Abstract
A previous study (i.e. Dickmann, L., et al. (2004) Mol. Pharmacol. 65, 842-850.) revealed some unusual properties of the R108H mutant of cytochrome P450 2C9 (CYP2C9), including elevated thermostability relative to CYP2C9, as well as a UV-visible absorbance spectrum that was indicative of nitrogenous ligation to the heme iron. In the present study, size-exclusion chromatography and UV-visible absorbance spectroscopy of CYP2C9 R108H monomers demonstrated that nitrogen ligation is indeed intramolecular. Pulsed electron paramagnetic resonance of CYP2C9 R108H monomers showed that a histidine is most likely bound to the heme as previously hypothesized. An energy-minimized model of the R108H mutant maintained a CYP fold, despite substantial movement of several loop regions of the mutant and, therefore, represents an extreme example of a closed conformation of the enzyme. Molecular dynamics (MD) simulations of CYP2C9 were performed in order to study the range of energetically accessible CYP2C9 conformations. These in silico studies showed that the B-C loop region of CYP2C9 moves away from the heme to a position resembling the putative open conformation described for rabbit CYP2B4. A model involving the movement of the B-C loop region and R108 between the open and closed conformations of CYP2C9 is presented, which helps to explain the enzyme’s ability to regio- and stereo-specifically metabolize some ligands, while allosterically activating others.
Keywords: CYP2C9, pulsed EPR, conformational changes, size-exclusion chromatography, HYSCORE, UV-visible spectroscopy
Human cytochrome P450 2C9 (CYP2C9) is found primarily in the liver and metabolizes a wide variety of xenobiotics and endogenous compounds (1). One of the intrinsic functions of this enzyme is to epoxidize arachidonic acid, which appears to be important for vascular homeostasis. CYP2C9 also hydroxylates numerous xenobiotics, including the oral anticoagulant S-warfarin, the hydantoin anticonvulsant phenytoin, a wide range of non-steroidal anti-inflammatory drugs (NSAIDs) and even some polycyclic aromatic hydrocarbons (2). Moreover, CYP2C9 displays atypical steady-state kinetics with several substrates, which has been interpreted as evidence for multiple ligand occupancy within the enzyme’s active site (3). Despite the recent availability of substrate-bound CYP2C9 X-ray crystal structures (4, 5), questions persist as to how this enzyme can accommodate such a broad range of substrates, and possibly even multiple ligands within the enzyme’s active site (3).
Site-directed mutagenesis of CYP2C9 and X-ray crystal structures have provided some clues as to how drugs and other ligands might interact with the enzyme. The first X-ray crystal structure of CYP2C9 (PDB ID: 1OG5 (4)) showed S-warfarin bound in a hydrophobic cleft that was unexpectedly distant from the heme center (4). However, the CYP2C9 construct had several mutations within the F-G region of the protein that may have impacted ligand binding (4, 6, 7). A more recent X-ray crystal structure of CYP2C9 and fluriprofen (FP) (PDB ID: 1R9O (5)) identified R108 as a critical residue for binding of acidic substrates because the carboxylate of FP was hydrogen bonded to R108 (5). There were also significant differences in overall structure between 1OG5 and 1R9O, implying intrinsic conformational flexibility of CYP2C9 (4, 5).
We previously reported unusual spectral characteristics of the R108H mutant of CYP2C9, wherein the mutant exhibited a red-shifted Soret absorbance spectrum (λmax=424 nm) that is characteristic of heme bound by a nitrogen-containing ligand. One possible explanation for this trait is an intermolecular interaction between two CYP2C9 R108H monomers. This scenario was observed for CYP2B4, which was originally crystallized as a dimer that forms when the hydrophobic G’ helix of one monomer inserts into the active site cavity of a second monomer, and is stabilized by a coordinate bond between residue H226 and the CYP2B4 heme iron (8). Another possibility is that ligation of H108 to the heme might proceed intramolecularly within a single CYP2C9 R108H monomer. Since the R108 residue is only 12 Å from the heme iron in the X-ray crystal structure of CYP2C9 with FP bound, conformational changes caused by mutating this amino acid might induce another nitrogen-containing residue, lysine or arginine, to bind to the heme. On the other hand, R108 is located on the enzyme’s flexible B-C loop, which may be sufficiently flexible to permit intramolecular histidine coordination. Indeed, intramolecular histidine coordination between H72 within the B-C loop of CYP105P1 and the heme has been observed in the substrate-free X-ray crystal structure of this bacterial cytochrome P450 (9). Still, for this to occur, the B-C loop would have to be folded over the heme, potentially disrupting the secondary structure of the protein. Conversely, if the secondary structure is not significantly perturbed, then the R108H mutant could provide valuable insights into the overall conformational flexibility of CYP2C9.
In the current study, the effect of detergents on both the aggregation state and the UV-visible absorbance spectrum of the R108H mutant of CYP2C9 was examined in order to test whether ligation of the heme occurs inter- or intramolecularly. The residue directly bound to the heme was then elucidated using the pulsed electron paramagnetic resonance (EPR) technique: hyperfine sublevel correlation spectroscopy (HYSCORE). These results were used to construct an energy-minimized model of the R108H mutant in order to determine the conformational changes necessary to accommodate this B-C loop mutation, and also to ascertain whether or not the model maintained a characteristic CYP-fold. Finally, the range of conformations energetically accessible by CYP2C9 was explored with molecular dynamics (MD) simulations to provide the basis for a substrate-binding model that incorporates open and closed conformations of the enzyme.
Materials and Methods
General chemicals and reagents
Oligonucleotide primers, AccuPrime Pfx DNA polymerase, T4 DNA Ligase, and E. coli MAX Efficiency® DH5αF’IQ™ competent cells were purchased from Invitrogen (Carlsbad, CA), and restriction endonucleases were from New England Biolabs (Beverly, MA). Imidazole hydrochloride, L-histidine, isopropyl β-D-1-thiogalactopyranoside (IPTG), dithiothreitol, ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail, δ-aminolevulinic acid (δ-ALA), sodium dithionite, β-NADPH, and trifluoroacetic acid were obtained from Sigma Chemical Co. (St. Louis, MO). Nickel-nitrilotriacetic acid (Ni-NTA) SuperFlow® resin was from Qiagen (Valencia, CA), and CM Sepharose Fast Flow® was obtained from Amersham Biosciences Corp. (Piscataway, NJ). The detergents, Emulgen 911 and 5-cyclohexyl-1-pentyl-β-D-maltoside (CYMAL-5), were purchased from Kao Corporation (Tokyo, Japan) and from Anatrace, Inc. (Maumee, OH), respectively. HPLC-grade solvents were purchased from Fisher Scientific Co. (Fair Lawn, NJ). All other supplies were reagent grade or higher.
CYP2C9 mutagenesis, protein expression and purification
Wild-type CYP2C9 was modified for high level expression in bacteria by deleting amino acid residues 3-19 (inclusive) and incorporating a G25S mutation (4). The R108H mutation was generated using the overlap extension polymerase chain reaction method and both genes (termed here, CYP2C9 and CYP2C9 R108H) cloned into pCWori (10). Transformed DH5αF’IQ™ colonies containing recombinant CYP2C9 constructs were grown in 0.5 L Terrific Broth containing supplements as previously described (11). Expression was induced with 1 mM IPTG and 0.5 mM δ-ALA and flasks were shaken at 160 rpm and 28°C for 48 hrs at which time, yields of the protein varied between 800 and 1400 nmol/L. Cultures were harvested by centrifugation at 5000 g for 10 min. in a Beckman GS-6R centrifuge (Beckman Coulter, Brea, CA) at 4°C, washed in Storage Buffer (i.e. 50 mM Potassium Phosphate, pH 7.4, 20% glycerol, and 1 mM EDTA), and stored at −80°C. CYP2C9 and CYP2C9 R108H cell pellets were thawed and resuspended in 15 ml of TES buffer (100 mM Tris-acetate buffer, pH 7.6, 500 mM sucrose, and 0.5 mM EDTA) per g original wet weight of cells. Cells were treated with 0.3 mg lysozyme per g cells and the suspension diluted 2-fold by the drop-wise addition of an equal volume of chilled H20 and stirred for 30 min. The resulting spheroplasts were then pelleted by centrifugation at 10,000 g for 10 min. Next, the cells were gently homogenized and resuspended in Sonication Buffer (500 mM Potassium Phosphate, pH 7.4, 20% glycerol, 10 mM β-mercaptoethanol, and EDTA-free protease inhibitor cocktail). The resuspension was sonicated with three 30 sec. pulses using a sonic demembrator with two min. cooling in between using ice-salt water baths. The sonicated samples were pooled and centrifuged at 110,000 g in a Beckman 50 Ti rotor for 1 hr. at 4°C using a Beckman L8-70 Ultracentrafuge. CYMAL-5 was added to the supernatant and the protein was purified sequentially by nickel affinity and cation exchange chromatography as described in (5). The proteins were then concentrated using spin columns and stored at −80°C. The final yields of purified CYP2C9 and CYP2C9 R108H were 70 and 150 nmoles per liter of cell culture, respectively. The purities of the enzymes was >95% as judged by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Size-exclusion chromatography
Size-exclusion chromatography was performed on a Shimadzu VP Series high performance liquid chromatography (HPLC) equipped with a Pharmacia Superose 6 HR10/300 column and EZStart version 7.2.1 software. The flow rate was maintained at 0.5 mL min.−1 with a maximum allowed pressure of 215 psi. The UV-visible absorbance was monitored between 200 and 600 nm. CYP2C9 samples were calibrated against the 670 kD, 158 kD, 44 kD, 17 kD and 1.35 kD gel filtration standards (Biorad, Hercules, CA).
UV-visible absorbance spectroscopy
UV-visible absorbance spectroscopy studies were performed at 25°C on a Cary 3E absorbance spectrophotometer (Varian Scientific Instruments, Inc., Lake Forest, CA) or an Olis Modernized Aminco® DW-2 (Olis, Inc., Bogart, GA), as previously described (12-14). All samples were in 100 mM potassium phosphate (pH 7.4).
Pulsed EPR Spectroscopy
Pulsed EPR experiments were conducted with an X-band Bruker ESP-380e spectrometer (Bruker-Biospin, Billerica, MA), equipped with an MD-5 dielectric resonator, and an Oxford Instruments CF935 helium cryostat and GFS600 Transfer line (Oxford Instruments, Oxfordshire, UK) as previously described (15). HYSCORE was used in this study to probe the interaction of nearby nuclei with the heme iron. This is a 2D correlation EPR pulsed spectroscopy technique that correlates ENDOR frequencies from the same nucleus. The HYSCORE measurements were performed using a four-pulse sequence, which correlates with the electron nuclear double resonance (ENDOR) frequencies, and disperses the spectrum in two-dimensions with increased resolution for small second-order shifts caused by anisotropic interactions (15-19). All HYSCORE spectra were measured at a magnetic field of 0.2865 Tesla, nominal microwave frequency of 9.690 GHz, 90° microwave pulse length of 16 ns, a separation t=296 ns between the first two microwave pulses, and temperatures between 10 and 11K. There were 256 points measured in each dimension with an increment of 24 ns between the points. Additional parameters are in the figure legends.
The individual HYSCORE spectra were processed identically in the Bruker Xepr package. Baseline correction by a second-order polynomial was applied to each slice parallel to the x-axis, followed by baseline correction by a second-order polynomial on each slice parallel to the y-axis. The resolution enhancement windowing function was applied with x-Max = 6120 and x0 = −4000 in both directions, followed by zero-filling to 1024 points in both directions and 2D-Fourier Transformation. The magnitude was taken to produce the final spectra in Figure 2.
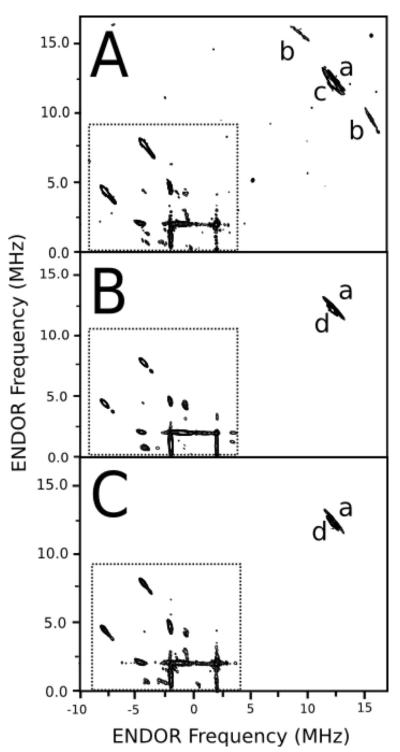
Figure 2. HYSCORE EPR spectra of A) CYP2C9, B) the R108H mutant, and C) CYP2C9 with 100 mM imidazole, respectively.
The ENDOR peaks labeled a-e) correspond to protons coupled to the heme iron. The nitrogen frequency region (< 9 MHz) is framed within the dashed rectangle. The location of a crosspeak indicates the ENDOR frequencies in different electron spin manifolds. The concentration of protein was 200 μM for all the samples and the spin echo-detected EPR spectra associated with the HYSCORE EPR spectra are found in Figure S1 of the Supplementary Materials. The x- and y- axes of the HYSCORE spectra correspond to the correlated 1H ENDOR frequencies.
The intensity of each HYSCORE spectrum was different due to small variations in the sample concentration, EPR spectrum shape, EPR relaxation times and number of acquisitions. The intense peak on the diagonal, which correspond to protons that are some distance away from the heme, were used to normalize the spectra. The effectiveness of the normalization is due to the relatively uniform proton density of proteins (20). Normalized spectra were subtracted from one another to produce the difference spectra in Figure 3.
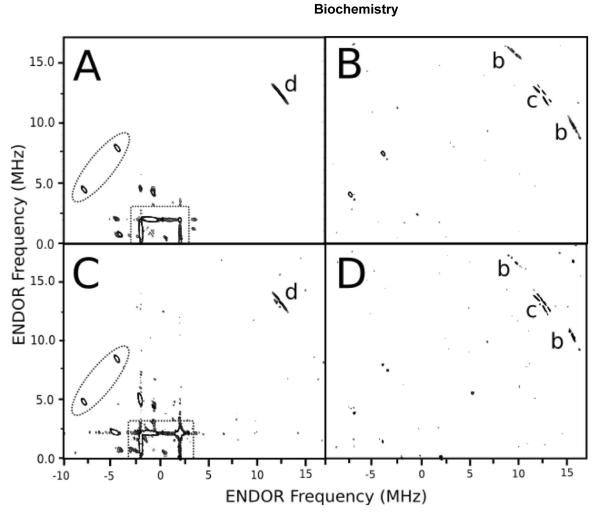
Figure 3. HYSCORE difference spectrum of the R108H mutant (A,B) and CYP2C9 with 100 mM imidazole (C,D), respectively.
The panels on the left (A and C) and right (B and D) are the positive and negative peaks from the HYSCORE difference spectra, respectively. The labeling is the same as in Figure 2, except that a dashed oval covers the peaks around 4.39 and 7.85 MHz, while the dashed box is around the ridges, found at ±1.91 MHz, in the nitrogen frequency region from both panels.
EPR theory
The low spin iron of the heme in the oxidized state has a spin quantum number S=½. The interaction of this electron with nearby nuclei shifts the NMR frequencies of these nuclei by the hyperfine coupling constant (A). The shifted frequencies are commonly known as ENDOR frequencies. For protons and other nuclei with a nuclear spin quantum number I= ½, the ENDOR frequencies are simply (ignoring second order effects):
| (Eq. 1) |
where vn is the NMR or nuclear Zeeman frequency, which is often much greater than A and νENDOR is the ENDOR frequency.
The 14N (I=1) of the pyrrole rings of the heme and the imidazole or histidine ligands to the heme are more complicated. They have a quadrupole moment (a characteristic of nuclei with I > ½) and typically have A>>νn. Their ENDOR frequencies (ignoring second order effects) are:
| (Eq. 2) |
where Q is the quadrupole splitting. Q, and often A, interactions are anisotropic, meaning that their values depend on the orientation of the heme, relative to the magnetic field of the EPR spectrometer. The anisotropy of A depends on the distance between the nucleus and the unpaired electron spin of the heme. Moreover, the anisotropy is very useful for locating protons, which are normally absent from protein structures determined by X-ray diffraction. The A at any orientation for an axial interaction can be written in terms of its isotropic part aiso (i.e. the isotropic hyperfine coupling constant) and its anisotropic part a⊥*(3 Cos2θ-1):
| (Eq. 3) |
where a⊥ is the perpendicular hyperfine coupling constant, and θ is the angle between the magnetic field and the unique axis of the hyperfine interaction. The anisotropic part of the hyperfine coupling were ascertained indirectly by measuring the second order shifts in the ENDOR frequencies of protons present in the HYSCORE spectra. These second-order shifts were then separated through a simple linear regression into the principal values of the hyperfine tensor as previously described (15-19).
The distance of the protons from the heme iron was calculated from the a⊥ using the standard formula for point dipoles.
| (Eq. 4) |
where μ0 is the magnetic permeability of free space, ge and gn are the electronic and nuclear g-factors, βe and βn are the electronic and nuclear Bohr magnetons, h is Planck’s constant and r is the electron-nuclear distance. This equation has already been used in earlier studies to estimate electron-proton distances in CYP101 (21, 22). The point dipole approximation is exact for measuring electron-nuclear distances, when the wavefunction of the unpaired electron spin does not overlap with the nucleus, has spherical symmetry and has a g-value that is isotropic. Our EPR results confirmed previous work on CYP101 (21, 22), showing a small isotropic hyperfine coupling with the protons of the axial water ligand and little wavefunction overlap. Goldfarb and coworkers found that the electron spin was largely confined to the Fe (III) d-orbital shell, which approaches spherical symmetry (22). The g-anisotropy of the system was about 15% (22), which gives a maximum error in the calculated distance of 5%.
Molecular dynamics simulations and energy minimization
MD simulations were performed on the X-ray crystal structure coordinates of CYP2C9 (PDB ID: 1R9O) and the R108H mutant (5), using the MD software package: Groningen MAchine for Chemical Simulation (GROMACS) Version 3.3.3 (23-25). Missing residues from the X-ray crystal structure of CYP2C9 and the R108H mutation were inserted using the “sculpt” and “mutate” features of the molecular modeling package: Open-Source PyMOL v0.99rev10 (DeLano Scientific LLC, Palo Alto, CA). The residues and connecting regions were then locally minimized, using the Dreiding minimization feature of DS Viewer Pro 6.0 (Accelrys Software Inc., San Diego, CA). The topology files, used in the energy minimization and MD simulation, were modified in order to reflect the cysteinyl and histidine ligation, respectively (26, 27, 28 and see Tables S1 and S2 of the Supplementary Materials). Both CYP2C9 and the R108H mutant structures were immersed in a simulated water box with 120 Å sides and ~70-80,000 waters, corresponding to twice the length of the longest diagonal of the protein, or ~65 Å. Both structures were energy minimized, by the method of steepest descent, in order to remove Van der Waals contact between overlapping waters and the amino acids of the protein. MD simulations were run with Berendsen temperature and pressure coupling (a.k.a. “bath”) (29) at a simulated temperature of 300K, using the GROMOS 53a6 force field (30, 31) and periodic boundary conditions in all directions. A derivation of the force field parameters and charges of the heme with a spherical iron in the GROMOS 53a6 forced field are described in (32). Electrostatics of the system were measured using the particle-mesh Ewald method (33). These simulations were performed on an IBM P655+ supercomputer at the Arctic Regional Supercomputer (Fairbanks, AK). During the MD simulation, the position of the protein structures was restrained for 50 ps in order to allow the waters to fill in the cavities. Next, MD simulations were continued for 11.5 ns. Analysis of CYP2C9 and CYP2C9 R108H structures was performed using GROMACS and software written in the Python programming language (Versions 2.4 and 2.6). The active site volumes were estimated using DS Viewer Pro 6.0 by taking the difference between the volume of a solvent accessible surface that excludes the active site (probe radius = 5 Å) and the volume of a solvent accessible surface that includes the active site (probe radius = 2 Å).
Results
Size-exclusion chromatography and UV-visible absorbance spectroscopy of the CYP2C9 R108H mutant show that heme ligation is intramolecular
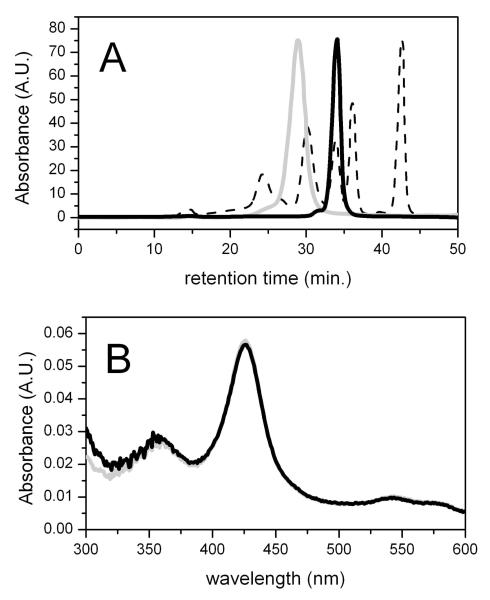
To determine if heme ligation was intermolecular or intramolecular, the CYP2C9 R108H mutant was treated with a variety of detergents to make the protein monomeric and analyzed by size-exclusion chromatography and UV-visible absorbance spectroscopy. Figure 1A shows the chromatogram of the CYP2C9 R108H mutant in the absence and presence of 0.5% cholate against the chromatogram of the protein standards. In the absence of 0.5% cholate, the mutant had a retention time of 28.8 min., which was near to the 158 kD protein standard and showed that CYP2C9 R108H was primarily a trimer or tetramer. After adding 0.5% cholate, the mutant’s retention time increased to 34.2 min, which was inline with the 44 kD protein standard and showed that the protein was monomeric. Figure 1B shows the UV-visible absorbance of the heme Soret region (300-600 nm) of the R108H mutant to assess the effects of monomerization on the heme absorbance spectrum. If the absorbance maximum shifted from 424 nm to 418 nm by cholate, this would indicate that heme ligation was intermolecular. Instead, the absorbance maximum remained at 424 nm in the presence of 0.5% cholate, and so conclusively, heme ligation is intramolecular.
Figure 1. The effect of 0.5% cholate on the aggregation state and the UV-visible Absorbance spectrum of the R108H mutant.
A) A size-exclusion chromatogram and B) an absolute absorbance spectrum with the R108H mutant with (thick black line) and without 0.5% cholate (thick gray line). The protein standard that was used in the size-exclusion chromatography is shown as a thin dotted line.
Pulsed EPR shows that the R108H heme iron is bound by a histidine
Pulsed EPR is a powerful technique for probing nuclei near paramagnetic centers (34-36). In this study, the pulsed EPR technique HYSCORE was used to determine the amino acid residue that was bound to the heme iron in the CYP2C9 R108H mutant. Figure 2 shows HYSCORE spectra of CYP2C9 and the R108H mutant. The spectra were collected at the magnetic field of the gz EPR peak (i.e. 0.2865 Tesla) with proton peaks labeled and the nitrogen frequencies shown (< 9 MHz) within the dashed box (21, 22, 37). In the absence of ligands, a set of proton peaks, labeled a in Figure 2A, were found at 11.68 and 13.23 MHz. The hyperfine coupling of these peaks were aiso=−0.76 ± 0.09 or −2.60 ± 0.20 MHz, and a⊥=3.40 ± 0.20 MHz. The distance from these protons to the heme iron was 3.03 ± 0.07 Å, using the a⊥ and equation 4. Flanking this peak were two peaks, labeled b, with hyperfine coupling constants of aiso=−1.16 ± 0.12 or −4.12 ± 0.05 MHz and a⊥=5.30 ± 0.10 MHz, which corresponds to a proton-heme iron distance of 2.62 ± 0.02 Å. These values are very similar to those reported for the axial water ligand to the Fe (III) in CYP101 (21, 22). Consequently, these peaks were assigned to the protons of the axial water ligand from CYP2C9. A third set of peaks, labeled c, had a aiso=−1.50 ± 0.80 or −0.90 ± 1.60 MHz and a⊥=2.50 ± 0.10 MHz with a calculated distance to the heme iron of 3.37 ± 0.70 Å.
The HYSCORE spectrum of the CYP2C9 R108H mutant is shown in Figure 2B. The hyperfine coupling constants for peak a in the mutant were aiso = −1.18 ± 0.09 or −2.20 ± 0.19 MHz, and a⊥=3.4 MHz. The calculated proton distance to the heme was 3.04 ± 0.07 Å, which was similar to peak a of CYP2C9 (Figure 2A). Unlike CYP2C9, there were no axial water proton peaks in the HYSCORE spectrum, suggesting that water had been displaced from the heme iron by some amino acid residue of the mutant. A set of R108H proton peaks, labeled d, was found near the position of peak c in CYP2C9 (Figure 2A). The hyperfine coupling of this peak was aiso = −1.27 ± 0.16 MHz, and a⊥=2.54 ± 0.22 MHz and the distance calculated, using the a⊥, was 3.34 ± 0.10 Å.
Figure 2C shows the HYSCORE spectrum of CYP2C9 in the presence of 100 mM imidazole to compare with the HYSCORE spectrum of the CYP2C9 R108H mutant. The proton peaks from the axial water ligand were also absent from this spectrum, which is consistent with the imidazole displacing the bound water. Peaks a and d were partially overlapping, so they could not be analyzed individually. The average hyperfine coupling constants estimated from the combined peaks were aiso=−1.68 ± 0.16 or −1.00 ± 0.30 MHz, and a⊥=2.71 ± 0.35 MHz. A distance of 3.27 ± 0.14 Å was calculated from the a⊥ and the peaks were assigned to both the protons on the imidazole ring and random protons of the protein, since they could not be assigned individually.
HYSCORE difference spectra are shown in Figure 3 to compare the CYP2C9 R108H mutant and CYP2C9 with imidazole. The difference spectra were calculated by subtracting the HYSCORE spectrum of CYP2C9 without ligands from the HYSCORE spectra of the CYP2C9 R108H mutant or CYP2C9 with 100 mM imidazole bound. The left and right panels show the positive and negative peaks from the HYSCORE difference spectrum, respectively. The positive peaks and spectral features in the left panels appear only from CYP2C9 with imidazole and the mutant, while the negative peaks in the right panels show the peaks that disappear from WT. In both of the left panels (i.e. Figures 3A and 3C), two peaks were found at (−4.39, +7.85 MHz) and (−7.85, +4.39 MHz) (dotted oval) from nitrogen nucleus with an AZ between 5.8 and 6.2 MHz. There were also extended ridges in these panels at ±1.91 MHz (dashed rectangle) with hyperfine coupling of ±1-2 MHz, which is roughly twice the nuclear Zeeman frequency of nitrogen (2νN). The negative difference spectra in the right panels were also similar, demonstrating that both the R108H mutant and CYP2C9 lack bound axial water proton peaks and c peaks. The positive and negative HYSCORE difference spectra were virtually identical and conclusively show that the hemes of the R108H mutant and CYP2C9 with imidazole are both bound by an imidazole functional group. Therefore, the peaks that appear in the nitrogen region of Figures 3A and 3B (i.e. < 9 MHz) were assigned to the nitrogens of an imidazole ring, and the peak d was assigned to protons on the imidazole ring.
Differences between the X-ray crystal structure of CYP2C9 and the energy-minimized R108H mutant
The foregoing UV-visible and pulsed EPR spectroscopy results were consistent with the R108H mutant maintaining at least some structural characteristics of a CYP fold. Nevertheless, these results did not exclude the possibility that the structure was severely disrupted by the mutation. To probe this possibility further, an energy-minimized model of the R108H mutant was generated with H108 bound to the heme iron in order to illuminate major structural differences between CYP2C9 and CYP2C9 R108H.
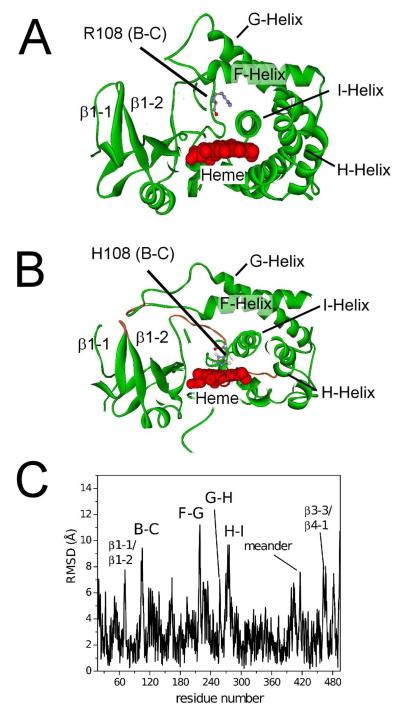
Figure 4 shows the structural differences between CYP2C9 and the R108H mutant. Figures 4A and 4B are cross-sections of CYP2C9 (PDB ID: 1R90 (5)), and the energy-minimized structure of the R108H mutant, respectively. In Figure 4C, the structural rearrangements required to energy minimize the mutant with the histidine ligated to the heme, are represented as the root mean square deviation (RMSD) of the CYP2C9 α-carbon backbone, as a function of amino acid number. Overall, the CYP fold of the R108H mutant remained intact with secondary structure elements largely unperturbed. The ε-nitrogen of H108 in the energy-minimized model was 2.4 Å away from the heme iron, while the two flanking protons were 3.5 and 3.7 Å distant. The latter values were similar to the distances calculated from the a⊥ (i.e. 3.04 and 3.34 Å) that were determined from HYSCORE. The average RMSD of the α-carbon backbone, between the R108H mutant and CYP2C9, was only 3.1 Å +/− 1.7 Å. The largest deviations were observed in the mobile loop regions and the N- and C-terminii. This correlates well with the X-ray crystal structure of CYP2C9 that either showed high b-factors or was disordered in those regions (5). Between the two structures, the F-G loop region showed the greatest deviation of 11 Å. The F-G loop was in direct contact with the β1-1/β1-2 loop, which was shifted 8 Å from CYP2C9 (Figure 4A). Also, there were shifts, of approximately 7 Å, in the nearby G-H loop region. Moreover, the B-C loop was abutted against the I-helix and moved the H-I loop by 9 Å. Some indirect effects in the meander region and β3-3/β4-1 loops were also noted. These results indicate that several regions within CYP2C9 are conformationally flexible, but provide limited information about their mobility. To investigate the intrinsic flexibility of CYP2C9, MD simulations were performed of the enzyme and compared to the open and closed conformations of CYP2B4 (38, 39).
Figure 4. Differences in the energy-minimized structures of CYP2C9 and the R108H mutant.
A) Cross-section of the X-ray crystal structure of CYP2C9 (PDB ID: 1R90 (5)) B) Cross-section of the hypothetical model of the R108H mutant, showing binding of H108 to the heme with secondary structures of highest mobility colored orange and labeled. C) The α-carbon backbone root mean square deviation (RMSD) between CYP2C9 (PDB ID: 1R9O) and the R108H mutant with the loops and the meander region, which had the highest deviations, labeled. For example, the labels B-C, β1-1/β1-2 and meander correspond to the B-C loop, β1-1/β1-2 loop and meander region, respectively.
Molecular dynamics simulations of CYP2C9 show that the B-C loop region and R108 are mobile
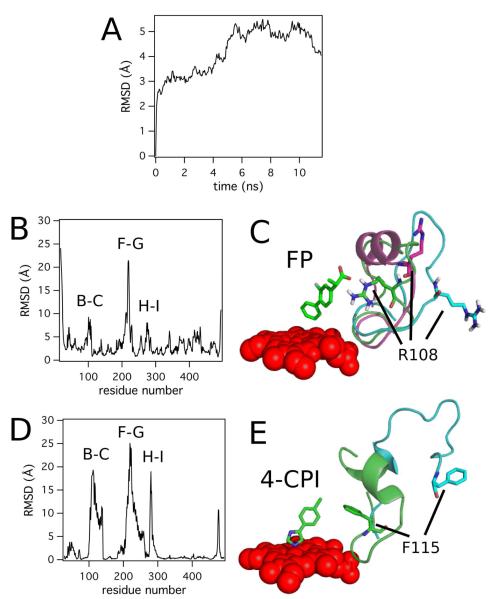
The MD simulations of CYP2C9 were used to probe its mobility, and to determine the energetically accessible conformations of the enzyme. Figure 5 shows the results and the analyses of the MD simulation with a comparison to the CYP2B4 X-ray crystal structures (38, 39). The RMSD of the α-carbon backbone, during the MD simulation from the starting structure, is illustrated in Figure 5A. The α-carbon backbone reached equilibrium at approximately 5 ns. In order to examine the specific conformational changes during the MD simulation, the average α-carbon backbone RMSD was calculated for each residue, during the 10-11.5 ns time period of the MD simulation, and compared to the starting structure in Figure 5B. The F-G loop region showed the largest displacement of 20 Å, while the B-C and H-I loop regions were only displaced ~10 Å. Figure 5C shows the B-C loop regions of CYP2C9 with FP (PDB ID: 1R9O), with warfarin (PDB ID: 1OG5) and CYP2C9 alone after the 11.5 ns MD simulation. In the FP-bound CYP2C9 structure, the distance between the α-carbon of R108 and the heme iron was 17 Å with the arginine interacting with FP. The distance between the α-carbon of R108 and the heme iron increased to 20 Å in warfarin-bound CYP2C9. By 11.5 ns in the MD simulation, the distance increased further to 22 Å. The angle formed between the heme plane and the R108 α-carbon also increased from 137° to 151°. With these increases in the distances and angles, the active site volume progressively increased between these three structures from approximately 15 nm3 to 17 nm3, according to our rough calculations. Thus, a range of conformations is energetically accessible for CYP2C9.
Figure 5. MD Simulation and Analysis of CYP2C9.
A) RMSD of the α-carbon backbone of CYP2C9 during the MD simulation B) Average α-carbon backbone RMSD per residue of CYP2C9 between 10 and 11.5 ns of the MD simulation from the starting structure. The B-C, F-G, and H-I loop regions with the highest deviations are labeled. C) The conformation and position of the B-C loop and R108 movement in the CYP2C9 structure with FP bound (PDB ID: 1R9O (5)) (green), warfarin-bound CYP2C9 structure (PDB ID: 1OG5,(4)) (magenta) and CYP2C9 structure after 11.5 ns MD simulation (blue). D) The α-carbon backbone RMSD per residue between CYP2B4 in the putative open conformation (PDB ID: 1PO5, (39)) and CYP2B4 with 4-(4-chlorophenyl)imidazole (4-CPI, PDB ID: 1SUO, (38)), and labeling similar to panel A. E) Conformational and positional differences in the B-C loop region and F115 of CYP2B4 with 4-CPI (green) and in the open conformation (blue).
There were striking similarities between the results of the MD simulation of CYP2C9 and the X-ray crystal structures of CYP2B4 in the closed (PDB ID: 1SUO (38)) and putative open (PDB ID: 1PO5 (39)) conformations. Figure 5D illustrates the backbone α-carbon RMSD per residue between the closed and open structures. Although they are not identical, CYP2B4 also showed similar large displacements of the B-C, F-G and H-I loop regions. Figure 5E shows the active site, and the B-C loop region of CYP2B4 with F115, in a position similar to that of R108 in the CYP2C9 structure. In the closed conformation, the F115 is in direct contact with 4-(4-chlorophenyl) imidazole (4-CPI) that is bound to the CYP2B4 heme. The distance from the α-carbon to the heme iron, in this conformation, is 12 Å. In the open conformation of CYP2B4, the F115 residue is significantly displaced from the heme to 23 Å, analogously to how the R108 residue was displaced in the MD simulation of CYP2C9. Additionally, the active site volume increased roughly from 14 nm3 to 17 nm3 in the open conformation. Overall, the similarities between CYP2C9 and CYP2B4 suggest that both enzymes can undergo similar conformational transitions.
Discussion
A number of investigations have suggested that conformational changes of CYPs play an important role in ligand binding and molecular recognition (e.g. 38, 39, 40, 41, 42). In this study, the conformations of the CYP2C9 R108H mutant, deduced from energy minimization, the X-ray crystal structures of CYP2C9 (4, 5) and MD simulations of CYP2C9 showed that this enzyme can assume a wide range of conformations. In the R108H mutant, the B-C loop was intramolecularly ligated to the heme, thus representing an extreme example of a closed conformation. Energy minimization of this mutant showed that the conformation can be accommodated without disrupting the overall protein fold of the enzyme, implying that several regions of the protein are quite flexible. Interestingly, this same conformation is evident in the ligand-free X-ray crystal structure of the bacterial CYP105P1, which has an analogous residue (i.e. H72) within the B-C loop, that is bound to the heme iron (9). Representing the opposite extreme, the B-C loop of CYP2C9 in the MD simulation (PDB ID: 1R9O (5)), and for comparison CYP2B4 X-ray crystal structures (PDB ID: 1SUO (38) and PDB ID: 1PO5 (39)), was shifted away from the heme and adopted a conformation that resembles an open conformation of the enzyme. In between these two extremes were the X-ray crystal structures of CYP2C9 with FP (PDB ID: 1R9O (5)) and warfarin bound (PDB ID: 1OG5 (4)) both of which demonstrated closed, ligand-bound conformations of CYP2C9.
Several P450 enzymes are believed to undergo open to closed conformational transitions during ligand binding, including CYP101 (43), CYP102A1 (44, 45), CYP105P1 (9), CYP2B4 (38, 39, 46, 47), CYP51(48), CYP2C9 (4, 5) and CYP3A4 (41). The transition typically occurs around secondary structures near the F-G loop, but may also involve the B-C loop region. The X-ray crystal structures of CYP2C9 with FP bound (PDBID: 1R9O (5)) and the chimeric X-ray crystal structures of CYP2C9 with warfarin (PDB ID: 1OG5 (4)) and ligand-free (PDB ID: 1OG2 (4)) showed significant conformational differences in the B-C and the F-G loop regions. The B-C loop region was a random coil in the 1R9O structure, and an α-helix (i.e. B’-helix) in both the 1OG5 and 1OG2 structures. The F-G loop regions were disordered in the 1R9O structure, while this region was helical in the 1OG5 structure. These differences suggest that CYP2C9 can undergo conformational transitions similar to those described for CYP2B4 (5). However, the backbone differences between the 1R9O and the 1OG5 B-C and F-G loop regions were actually quite modest with an average α-carbon backbone displacement of ~2 Å compared to the ~10 Å displacement observed for CYP2B4 (5, 8). In order to test the range of conformations that CYP2C9 can assume, MD simulations were initially performed with the FP bound CYP2C9 structure as the putative closed conformation. The B-C loop region moved more than 20 Å from the heme in the absence of FP, which parallels the enzyme adopting the open conformation seen in the CYP2B4 X-ray crystal structure (39).
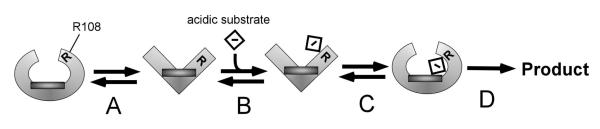
A model of substrate binding, based on the range of conformational changes that CYP2C9 can assume, is shown in Scheme 1. Size-exclusion chromatography, HYSCORE spectroscopy and energy minimization results provided experimental support for the closed conformation, whereas the open conformation was deduced from the MD simulations. Therefore, in the absence of substrates, we propose that the closed and open structures are energetically accessible conformations of CYP2C9 (Scheme 1A). In the closed conformation, R108 within the B-C loop is folded over, and relatively close to the heme, as seen in the X-ray crystal structure of CYP2C9 (PDB ID: 1R9O (5)) with FP bound, or (a more extreme example) the position of H108 in the CYP2C9 mutant (Figure 4B) or H72 in CYP105P1 (9). Conversely, in the open conformation, the R108 residue is facing away from the heme similar to F115 in the X-ray crystal structure of CYP2B4 (Figure 5E, PDB ID: 1PO5 (39)).
Scheme 1. Substrate binding model of CYP2C9.
A) The closed and open conformations of CYP2C9. B) Binding of an acidic substrate to R108 (R) in the open conformation CYP2C9. C) Trapping of the acidic substrate in the closed conformation of CYP2C9 followed by D) product formation.
Scheme 1B shows acidic substrates binding to CYP2C9 in the open conformation, through hydrogen bonding to R108 (5). This residue hydrogen bonds to FP in the X-ray crystal structure of CYP2C9 (5) and an R108F mutation precludes metabolism of S-warfarin and diclofenac (6), suggesting that it is critical for binding. The R108 residue is oriented out into the bulk solvent after the 10 ns MD simulation (Figure 5C), where it can readily access substrates. In this position, the drug substrate is too far from the heme to undergo productive catalysis, and so the B-C loop folds over and “traps” the substrate within the active site, near the heme (Scheme 1C). The energy-minimized model of the CYP2C9 R108H mutant suggests that CYP2C9 can form a closed compact structure that can prevent the release of bound substrates. We propose that this conformation of the enzyme resembles the X-ray crystal structure of CYP2C9 with FP bound, where the drug is “trapped” by a hydrogen-bond to R108 and hydrophobic interactions with residues V113, F114, A297, T301 and L366 (5). The importance of several of these hydrophobic residues in CYP2C enzymes has been revealed in mutagenesis studies (49, 50). By trapping the substrate, CYP2C9 can then undergo efficient catalysis to products, which is shown in Scheme 1D.
This model helps to explain the complex substrate binding behavior of CYP2C9. Hydrogen bonding of weak acids, to R108 of CYP2C9, provides the enzyme with ligand specificity. The mobility and the flexibility of the B-C loop region permits CYP2C9 to accept a wide range of substrates or even multiple substrates at the same time. This scenario also helps to explain how the enzyme can regio- and stereo-specifically hydroxylate a substrate like phenytoin (51), yet possess the flexibility to allosterically activate the metabolism of other ligands (3).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Jill L. Hartmann for editing the manuscript. A portion of the research was performed using the Environmental Molecular Sciences Laboratory (EMSL), which is a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL).
ABBREVIATIONS
- δ-ALA
δ-aminolevulinic acid
- CYP
cytochrome P450
- 4-CPI
4-(4-chlorophenyl) imidazole
- CYMAL-5
5-Cyclohexyl-1-pentyl-β-D-maltoside
- DLPC
dilaurylphosphatidylcholine
- EDTA
ethylenediaminetetraacetic acid
- EMSL
Environmental Molecular Sciences Laboratory
- EPR
electron paramagnetic resonance
- ENDOR
electron nuclear double resonance
- FP
flurbiprofen
- HPLC
high-performance liquid chromatography
- HYSCORE
hyperfine sublevel correlation spectroscopy
- MD
molecular dynamics
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- Ni-NTA
Nickel-nitrilotriacetic acid
- NIH
National Institute of Health
- NMR
nuclear magnetic resonance
- NSAID
non-steroidal anti-inflammatory drug
- PNNL
Pacific Northwest National Laboratory
- RMSD
root mean square deviation
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
This study was supported by National Institute of Health (NIH) grants GM32165 and GM61904.
SUPPORTING INFORMATION STATEMENT: Figure S1 shows the spin echo-detected EPR spectra that were associated with the HYSCORE spectra in this study. The Tables S1 and S2 of the supporting materials are additional force field parameters that were used for the MD simulations and the energy minimization to ligate the C435 of heme and R108 of the CYP2C9 R108H mutant to the heme. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ortiz de Montellano PR. Cytochrome P450 : Structure, Mechanism, and Biochemistry. 3rd ed Plenum Publishing; New York: 2004. [Google Scholar]
- 2.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br. J. Clin. Pharmacol. 1998;45:525–538. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 4.Williams PA, Cosme J, Ward A, Angove HC, Vinkovic D. Matak, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 5.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-Å resolution. J. Biol. Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 6.Dickmann LJ, Locuson CW, Jones JP, Rettie AE. Differential roles of Arg97, Asp293, and Arg108 in enzyme stability and substrate specificity of CYP2C9. Mol. Pharmacol. 2004;65:842–850. doi: 10.1124/mol.65.4.842. [DOI] [PubMed] [Google Scholar]
- 7.Ridderstrom M, Masimirembwa C, Trump-Kallmeyer S, Ahlefelt M, Otter C, Andersson TB. Arginines 97 and 108 in CYP2C9 are important determinants of the catalytic function. Biochem. Biophys. Res. Commun. 2000;270:983–987. doi: 10.1006/bbrc.2000.2538. [DOI] [PubMed] [Google Scholar]
- 8.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu LH, Fushinobu S, Ikeda H, Wakagi T, Shoun H. Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state. J. Bacteriol. 2009;191:1211–1219. doi: 10.1128/JB.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosher CM, Hummel MA, Tracy TS, Rettie AE. Functional analysis of phenylalanine residues in the active site of cytochrome P450 2C9. Biochemistry. 2008;47:11725–11734. doi: 10.1021/bi801231m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheesman MJ, Baer BR, Zheng YM, Gillam EM, Rettie AE. Rabbit CYP4B1 engineered for high-level expression in Escherichia coli: ligand stabilization and processing of the N-terminus and heme prosthetic group. Arch. Biochem. Biophys. 2003;416:17–24. doi: 10.1016/s0003-9861(03)00278-9. [DOI] [PubMed] [Google Scholar]
- 12.Jefcoate CR. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 1978;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrandt A, Remmer H, Estabrook RW. Cytochrome P-450 of liver microsomesone pigment or many. Biochem. Biophys. Res. Commun. 1968;30:607–612. doi: 10.1016/0006-291x(68)90555-x. [DOI] [PubMed] [Google Scholar]
- 14.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II. solubilization, purification, and properties. J. Biol. Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 15.Dikanov SA, Bowman MK. Determination of ligand conformation in reduced [2Fe-2S] ferredoxin from cysteine β-proton hyperfine couplings. Journal of Biological Inorganic Chemistry. 1998;3:18–29. [Google Scholar]
- 16.Dikanov SA, Tyryshkin AM, Bowman MK. Intensity of cross-peaks in HYSCORE spectra of S = 1/2, I = 1/2 spin systems. J. Magn. Reson. 2000;144:228–242. doi: 10.1006/jmre.2000.2055. [DOI] [PubMed] [Google Scholar]
- 17.Dikanov SA, Bowman MK. Cross-peak lineshape of two-dimensional ESEEM spectra in disordered S=1/2, I=1/2 spin systems. Journal of Magnetic Resonance A. 1995;116:125–128. [Google Scholar]
- 18.Maryasov AG, Bowman MK. Hyperfine sublevel correlation (HYSCORE) spectra for paramagnetic centers with nuclear spin I=1 having hyperfine interactions. Journal of Physical Chemistry B. 2004;108:9412–9420. [Google Scholar]
- 19.Maryasov AG, Bowman MK. Two-dimensional hyperfine sublevel correlation spectroscopy: powder features for S=1/2, I=1. J. Magn. Reson. 2006;179:120–135. doi: 10.1016/j.jmr.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Stuhrmann HB. Neutron small-angle scattering of biological macromolecules in solution. J. Appl. Crystallogr. 1974;7:173–178. [Google Scholar]
- 21.LoBrutto R, Scholes CP, Wagner GC, Gunsalus IC, Debrunner PG. Electron Nuclear Double Resonance of Ferric Cytochrome P450CAM. J. Am. Chem. Soc. 1980;102:1167–1170. [Google Scholar]
- 22.Goldfarb D, Bernardo M, Thomann H, Kroneck PMH, Ullrich V. Study of water binding to low-spin Fe(III) in cytochrome P450 by pulsed ENDOR and four-pulse ESEEM spectroscopies. J. Am. Chem. Soc. 1996;118:2686–2693. [Google Scholar]
- 23.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 24.Berendsen HJC, van der Spoel D, Drunen R. v. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 25.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 26.Autenrieth F, Tajkhorshid E, Baudry J, Luthey-Schulten Z. Classical force field parameters for the heme prosthetic group of cytochrome c. J. Comput. Chem. 2004;25:1613–1622. doi: 10.1002/jcc.20079. [DOI] [PubMed] [Google Scholar]
- 27.Oda A, Yamaotsu N, Hirono S. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J. Comput. Chem. 2005;26:818–826. doi: 10.1002/jcc.20221. [DOI] [PubMed] [Google Scholar]
- 28.Henry ER, Levitt M, Eaton WA. Molecular dynamics simulation of photodissociation of carbon monoxide from hemoglobin. Proc. Natl. Acad. Sci. U. S. A. 1985;82:2034–2038. doi: 10.1073/pnas.82.7.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berendsen HJC, Postma JPM, van Gunsteren W, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 30.Oostenbrink C, Soares TA, van der Vegt NF, van Gunsteren WF. Validation of the 53A6 GROMOS force field. Eur. Biophys. J. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 31.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 32.Gunsteren W. F. v., Billeter SR, Eising AA, Hunenberger PH, Kruger P, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Verlag der Fachvereine Hochschulverlag AG an der ETH Zurich; Zurich, Switzerland: 1996. [Google Scholar]
- 33.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 34.Kevan L, Bowman MK. Modern pulsed and continuous-wave electron spin resonance. Wiley; New York: 1990. [Google Scholar]
- 35.Prisner T, Rohrer M, MacMillan F. Pulse EPR spectroscopy: biological applications. Annu. Rev. Phys. Chem. 2003;52:279–313. doi: 10.1146/annurev.physchem.52.1.279. [DOI] [PubMed] [Google Scholar]
- 36.Bowman MK. Pulsed electron paramagnetic resonance. In: Brustolon M, Giamello E, editors. Electron Paramagnetic Resonance: A Practitioners Toolkit. John Wiley & Sons, Hoboken; New Jersey: 2009. pp. 159–194. [Google Scholar]
- 37.Fann Y-C. ENDOR determination of heme ligation in chloroperoxidase and comparison with cytochrome P-450cam. J. Am. Chem. Soc. 1994;116:5989–5990. [Google Scholar]
- 38.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 Å resolution: Insight into the range of P450 conformations and coordination of redox partner binding. J. Biol. Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 39.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts AG, Atkins WM. Energetics of heterotropic cooperativity between α-naphthoflavone and testosterone binding to CYP3A4. Arch. Biochem. Biophys. 2007;463:89–101. doi: 10.1016/j.abb.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davydov DR, Botchkareva AE, Kumar S, He YQ, Halpert JR. An electrostatically driven conformational transition is involved in the mechanisms of substrate binding and cooperativity in cytochrome P450eryF. Biochemistry. 2004;43:6475–6485. doi: 10.1021/bi036260l. [DOI] [PubMed] [Google Scholar]
- 43.Yao H, McCullough CR, Costache AD, Pullela PK, Sem DS. Structural evidence for a functionally relevant second camphor binding site in P450cam: model for substrate entry into a P450 active site. Proteins. 2007;69:125–138. doi: 10.1002/prot.21508. [DOI] [PubMed] [Google Scholar]
- 44.Chang YT, Loew GH. Molecular dynamics simulations of P450 BM3--examination of substrate-induced conformational change. J. Biomol. Struct. Dyn. 1999;16:1189–1203. doi: 10.1080/07391102.1999.10508327. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Poulos TL. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat. Struct. Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 46.Muralidhara BK, Negi S, Chin CC, Braun W, Halpert JR. Conformational flexibility of mammalian cytochrome P450 2B4 in binding imidazole inhibitors with different ring chemistry and side chains. Solution thermodynamics and molecular modeling. J. Biol. Chem. 2006;281:8051–8061. doi: 10.1074/jbc.M509696200. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J. Biol. Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 48.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haining RL, Jones JP, Henne KR, Fisher MB, Koop DR, Trager WF, Rettie AE. Enzymatic determinants of the substrate specificity of CYP2C9: role of B’-C loop residues in providing the π-stacking anchor site for warfarin binding. Biochemistry. 1999;38:3285–3292. doi: 10.1021/bi982161+. [DOI] [PubMed] [Google Scholar]
- 50.Imai Y, Nakamura M. Point mutations at threonine-301 modify substrate specificity of rabbit liver microsomal cytochromes P-450 (laurate (omega-1)-hydroxylase and testosterone 16 alpha-hydroxylase) Biochem. Biophys. Res. Commun. 1989;158:717–722. doi: 10.1016/0006-291x(89)92780-0. [DOI] [PubMed] [Google Scholar]
- 51.Mosher CM, Tai G, Rettie AE. CYP2C9 amino acid residues influencing phenytoin turnover and metabolite regio- and stereochemistry. J. Pharmacol. Exp. Ther. 2009;329:938–944. doi: 10.1124/jpet.109.150706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.