Abstract
Background
Aging is a complex and challenging phenomenon that requires interdisciplinary efforts to unravel its mystery. Insight into genes relevant to the aging process would offer the chance to delay and avoid some of deteriorative aspects of aging through the use of preventive methods. To assist basic research on aging, a comprehensive database and analysis platform for aging-related genes is required.
Results
We developed a web-based database server, called Gerontome that contains aging-related gene information and user-friendly analysis pipelines. To construct the Gerontome database, we integrated aging-related genes and their annotation data. The aging-related genes were categorized by a set of structural terms from Gene Ontology (GO). Analysis pipelines for promoter analysis and protein-ligand docking were developed. The promoter analysis pipeline allows users to investigate the age-dependent regulation of gene expression. The protein-ligand docking pipeline provides information on the position and orientation of a ligand in an age-related protein surface.
Conclusion
Gerontome can be accessed through web interfaces for querying and browsing. The server provides comprehensive age-related gene information and analysis pipelines. Gerontome is available free at http://gerontome.kobic.re.kr.
Background
Aging is universal phenomenon among all organisms. Because the processes underlying aging are controversial and it is a poorly understood biological problem, aging-related genes have attracted a fair amount of attention from both the academic community, the medicinal community and the public in general [1]. Aging is a risk factor for many diseases [2]. Many studies have been performed in several model organisms, including humans, to obtain new insights into the process of aging and to identify aging-related genes by comparing young and old tissues or by comparing samples across a lifespan [3]. Information on genetic links to cellular aging suggests new treatments for a variety of age-related diseases and cancers [4].
A collection of age-related information in multiple organisms is important to understand complicated age phenomenon and to identify new age-related data. Several age-related databases have been constructed based on gene, protein, or microarray experiments. The Human Aging Genomic Resource (HAGR) [5] provides manually-curated aging genes in human and model animals. Gene Aging Nexus (GAN) [6] contains aging-related gene expression patterns in multiple organisms under different conditions. The aging genes and interventions database (AGEID) [7] provides experimental results related to aging and information on genes that influence the incidence of age-associated disorders such as Alzheimer's disease. However, the efficient exploitation of this large data set is hampered by the lack of an integrated database and data analysis platform.
Here we have constructed a database server, called Gerontome, to provide comprehensive information on aging-related genes and analysis interfaces. We integrated aging-related resources and developed automated analysis pipelines to provide transcription factor binding sites of regulatory regions and docking information between proteins and ligands in aging-related genes. We categorized aging-related genes by a set of structural terms from Gene Ontology (GO). Our aim in building Gerontome is to provide researchers with a comprehensive online resource and a user-friendly analysis interface to study the genetic basis of aging.
Methods
Data sources
Aging-related gene information was obtained from HAGR (http://genomics.senescence.info/), AGEID (http://uwaging.org/genesdb/index.php), the meta-analysis of age-related gene expression Profiles [3], and aging-related yeast2hybrid experiment [8]. From the downloaded data, we created a non-redundant gene set by removing the redundancy in the three databases. As of April 1, 2010, the Gerontome database had 848 non-redundant aging-related genes.
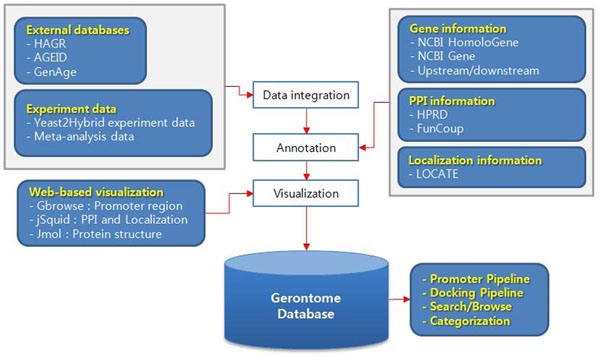
Gerontome uses data from a number of other databases. Human homologs were downloaded from NCBI’s Homologene [9]. Promoter sequences of human genes were obtained from the UCSC genome browser (hg18) [10]. Transcriptional profiles and protein-protein interactions were taken from the Transcription Factor Binding Site (TFBS) conserved track in the UCSC genome browser [10] and HPRD [11] databases, respectively. We used the LOCATE database [12] to identify localization information and the Funcoup database [13] to obtain confidence scores of protein-protein interactions. These data were mapped into aging-related genes and integrated into the Gerontome database (Fig. 1).
Figure 1.
Flowchart of Gerontome database construction.
We used Gene Ontology (GO) annotation, which describes how gene products behave in a cellular context [14]. GO is composed of three subdivisions covering basic areas of biological research: molecular function, biological process, and cellular function. To identify GO categories that tend to be associated with aging genes, we used files downloaded from Entrez Gene database [15]. Through the categorization, we were able to assign 848 genes to GO accession numbers.
Analysis pipelines
Gerontome provides information regarding the molecular features of aging-related genes such as transcription factor binding sites and protein-ligand docking. To provide this information, we developed two analysis pipelines: promoter analysis and protein-ligand docking.
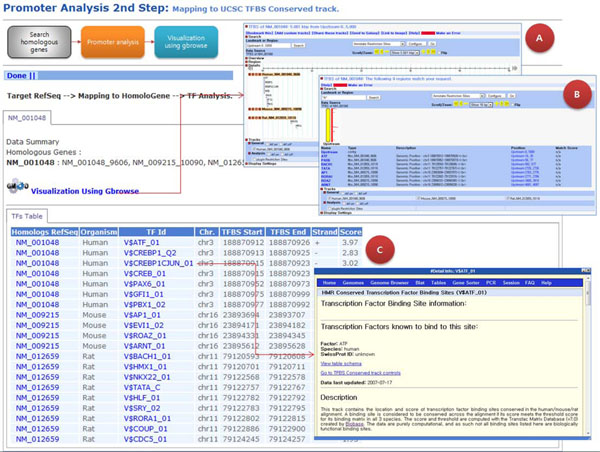
The promoter analysis pipeline allows users to investigate the age-dependent regulation of gene expression through the identification of transcription factors and their binding sequences (Fig. 2). Identification of transcriptional regulation of age-related genes is generally the most important step in aging research. In the pipeline, homologous genes to the query identifiers were first extracted from NCBI's Homologene. Second, upstream sequences of the extracted homologous genes were obtained. The default length of upstream sequences was set at 1000 bases. Third, the server scanned transcription factor binding sites in the upstream sequences using the TFBS conserved information from the UCSC genome browser. Lastly the server provided comparative visualization of homologous genes, TFBSs information, and known genes. We used Gbrowse [16] to visualize the results. From the pipeline, users can find a correlation between age-related genes and transcription factor binding sites [17-19].
Figure 2.
Comparative transcriptional regulatory network by promoter pipeline. The pipeline provides step-by-step processes with the user’s own data. In the pipeline, a user can obtain information such as A) a visualization of promoter regions, B) a list of TFBS, and C) detailed information about TFBS.
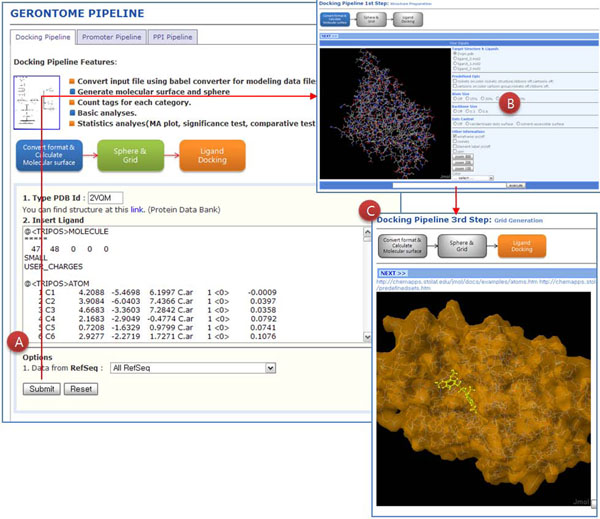
The identification of protein structure is a key step to understanding the biological function and biomolecular interactions of proteins. Docking between proteins and ligands is important in the development of anti-aging drugs. Docking is the identification of the low-energy binding modes of a small molecule or ligand within the active site of a macromolecule or receptor whose structure is known. In the protein-ligand docking pipeline, the positions and orientations of ligands in protein surfaces were predicted using a geometric matching algorithm in the Dock Program [20] (Fig. 3). Users can dock their ligands to surfaces of protein structures. To view ligand positions on protein structures, we used a JMol program [21]. The protein-ligand docking pipeline enables users to simulate interaction affinity without ligand information or a specific protein structure.
Figure 3.
Docking analysis between proteins and ligands. A) PDB number and ligand text format were used to run docking simulations and visualization. B) Docking between protein structures and ligands were visualized by the Jmol program. C) Users can find additional information about the affinity and activity of small molecules.
Web-based server
We developed a web-based server to provide a back-end pipeline for aging analysis and to allow users to compare their genes and proteins with the Gerontome database. The Gerontome database server is composed of a wiki-based web interface and a MySQL 5.0 database management system. The web interface is implemented in static HTML pages, PHP, and JavaScript under an Apache 2.2 web server. MySQL is used to store the age-related gene information and their annotations and analysis data.
Results and discussion
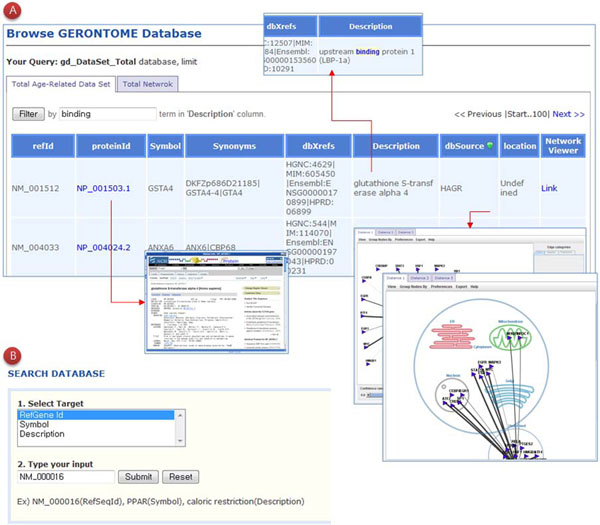
Gerontome can be accessed through a web interface for querying and browsing (Fig. 4). The querying interface allows the user to search against age-related genes and their annotation data. Age-related genes can be searched by RefSeq [22] number, gene symbol, and description. The search results contain basic information, observations, phenotypes, and gene expression data for aging. In the browsing interface, the user can select an aging data source and then see all of the aging-related data. In the browsing results, the user can filter by description term.
Figure 4.
User interface for search and browse. A) Users can choose a database source from Total, AGEID, HAGR, and Y2H. Users can filter by term, network visualization, and other options. B) Gerontome also provides various keyword inputs such as RefSeq number, gene symbol, and description.
We also categorized the aging-related genes by a set of structural terms from Gene Ontology (GO). The user can see the categorized genes by clicking on ‘categorization of age-related data’ and download GO mapping results for biological process, cellular component and molecular function at our site. From the GO mapping results, we found that age-related genes are highly related to 'regulation of transcription', 'anti-apoptosis', 'apoptosis', and 'response to DNA damage stimulus' in the biological process category (Table 1). In addition we used the Gene Ontology Enrichment Analysis Software Toolkit (GOEAST) [23] for GO enrichment analysis. Users can browse the GO enrichment analysis results.
Table 1.
Classification of age-related genes according to Gene Ontology(GO) terms.
| Order | Cellular component | No. genes | Molecular function | No. genes | Biological process | No. genes |
| 1 | Nucleus | 408 | protein binding | 435 | regulation of transcription, DNA-dependent | 117 |
| 2 | cytoplasm | 391 | nucleotide binding | 160 | signal transduction | 98 |
| 3 | membrane | 163 | metal ion binding | 137 | transcription | 82 |
| 4 | plasma membrane | 155 | zinc ion binding | 131 | anti-apoptosis | 48 |
| 5 | extracellular region | 150 | ATP binding | 123 | cell cycle | 47 |
| 6 | cytosol | 137 | transferase activity | 95 | interspecies interaction between organisms | 44 |
| 7 | mitochondrion | 107 | transcription factor activity | 85 | apoptosis | 44 |
| 8 | integral to membrane | 107 | DNA binding | 78 | response to DNA damage stimulus | 43 |
| 9 | intracellular | 106 | calcium ion binding | 64 | multicellular organismal development | 42 |
| 10 | nucleolus | 86 | hydrolase activity | 56 | cell adhesion | 38 |
Gerontome provides several viewers for the TFBSs position, protein structure, and protein interaction of each entry by using Gbrowse, jSquid [24], and JMol programs. In the Gbrowse interface, users can compare biological features between homologous genes and proteins which represent relatively closed protein groups. jSquid displays the protein-protein interaction network among age-related proteins. In the jSquid search results, users can modify subgroups of network elements based on the annotation information on protein localization and the confidence score of protein-protein interaction. After docking between aging-related protein and ligands, users can see the position and orientation of a ligand in an age-related protein surface by using JMol, which is a Java viewer for chemical structures in 3D with features of bio-molecules and materials.
In addition, we developed a wiki site for sharing information about Gerontome. The wiki aims to promote sharing information and knowledge among researchers. The wiki also includes detailed information on the analysis pipelines, the parameters of programs, and a data summary of our database. The Gerontome wiki is available at http://www.gerontome.info/wiki/index.php.
Conclusion
We developed a database and tools that will be useful to researchers working on the science of aging. Our aim is for Gerontome to become a major resource for understanding the systematic mechanisms of human aging. To facilitate the integrative analysis of aging genes, we constructed a comprehensive aging gene database and developed a web-based analysis platform, which is freely accessible to the research community to query, analyze, and visualize age-related genes. The database also has links to genomic information from different species to facilitate the discovery of candidate genes that are involved in aging through a genome-wide comparative analysis. The analysis pipelines in Gerontome are useful to predict regulatory networks of homologous genes, docking simulations between protein structures and ligands, and protein interaction networks.
In the future, we will upgrade, update and expand the resources in Gerontome as well as develop new tools that can benefit the gerontology community. The aging gene information in the Gerontome will be useful when trying to identify new treatments and drugs for a variety of age-related diseases. We would like Gerontome to become a general platform for bio-gerontologists and bioinformaticians.
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
JK and BL were responsible for development of the web interface and web-based pipelines. HC launched the GERONTOME project and supervised it. HC provided useful information about the needs of age-related biology research. JK and BL wrote the draft of manuscript. All authors read and approved the final manuscript.
Contributor Information
Jekeun Kwon, Email: kwon@kribb.re.kr.
Byungwook Lee, Email: bulee@kribb.re.kr.
Haeyoung Chung, Email: hyung@pusan.ac.kr.
Acknowledgements
The work was supported by the KRIBB Research Initiative Program, the Ministry of Education, Science and Technology (under grant number 20100002064), and a National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2009-0083538). We thank the Aging Tissue Bank for providing research resources.
This article has been published as part of BMC Genomics Volume 11 Supplement 4, 2010: Ninth International Conference on Bioinformatics (InCoB2010): Computational Biology. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2164/11?issue=S4.
References
- de Magalhaes JP. et al. The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell. 2009;8(1):65–72. doi: 10.1111/j.1474-9726.2008.00442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Functional genomics of ageing. Mech Ageing Dev. 2003;124(1):3–8. doi: 10.1016/S0047-6374(02)00163-X. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25(7):875–81. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Toussaint O. GenAge: a genomic and proteomic network map of human ageing. FEBS Lett. 2004;571(1-3):243–7. doi: 10.1016/j.febslet.2004.07.006. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33(Database issue):D537–43. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F. et al. Gene Aging Nexus: a web database and data mining platform for microarray data on aging. Nucleic Acids Res. 2007;35(Database issue):D756–9. doi: 10.1093/nar/gkl798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Jegalian B, McVey M. AGEID: a database of aging genes and interventions. Mech Ageing Dev. 2002;123(8):1115–9. doi: 10.1016/S0047-6374(02)00011-8. [DOI] [PubMed] [Google Scholar]
- Bell R. et al. A human protein interaction network shows conservation of aging processes between human and invertebrate species. PLoS Genet. 2009;5(3):e1000414. doi: 10.1371/journal.pgen.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38(Database issue):D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B. et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38(Database issue):D613–9. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TS, Kandasamy K, Pandey A. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. full_text. [DOI] [PubMed] [Google Scholar]
- Sprenger J. et al. LOCATE: a mammalian protein subcellular localization database. Nucleic Acids Res. 2008;36(Database issue):D230–3. doi: 10.1093/nar/gkm950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19(6):1107–16. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38(Database issue):D331–5. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D. et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35(Database issue):D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin MJ. Using the Generic Genome Browser (GBrowse) Curr Protoc Bioinformatics. 2009;Chapter 9:Unit 9–9. doi: 10.1002/0471250953.bi0909s28. [DOI] [PubMed] [Google Scholar]
- Southworth LK, Owen AB, Kim SK. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5(12):e1000776. doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler AS. et al. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21(24):3244–57. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ. et al. Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8(7):R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miteva MA. et al. Fast structure-based virtual ligand screening combining FRED, DOCK, and Surflex. J Med Chem. 2005;48(19):6012–22. doi: 10.1021/jm050262h. [DOI] [PubMed] [Google Scholar]
- Cammer S. SChiSM2: creating interactive web page annotations of molecular structure models using Jmol. Bioinformatics. 2007;23(3):383–4. doi: 10.1093/bioinformatics/btl603. [DOI] [PubMed] [Google Scholar]
- Pruitt KD. et al. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37(Database issue):D32–6. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36(Web Server issue):W358–63. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klammer M, Roopra S, Sonnhammer EL. jSquid: a Java applet for graphical on-line network exploration. Bioinformatics. 2008;24(12):1467–8. doi: 10.1093/bioinformatics/btn213. [DOI] [PubMed] [Google Scholar]