibrils. Proc. Nat Acad. Sci. U. S. A. 2006;103:17249–17254. doi: 10.1073/pnas.0608417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaikaran ET, Clark A. Islet amyloid and type 2 diabetes: From molecular misfolding to islet pathophysiology. Biochim. Biophys. Acta. 2001;1537:179–203. doi: 10.1016/s0925-4439(01)00078-3. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: the parallel superpleated β-structure. Proc. Natl. Acad. Sci. U. S. A. 2004a;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava A,V, Potekhin S,A, Corradin G, Leapman Richard D. Organization of designed nanofibrils assembled from alpha-helical peptides as determined by electron microscopy. J. Pept. Sci. 2004b;10:291–297. doi: 10.1002/psc.520. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Aebi U, Steven AC. The parallel superpleated β-structure as a model for amyloid fibrils of human amylin. J. Mol. Biol. 2005;348:247–252. doi: 10.1016/j.jmb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 2001;42:79–92. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Baxa U, Steven AC. β-arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. FASEB J. 2009 doi: 10.1096/fj.09-145979. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer RA, Kostrewa D, Zurdo J, Detken A, Garcia-Echeverria C, Green JD, Müller SA, Meier BH, Winkler FK, Dobson CM, Steinmetz MO. Exploring amyloid formation by a de novo design. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4435–4440. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A, McDonald M, Bian W, Bowles T, Baumgarten SC, Shi J, Stewart PL, Bullitt E, Gore D, Irving TC, Havens WM, Ghabrial SA, Wall JS, Stubbs G. Structure of flexible filamentous plant viruses. J. Virol. 2008;82:9546–9554. doi: 10.1128/JVI.00895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Köhler R, Schaefer K, Müller S, Vignon G, Diederichs K, Philippsen A, Ringler P, Pugsley AP, Engel A, Welte W. Structure and assembly of the pseudopilin PulG. Molec. Microbiol. 2004;54:647–664. doi: 10.1111/j.1365-2958.2004.04307.x. [DOI] [PubMed] [Google Scholar]
- Krzyžáneka V, Müller SA, Engel A, Reichelt R. MASDET — A fast and user-friendly multiplatform software for mass determination by dark-field electron microscopy. J. Struct. Biol. 2009;165:78–87. doi: 10.1016/j.jsb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Ksiezak-Reding H, Wall JS. Characterization of paired helical filaments by scanning transmission electron microscopy. Microsc. Res. Tech. 2005;67:126–140. doi: 10.1002/jemt.20188. [DOI] [PubMed] [Google Scholar]
- Ksiezak-Reding H, Wall JS. Mass and physical dimensions of two distinct populations of paired helical filaments. Neurobiol. Aging. 1994;15:11–19. doi: 10.1016/0197-4580(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Ksiezak-Reding H, Yang G, Simon M, Wall JS. Assembled tau filaments differ from native paired helical filaments as determined by scanning transmission electron microscopy (STEM) Brain Res. 1998;814:86–98. doi: 10.1016/s0006-8993(98)01052-x. [DOI] [PubMed] [Google Scholar]
- Ksiezak-Reding H, Tracz E, Yang LS, Dickson DW, Simon M, Wall JS. Ultrastructural instability of paired helical filaments from corticobasal degeneration as examined by scanning transmission electron microscopy. Am. J. Path. 1996;149:639–651. [PMC free article] [PubMed] [Google Scholar]
- King ME, Ghoshal N, Wall JS, Binder LI, Ksiezak-Reding H. Structural analysis of Pick’s disease-derived and in vitro-assembled tau filaments. Am. J. Pathol. 2001;158:1481–1490. doi: 10.1016/S0002-9440(10)64099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarkowska B, Aitken JF, Kistler J, Zhang S, Cooper GJ. The aggregation potential of human amylin determines its cytotoxicity towards islet β-cells. FEBS. J. 2006;273:3614–3624. doi: 10.1111/j.1742-4658.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- Krzyžáneka V, Müller SA, Engel A, Reichelt R. MASDET — A fast and user-friendly multiplatform software for mass determination by dark-field electron microscopy. J. Struct. Biol. 2009;165:78–87. doi: 10.1016/j.jsb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Aβ1-42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MP, Velasco PT, Chang L, Viola KL, Fernandez S, Lacor PN, Khuon D, Gong Y, Bigio EH, Shaw P, De Felice FG, Krafft GA, Klein WL. Monoclonal antibodies that target pathological assemblies of Aβ. J. Neurochem. 2007;100:23–35. doi: 10.1111/j.1471-4159.2006.04157.x. [DOI] [PubMed] [Google Scholar]
- Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Hartley DM, Petre BM, Wall JS, Simon MN, Walz T, Lansbury PT., Jr. Mixtures of wild-type and a pathogenic (E22G) form of Aβ40 in vitro accumulate protofibrils, including amyloid pores. J. Mol. Biol. 2003;332:795–808. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Petre BM, Wall J, Simon M, Nowak RJ, Walz T, Lansbury PT., Jr. Alpha-synuclein, especially the Parkinson’s disease-associated mutants, forms pore-like annular and tubular protofibrils. J. Mol. Biol. 2002;322:1089–102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Wall JS. Molecular electron microscopy approaches to elucidating the mechanisms of protein fibrillogenesis. Methods Mol. Biol. 2005;299:81–101. doi: 10.1385/1-59259-874-9:081. [DOI] [PubMed] [Google Scholar]
- Leapman RD, Gallant PE, Reese TS, Andrews SB. Phosphorylation and subunit organization of axonal neurofilaments determined by scanning transmission electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7820–7824. doi: 10.1073/pnas.94.15.7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer’s disease β-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–7. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Mandelkow E, von Bergen M, Biernat J, Mandelkow EM. Structural principles of tau and the paired helical filaments of Alzheimer’s disease. Brain Pathol. 2007;17:83–90. doi: 10.1111/j.1750-3639.2007.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, Vigouret JM, Paganetti P, Walsh DM, Mathews PM, Ghiso J, Staufenbiel M, Walker LC, Jucker M. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- Mocanu MM, Nissen A, Eckermann K, Khlistunova I, Biernat J, Drexler D, Petrova O, Schönig K, Bujard H, Mandelkow E, Zhou L, Rune G, Mandelkow EM. The potential for β-structure in the repeat domain of tau protein determines aggregation, synaptic decay, neuronal loss, and coassembly with endogenous Tau in inducible mouse models of tauopathy. J. Neurosci. 2008;28:737–748. doi: 10.1523/JNEUROSCI.2824-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller SA, Engel A. Structure and mass analysis by scanning transmission electron microscopy. Micron. 2001;32:21–31. doi: 10.1016/s0968-4328(00)00022-6. [DOI] [PubMed] [Google Scholar]
- Müller SA, Engel A. Biological scanning transmission electron microscopy: imaging and single molecule mass determination. CHIMIA. 2006;60:749–753. [Google Scholar]
- Müller SA, Goldie KN, Bürki R, Häring R, Engel A. Factors influencing the precision of quantitative scanning transmission electron microscopy. Ultramicroscopy. 1992;46:317–334. [Google Scholar]
- Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- Novitskaya V, Bocharova OV, Bronstein I, Baskakov IV. Amyloid fibrils of mammalian prion protein are highly toxic to cultured cells and primary neurons. J. Biol. Chem. 2006;281:13828–13836. doi: 10.1074/jbc.M511174200. [DOI] [PubMed] [Google Scholar]
- Nybo M, Svehag SE, Holm Nielsen E. An ultrastructural study of amyloid intermediates in Aβ1-42 fibrillogenesis. Scand. J. Immunol. 1999;49:219–223. doi: 10.1046/j.1365-3083.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Miranker AD. Islet amyloid polypeptide: identification of long-range contacts and local order on the fibrillogenesis pathway. J. Mol. Biol. 2001;308:783–94. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of β-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc. Nat. Acad. .Sci. U.S.A. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- Pollanen MS, Bergeron C. Modeling of a periodic instability in paired helical filaments reveals an axial repeat. Acta Neuropathol. Berl. 2000;99:534–538. doi: 10.1007/s004010051157. [DOI] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jr., Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell. Biol. 1990;10:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter C, Maddelein ML, Siemer AB, Lührs T, Ernst M, Meier BH, Saupe SJ, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl.):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Sachse C, Richter W, Xu C, Fändrich M, Grigorieff N. Comparison of Alzheimer Abeta(1-40) and Abeta(1-42) amyloid fibrils reveals similar protofilament structures. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19813–19818. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Baxa U, Simon MN, Wall JS, Sabate R, Saupe SJ, Steven AC. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J. Biol. Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt MJ, Graham HK, Kielty CM, Holmes DF. Mass-mapping of ECM macromolecules by scanning transmission electron microscopy. Methods Mol. Biol. 2009;522:151–161. doi: 10.1007/978-1-59745-413-1_9. [DOI] [PubMed] [Google Scholar]
- Steinmetz MO, Gattin Z, Verel R, Ciani B, Stromer T, Green JM, Tittmann P, Schulze-Briese C, Gross H, van Gunsteren WF, Meier BH, Serpell LC, Müller SA, Kammerer RA. Atomic models of de novo designed cc β-Met amyloid-like fibrils. J. Mol. Biol. 2008;376:898–912. doi: 10.1016/j.jmb.2007.11.100. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–238. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- von Bergen M, Barghorn S, Müller SA, Pickhardt M, Biernat J, Mandelkow E-M, Davies P, Aebi U, Mandelkow E. The core of tau-paired helical filaments studied by scanning transmission electron microscopy and limited proteolysis. Biochemistry. 2006;45:6446–6457. doi: 10.1021/bi052530j. [DOI] [PubMed] [Google Scholar]
- Wall JS, Hainfeld JF. Mass mapping with the scanning transmission electron microscope. Annu. Rev. Biophys. Biophys. Chem. 1986;15:355–76. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- Wall JS, Simon MN, Lin BY, Vinogradov SN. Mass mapping of large globin complexes by scanning transmission electron microscopy. Methods Enzymol. 2008;436:487–501. doi: 10.1016/S0076-6879(08)36027-3. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB. Amyloid beta-protein fibrillogenesis. Detection of a protofibrillar intermediate. J. Biol. Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- White HE, Hodgkinson JL, Jahn TR, Cohen-Krausz S, Gosal WS, Müller S, Orlova EV, Radford SE, Saibil HR. Globular tetramers of β(2)-microglobulin assemble into elaborate amyloid fibrils. J. Mol. Biol. 2009;389:48–57. doi: 10.1016/j.jmb.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius JJ, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Atomic structure of the cross-β spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischik CM, Novak M, Edwards PC, Klug A, Tichelaar W, Crowther RA. Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4884–4888. doi: 10.1073/pnas.85.13.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogulis M, Wright S, Cunningham D, Chilcote T, Powell K, Rydel RE. Nucleation-dependent polymerization is an essential component of amyloid-mediated neuronal cell death. J. Neurosci. 2005;25:1071–80. doi: 10.1523/JNEUROSCI.2381-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S, Urbanc B, Cruz L, Bitan G, Teplow DB, Stanley HE. Role of electrostatic interactions in amyloid β-protein (A) oligomer formation: a discrete molecular dynamics study. Biophys. J. 2007;92:4064–4077. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.