Abstract
Mutations of transmembrane channel-like gene 1 (TMC1) cause hearing loss in humans and mice. TMC1 is the founding member of a family of genes encoding proteins of unknown function that are predicted to contain multiple transmembrane domains. The goal of our study was to define the topology of mouse TMC1 expressed heterologously in tissue culture cells. TMC1 was retained in the endoplasmic reticulum (ER) membrane of five tissue culture cell lines that we tested. We used anti-TMC1 and anti-HA antibodies to probe the topologic orientation of three native epitopes and seven HA epitope tags along full-length TMC1 after selective or complete permeabilization of transfected cells with digitonin or Triton X-100, respectively. TMC1 was present within the ER as an integral membrane protein containing six transmembrane domains and cytosolic N- and C- termini. There is a large cytoplasmic loop, between the fourth and fifth transmembrane domains, with two highly conserved hydrophobic regions that might associate with or penetrate, but do not span, the plasma membrane. Our study is the first to demonstrate that TMC1 is a transmembrane protein. The topologic organization revealed by this study shares some features with the shaker-TRP superfamily of ion channels.
Keywords: deafness, endoplasmic reticulum, hearing, topology, transmembrane
Mutations in transmembrane channel-like gene 1 (TMC1/Tmc1) can cause dominant or recessive hearing loss in humans and mice (1, 2). Tmc1 mRNA is specifically expressed in neurosensory hair cells of the inner ear (1, 2). Cochlear neurosensory hair cells of Tmc1 mutant mice fail to mature into fully functional sensory receptors (3) and exhibit concomitant structural degeneration that could be a cause or an effect of the maturational defect (2). The molecular and cellular functions of TMC1 protein remain unknown due, at least in part, to in situ expression levels that are prohibitively low for direct biochemical analysis.
There are seven additional mammalian TMC paralogs whose structure and function are also unknown. There are no significant sequence similarities of any TMC protein with other proteins of known function. An initial PSORT-II analysis of human and mouse TMC proteins did not detect any N-terminal signal sequences or other trafficking signals, but it did predict that TMC proteins reside in the plasma membrane (4). The TMC proteins are all predicted to contain six to ten transmembrane domains (TMDs) and a novel, conserved region, which we termed the TMC domain (4). TMHMM2.0 analysis of mouse and human TMC1 predicts cytoplasmically oriented N- and C-termini and six TMDs that are also predicted for the other paralogs (4). Other algorithms such as PSORTII and TopPred predict two to four additional TMDs, for a total of eight to ten TMDs, per TMC homolog (2, 5). PROSITE and NetNGlyc identified several TMC sequence sites with varying probabilities of glycosylation, but neither PSORT II nor SignalP detected an N-terminal signal peptide sequence (4). The in situ cellular location of TMC proteins is unknown, but human TMC6 (also known as EVER1) and TMC8 (EVER2) proteins expressed in transiently transfected human HaCaT keratinocyte cells appear to be retained in the endoplasmic reticulum (6). Truncating mutations of EVER1 and EVER2 cause epidermodysplasia verruciformis (EV; MIM 226400), characterized by susceptibility to cutaneous human papilloma virus infections and associated non-melanoma skin cancers (6).
The purpose of our study was to determine the transmembrane topology of TMC1. We performed our experiments on mouse TMC1 (mTMC1) expressed in transiently transfected COS-7 and HeLa cells. We used differential detergent treatment to distinguish cytoplasmic from intraluminal epitopes of transmembrane proteins in the endoplasmic reticulum (ER). Our results indicate that heterologously expressed mTMC1 is an integral membrane protein with six TMDs and cytoplasmically oriented N- and C- termini.
EXPERIMENTAL PROCEDURES
Antibodies
We derived polyclonal antisera #272, #277, #274, and #255 from rabbits immunized with keyhole limpet hemocyanin (KLH)-conjugated synthetic peptides corresponding to mTMC1 amino acids 21–39 (EEDKLPRRESLRPKRKRTR), 53–72 (DEETRKAREKERRRRLRRGA), 216-236 (GSLPRKTVPRAEEASAANFGV), and 731-747 (MKQQALENKMRNKKMAA), respectively. We ordered peptides from Princeton BioMolecules (Langhorne, PA) and antibodies from Covance Research Products (Denver, PA). We purchased polyclonal anti-β-tubulin and monoclonal anti-PDI (Abcam, Cambridge, MA), monoclonal anti-α-tubulin (Molecular Probes, Carlsbad, CA), polyclonal anti-GRP94, monoclonal anti-KDEL (Stressgen, San Diego, CA). Monoclonal anti-hemagglutinin (HA) antibodies were from Abcam and polyclonal anti-HA antibodies were from Covance.
Plasmids
We PCR-amplified the full-length mouse Tmc1 open reading frame from a previously reported cDNA clone in pGEM T-easy (1). Our sense (5’-GCT AGC ATG TTG CAA ATC CAA GTG-3’) and antisense (5’-GGA TCC CTG GCC ACC AGC AGC TGC-3’) amplification primers contained NheI and BamHI restriction sites, respectively, for subsequent cloning. We used site-directed mutagenesis (QuickChange, Stratagene, La Jolla, CA) to insert one HA epitope tag (YPYDVPDYA) (7) per expression construct at each of seven sites. Each pair of 67-bp mutagenic primers contained 27 bp (5’-TAC CCA TAT GAC GTC CCG GAC TAC GCC-3’) encoding the HA tag, flanked by two 20-bp Tmc1 sequences encoding each side of the target insertion site. The HA tag was inserted between amino acids 237 and 238 (HA1), 327 and 328 (HA2), 402 and 403 (HA3), 510 and 511 (HA4), 568 and 569 (HA5), 616 and 617 (HA6), and 671 and 672 (HA7) (Fig. 1C). Clones were sequenced, to verify correct insertion of the HA-tag sequence without unwanted mutagenic events, and digested with NheI and BamHI. The cDNA inserts were purified by 1% agarose gel electrophoresis and QIAquick Gel Extraction (QIAGEN, Valencia, CA) and subcloned into NheI- and BamHI-digested pcDNA3.1 (-) (Invitrogen, Carlsbad, CA).
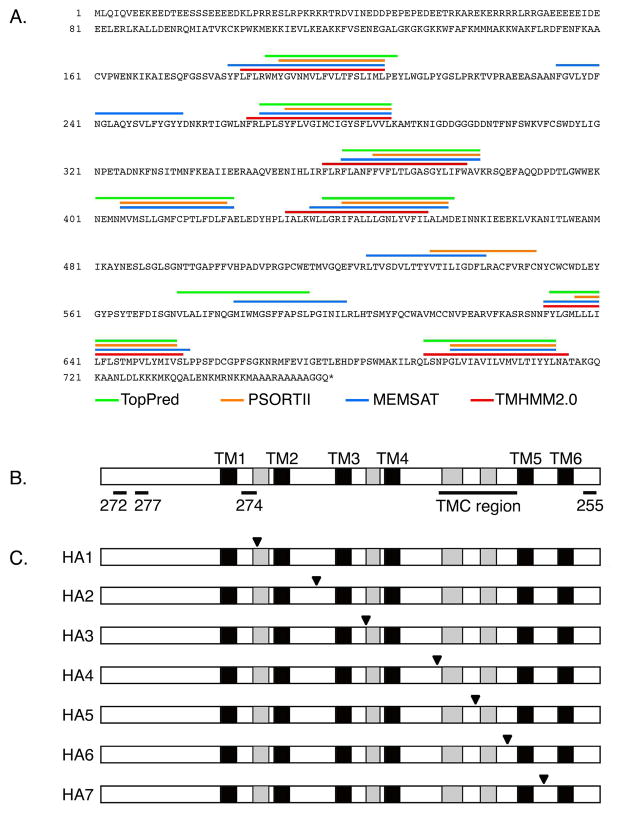
Fig. 1.
Potential models of membrane topology, immunogenic peptides and epitope tags of mTMC1. (A) Deduced amino acid sequence of mTMC1 and transmembrane domains predicted by TMHMM2.0 (red (17)), MEMSAT (blue (27)), PSORT II (orange (28)), and TopPred (green (29)). Each algorithm used standard parameters for eukaryotic membrane proteins. (B) Schematic illustration of mTMC1 showing six regions (black: TM1 to TM6) with hydropathy values higher than 1.6 and four regions (grey) with hydropathy values between zero and 1.6. Also shown are the positions of the TMC domain and of the four synthetic peptides used to generate anti-TMC1 antisera #272, #274, #277, and #255. (C) Schematic illustration of HA-tagged mTMC1 expression constructs with tag insertion positions (black triangles).
Tissue Culture and Transient Transfection
COS-7 and HeLa cells were grown in DMEM (Invitrogen) with 10% fetal bovine serum in an incubator at 37°C with 5% CO2. For immunofluorescence assays, cells were grown on glass coverslips in six-well plates (Corning Glass, Lowell, MA) and transfected with the expression construct using Lipofectamine 2000 (Invitrogen) for COS-7 cells or FuGENE HD (Roche, Indianapolis, IN) for HeLa cells. COS-7 and HeLa cells were incubated in the transfection mix in a 5% CO2 incubator at 37°C for 18–24 h. Cells were grown in T75 flasks (Corning Glass, Lowell, MA) and incubated with transfection mix for 18–24 h for protein extraction.
Protein extraction
Transiently transfected cells were detached with PBS containing 1mM EDTA for 5–10 minutes at 37°C. All of the following procedures were done on ice or at 4°C. Cells were washed twice in PBS, resuspended in MB buffer (210mM mannitol, 70mM sucrose, 1mM EGTA, and 10mM Hepes pH 7.5, protease inhibitor cocktail III (Calbiochem, Gibbstown, NJ)), and incubated for 1 h. Cells were lysed by 20 passages through a 29G x 1/2” (0.34mmx13mm) needle on a 1ml syringe (Kendall, Tyco Healthcare Group, Mansfield, MA). Microscopic examination assured that cell lysis was 99% complete. The lysed cell suspension was centrifuged at 600g for 10 min to pellet the nuclei. The supernatant was regarded as a whole cell lysate. Microsomes were prepared by a 20-min centrifugation of the whole cell lysate at 6,800g to remove mitochondria. The resulting supernatant was centrifuged for 1 h at 100,000g to obtain a microsomal pellet, which was resuspended in MB buffer.
Western blot analysis
Whole cell extracts and microsomal preparations were denatured by boiling in NuPAGE LDS sample buffer (Invitrogen, Carlsbad, CA) for 5 min. Samples were separated by SDS-PAGE on 4–12% Bis-Tris gels using either MOPS or MES buffer (Invitrogen). Proteins were transferred to PVDF membranes (Millipore, Billerica, MA), blocked overnight with 5% nonfat dry milk in TBST (10mM Tris-HCL pH 7.5, 150 mM NaCl, and 0.05% Tween-20), and probed with either anti-TMC1 antibody #277 or polyclonal anti-HA antibody (both at 1:400 dilution). Proteins were detected by ECL using a 1:10,000 dilution of horseradish peroxidase-conjugated anti-rabbit IgG (Amersham, Piscataway, NJ) or with the Typhoon Trio Plus imaging system (GE Healthcare Life Sciences, Piscataway, NJ) using a 1:2,500 dilution of ECL Plex Cy5 fluorescent anti-rabbit secondary antibody (Amersham).
Immuno-fluorescence
All procedures were performed at room temperature. Cells on coverslips were washed three times with PBS, fixed with 4% paraformaldehyde in PBS for 10 min, and washed again three times with PBS for 10 min each. The plasma membrane was selectively permeabilized with 5-μg/ml (0.0005%) digitonin (Sigma, St. Louis, MO) in incubation buffer (0.3M sucrose, 2.5mM MgCl2, 0.1M KCl, 1mM EDTA, 10mM Pipes, pH 6.8) for 4 min. Alternatively, permeabilization of the plasma and ER membranes was achieved by incubation with 0.25% Triton X-100 in PBS for 10 min. Immediately following permeabilization, cells were washed three times with PBS and blocked with 2% bovine serum albumin (Roche) and 5% goat serum (Invitrogen) in PBS for 30 min. Cells were then incubated for 1 h with primary antibodies: polyclonal antisera #272, #255, or #274, or a monoclonal antibody to hemagglutinin. Cells were co-stained with antibodies to cytosolic tubulin, and ER luminal proteins PDI or GRP94 or soluble ER resident proteins carrying the KDEL retention signal. We initially used an anti-PDI antibody to stain COS-7 cells, but it produced high background staining with HeLa cells. We also tried antibodies against KDEL and GRP94, and found that anti-GRP94 gave the best results with HeLa cells. Cells were washed three times with PBS and incubated with secondary antibodies (Alexa Fluor, Invitrogen) for 30 min. After three 15-min washes with PBS, we mounted the coverslips onto glass slides and added ProLong Gold antifade reagent (Invitrogen). We randomly chose approximately 20 to 30 different areas of each sample for visualization with a Zeiss LSM510 confocal microscope, 63× apochromat oil immersion phase-contrast objective, and Texas Red or FITC optics.
RESULTS
Potential mTMC1 Transmembrane Topologic Models
We compared the topologies of mTMC1 predicted by four different algorithms (Fig. 1A). Although the number of predicted TMDs varies among algorithms, all of the algorithms predicted the N-terminus of mTMC1 to be cytoplasmic. A Kyte-Doolittle hydropathy plot detects ten hydrophobic regions, including six regions with a hydropathy score larger than 1.6 (Fig. 1B) and, thus, a higher probability to span the membrane (8). The latter six regions are predicted to be TMDs by all of the algorithms, whereas the four regions with lower hydropathy scores are predicted to be TMDs by only some of the algorithms (Fig. 1A, 1B).
Validation of mTMC1 antibodies
We tested the specificity of mTMC1 antibodies #272, #274, and #255 in immunofluorescence staining. We expressed HA epitope-tagged mTMC1 (construct HA2) in COS-7 cells. Cells were permeabilized with Triton x-100 and co-stained with anti-HA antibody and one of each of the three anti-TMC1 antisera (Fig. 2A), or co-stained with anti-tubulin and one of each of the anti-TMC1 antisera (Fig. 2B). Immunoreactivity with anti-HA completely overlaps with that of each of the anti-TMC1 antibodies, confirming that the TMC1 antibodies specifically recognize TMC1 in this assay (Fig. 2A). Immunoreactivity with anti-TMC1 antibody is only detected in transfected cells, further validating the specificity of anti-TMC1 antibodies in this assay (Fig. 2B).
Fig. 2.
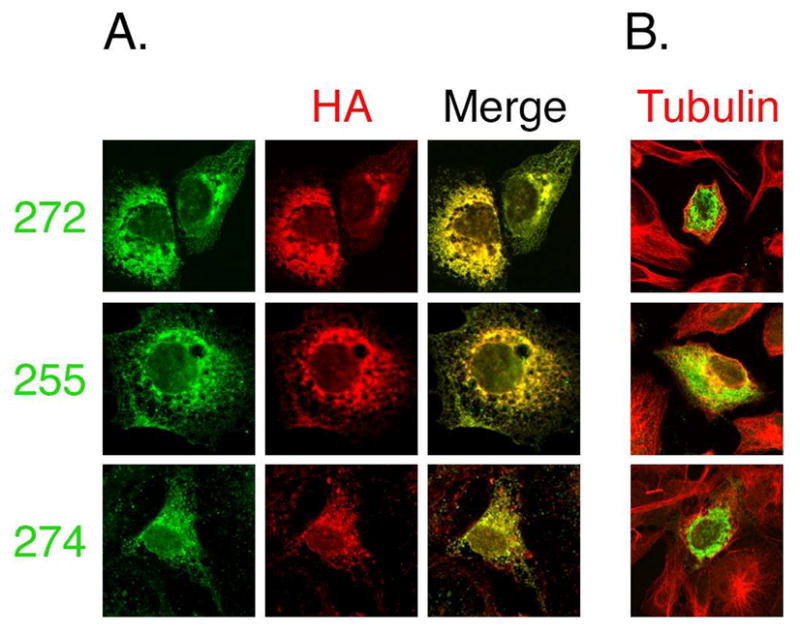
Specificity of mTMC1 antibodies. COS-7 cells were transfected with HA-tagged mTMC1 (construct HA2) and co-stained with either anti-HA or anti-tubulin antibody and each of three different anti-TMC1 antisera. (A) Left panels, immunoreactivity with anti-TMC1 antisera #272, #255 or #274 (green). Middle panels, immunoreactivity with anti-HA antibody (red). Right panels, merge of left and middle panels. Immunoreactivity with anti-HA completely overlaps with that of each of the different anti-TMC1 antibodies. (B) Merge of anti-tubulin (red) immunoreactivity used to visualize all cells and antisera #272, #255 or #274 (green) immunoreactivity. Anti-TMC1 immunoreactivity is seen only in transfected cells.
Heterologous expression and localization of mTMC1
We repeated the PSORT II analysis of TMC proteins. The results indicate that all human and mouse TMC proteins might reside in the ER membrane, albeit at a lower probability (18–39%) than in the plasma membrane (56–72%). Immunofluorescence analysis of epitope-tagged mTMC1 expressed in a variety of different tissue culture cell lines (COS-7, HeLa, SHSY5Y, HEK293, MDCK) results in a reticular pattern consistent with the location of the ER and overlapping with staining for ER-specific proteins (Fig. 2 and 3). We could not detect plasma membrane localization by staining unfixed, non-permeabilized tissue culture cells expressing mTMC1 (results not shown). In contrast, other multi-pass transmembrane proteins (TRPV4, pendrin) traffic normally to the plasma membrane in this expression system (9). We identified several known (amino acids 35–37 and 64–67) as well as putative (amino acids 37–39 and 67–69) arginine-based ER retention signals of the RXR type (10–12) in the N-terminal region of mTMC1. Disrupting these retention signals by alanine mutagenesis does not affect ER retention (Results not shown).
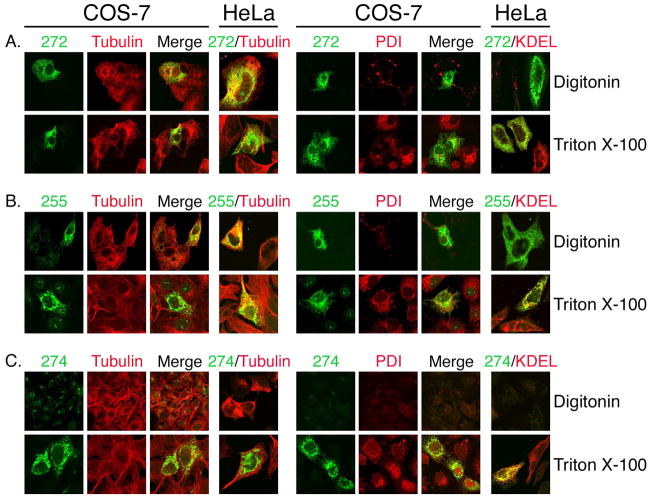
Fig. 3.
Topology probed by mTMC1 epitope accessibility. COS-7 or HeLa cells transfected with native mTMC1 were either selectively or fully permeabilized with digitonin or Triton X-100, respectively. Cells were stained with each of the anti-TMC1 antisera as well as antibodies against ER luminal protein PDI or soluble ER resident proteins carrying the KDEL retention signal (right panels) or cytosolic tubulin (left panels) to evaluate the extent of permeabilization. Only merged images are shown for HeLa cells as similar results were obtained for both cell lines. (A and B) the N- and C- terminal epitopes (recognized by #272 and #255, respectively) and tubulin are readily detected following digitonin permeabilization (upper rows), whereas PDI and KDEL are only accessible following full permeabilization by Triton X-100 (lower rows). (C) PDI and KDEL and the epitope recognized by antiserum #274 are only accessible after Triton X-100 permeabilization (lower row).
While the ER could be the in situ location of mTMC1, our results may also reflect retention of overexpressed and misfolded mTMC1 in the ER, a lack of appropriate trafficking signals or partners to deliver mTMC1 to the plasma membrane, or a combination of these mechanisms.
Topologic Structure Determined by Native mTMC1 Epitope Accessibility
We used polyclonal antisera #272, #255, and #274 to determine the epitope accessibility, and thus topologic orientation, of amino acids 21–39 in the N-terminus, amino acids 731–747 in the C- terminus, and amino acids 216–236 in the first predicted internal loop, respectively, in native mTMC1 protein (Fig. 1B). We either permeabilized all plasma and intracellular membranes with 0.25% Triton X-100 or selectively permeabilized only the plasma membrane while leaving the ER membrane intact with 5-μg/ml digitonin (13, 14). We used antibodies against ER luminal protein PDI or soluble ER resident proteins carrying the KDEL retention signal, or against cytosolic tubulin to evaluate the extent of permeabilization. The native mTMC1 N- and C-terminal epitopes and tubulin were accessible for binding to their respective antibodies with either Triton X-100 or digitonin permeabilization of either COS-7 or HeLa cells (Fig. 3A, B). In contrast, amino acids 216–236 of mTMC1, and the ER luminal proteins, were accessible to antibodies only when cells were exposed to Triton X-100 (Fig. 3C). These results demonstrate that the N- and C- termini of native mTMC1 are oriented toward the cytoplasm and the predicted first internal loop is oriented toward the ER lumen.
mTMC1 Topology Determined by HA Epitope Accessibility
We designed a series of HA-tagged mTMC1 expression constructs to differentiate among the possible mTMC1 topological models (Fig. 1C). The topologic orientations of tagged regions were determined by their accessibility to anti-HA antibodies after differential detergent treatment. The insertion of the 9-amino acid HA tag into hydrophilic loops of other proteins typically has little or no effect on their structure and function (15, 16). The HA-tagged constructs were transiently expressed in either HeLa or COS-7 cells to yield HA-tagged mTMC1 proteins of the predicted size (Fig. 4).
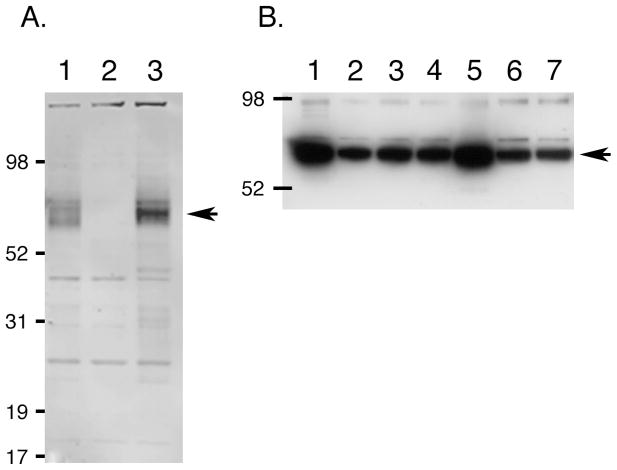
Fig. 4.
Western blot analysis of native and HA-tagged mTMC1. (A) Whole cell lysates of COS-7 cells transiently transfected with a native mTMC1 expression construct (lane 1), empty pcDNA 3.1 (−) vector (lane 2), and a representative HA (HA4) expression construct (lane 3) were resolved on a 4–12% Bis-Tris gel using MOPS running buffer. The immunoblot was probed with anti-TMC1 antibody #277. An arrow indicates a strong band of the expected size (approximately 87 kDa) in lanes 1 and 3, but not in lane 2. (B) Microsomal extractions from COS-7 cells transiently transfected with each of the HA-tagged constructs HA1-HA7 (lanes 1–7, respectively) were resolved on a 4–12% Bis-Tris gel using MES running buffer. The blot was probed with anti-HA antibody. Arrow indicates a band of the expected size in all lanes.
The HA epitopes encoded by HA1, HA3 and HA7 and the ER luminal protein GRP94 were accessible to antibodies only after permeabilization with 0.25% Triton X-100 (Fig. 5). These results demonstrate that the TMHMM2.0-predicted first, third and fifth loops of mTMC1 flanked by transmembrane domains TM1 and TM2, TM3 and TM4, and TM5 and TM6, respectively, are all oriented toward the ER lumen. In contrast, the HA epitope encoded by HA2 and the cytosolic marker tubulin were accessible for antibody binding after either Triton X-100 or digitonin treatment (Fig. 5).
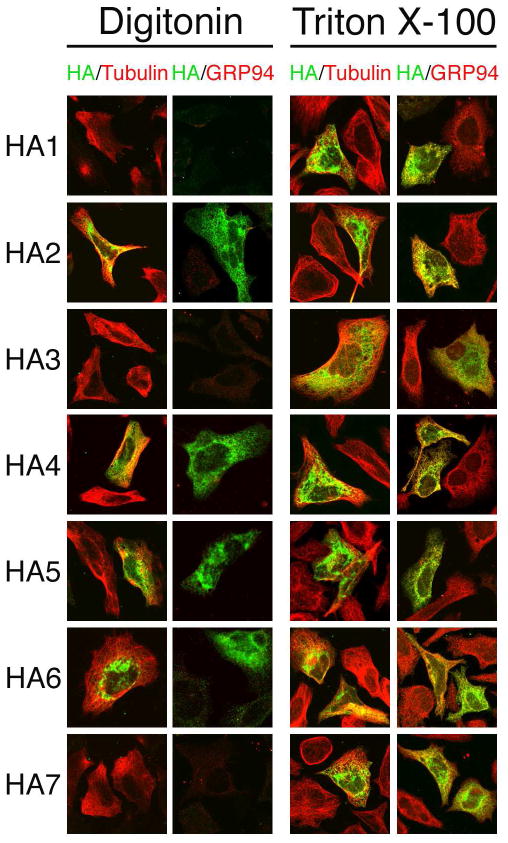
Fig. 5.
Topology probed by HA epitope tag accessibility. HeLa cells transfected with each of the HA-tagged mTMC1 expression constructs were permeabilized with either digitonin (left panels) or Triton X-100 (right panels). With each treatment, cells were co-stained with anti-HA (green) and anti-tubulin (red) antibodies or with anti-HA (green) and anti-GRP94 (red) antibodies. Tubulin and the HA epitope tags of HA2, HA4, HA5, and HA6 were accessible to antibodies after selective permeabilization with digitonin. In contrast, GRP94 and the HA epitope tags of HA1, HA3, and HA7 were stained only after full permeabilization with Triton X-100.
The TMHMM2.0-predicted loop flanked by TM4 and TM5 contains a region of hydrophobic residues predicted by other algorithms (MEMSAT, PSORT II, and TopPred) to include one or two additional TMDs (Fig. 1A, 1B). We designed and expressed constructs HA4, HA5, and HA6 to differentiate among these models. Their HA epitopes as well as tubulin were stained by antibodies after either Triton X-100 or digitonin treatment, whereas GRP94 was stained only after Triton X-100 (Fig. 5). These results confirm that all three HA epitope tags inserted into the loop flanked by TM4 and TM5 are oriented toward the cytoplasm.
The topologic orientations of the HA epitopes in the seven different HA-tagged mTMC1 expression constructs confirm the TMHMM2.0-predicted model of six membrane-spanning domains and rule out the existence of the additional TMDs predicted by other algorithms. Accessibility of the HA epitope tag encoded by HA1 only after Triton X-100 permeabilization is consistent with the result with native mTMC1 and antibody #274. This indicates that the insertion of the HA tag into the TMHMM2.0-predicted first loop of mTMC1 had no effect on its orientation. Furthermore, the insertion of HA epitope tags did not disrupt the cytoplasmic orientation of the N- and C- termini that was observed for both the HA-tagged expression constructs and native mTMC1 (Fig. 6).
Fig. 6.
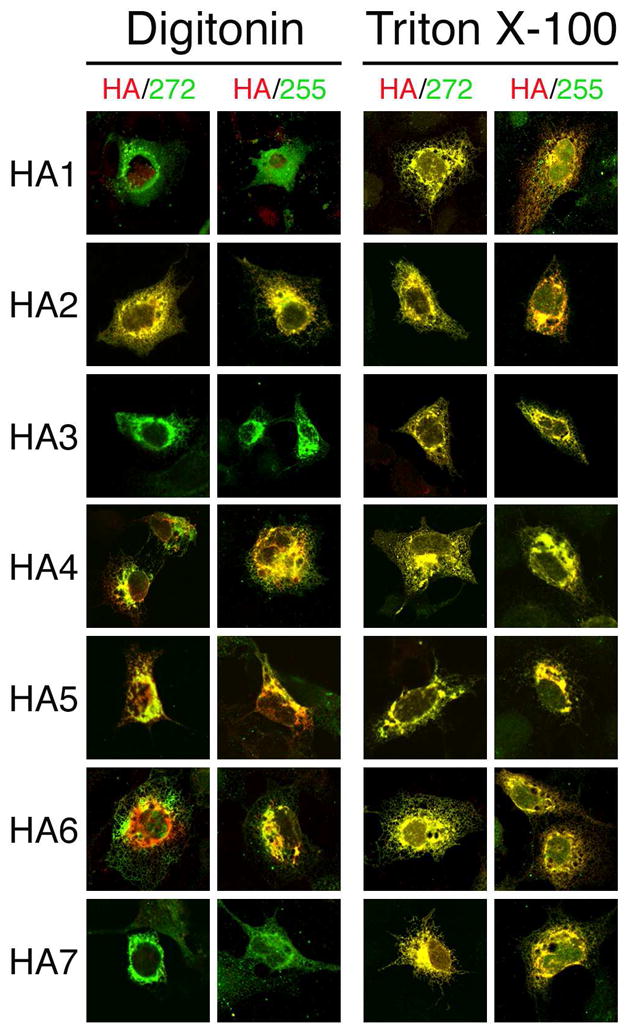
Preservation of N- and C- terminal topology in HA epitope-tagged mTMC1. COS-7 cells transfected with each of the HA-tagged mTMC1 expression constructs were permeabilized with either digitonin (left panels) or Triton X-100 (right panels). Cells were stained with anti-HA antibody (red) and with either antisera #272 or #255 (green). The N- and C- termini epitopes of each expression product are accessible for staining with their respective antibodies following digitonin permeabilization. Anti-TMC1 staining of HA1, HA3, and HA7 appears green because the HA tags inserted in the internal loops are not accessible for staining. For HA2, HA4, HA5, and HA6, the HA tag is inserted in an accessible cytoplasmic loop and its (red) staining overlaps the green staining of N- and C- terminal epitopes and appears yellow.
DISCUSSION
In this study, we investigated the topological organization of mTMC1 heterologously expressed in COS-7 or HeLa cells. The reticular expression pattern of heterologous mTMC1 indicates that it was localized within or associated with the endoplasmic reticulum membranes. TMHMM2.0 was previously reported to be the best-performing topology prediction algorithm (17–19). Our results are consistent only with the TMHMM2.0-predicted model of mTMC1 having cytoplasmically oriented N- and C- termini and six transmembrane domains (Fig. 7).
Fig. 7.
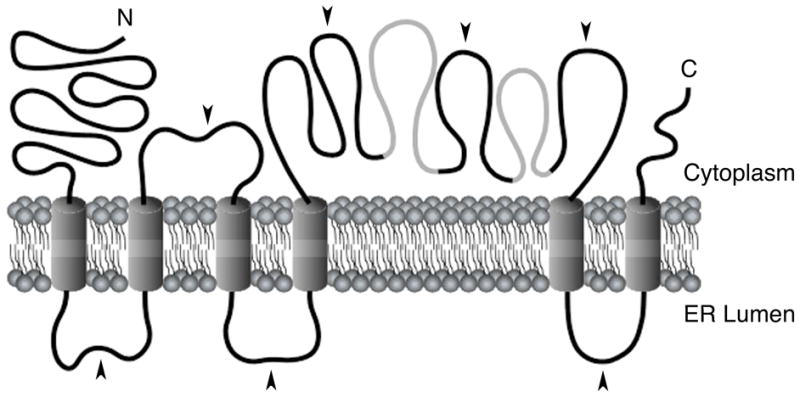
Model for the membrane topology of mTMC1. Our results show that mTMC1 expressed in COS-7 or HeLa cells contains six membrane-spanning domains with N- and C- termini oriented towards the cytoplasm. The model is shown with TM4 and TM5 flanking a long cytoplasmic loop. The two grey regions in this loop are predicted to be TM domains at very low probability and might be membrane-associated or re-entrant domains. Black arrowheads indicate HA tag insertion sites.
Although some algorithms predict the existence of additional TMDs in mTMC1 and other TMC proteins, only the domains homologous to the six TMDs of mTMC1 are predicted to span membranes by all algorithms. Furthermore, in silico hydrophobicity analyses of other TMC proteins indicate that these homologous domains consistently have the highest hydropathy values (>1.6). The six-pass transmembrane topology of mTMC1 may therefore be a shared structure with all TMC proteins.
The large cytoplasmic loop flanked by TM4 and TM5 includes the TMC domain (Fig. 1B), which is highly conserved among all TMC proteins, including TMC homologs in Drosophila melanogaster and Caenorhabditis elegans (4). Although the function of this region is unknown, its importance is highlighted by an in-frame deletion of 57 amino acids of the large cytoplasmic loop, including part of the TMC domain, encoded by the Tmc1dn allele of the deafness (dn) mutant mouse strain (1). This region contains eight cysteine residues, including at least three (amino acids 553, 555 and 610) of which are highly conserved and one (amino acid 512) which is conserved among all known TMC homologs (4, 5). These residues are likely to be required for TMC1 function and structure and thus may confound a cysteine accessibility approach to probe the topology of this region in the absence of a functional assay to rule out misfolding. The TMC domain also contains two mildly hydrophobic regions that are predicted, at low probability, to span the membrane by some algorithms, but not by TMHMM2.0 (Fig. 7). Since the three HA epitope tags flanking these regions of mTMC1 were all oriented toward the cytoplasm (Fig. 5, 7), these two segments might be membrane-associated or re-entrant domains. This raises the possibility that these mildly hydrophobic segments contribute to a pore-forming structure of TMC1 if it is a channel or transporter. Indeed, ProtFun 2.2 (20, 21) predicts that all human and mouse TMC proteins are transporters or, at a lower probability, ion channels.
We do not have a functional assay for TMC1 and we cannot rule out the possibility that mTMC1 expression products are retained in the ER due to incomplete folding or misfolding. Polytopic membrane proteins depend on specialized membrane-localized chaperones to prevent inappropriate interactions between membrane-spanning segments as they insert and fold in the ER membrane (22). In the absence of these chaperones, membrane proteins are retained in the ER despite correct insertion of their membrane-spanning segments. A lack of hair cell-specific chaperones, trafficking signals or protein partners in our heterologous expression systems could also lead to ER retention of properly folded TMC1 protein.
The topology of mTMC1 resembles that of a large super-family of proteins that includes TRP channels and voltage-gated K channels. These proteins also have six membrane-spanning segments (S1–S6) and cytoplasmic N- and C- termini (23, 24). However, we cannot detect any specific sequence similarities of TMC proteins with these other channels (4). Moreover, the pore-forming loop of the channels is located between S5 and S6 (24–26) whereas our in silico hydropathy analyses imply that any potential pore-forming loop of TMC1 would be located between TM4 and TM5. We, and others, have been unable to directly attribute any channel or transporter activity to TMC1 by electrophysiological methods (unpublished observations; 2, 3), but its topology justifies continued consideration of this possibility. Finally, the topology of TMC1 provides a foundation for the design and interpretation of experiments to identify interacting protein partners that might give us insight into the possible function of TMC1 and other TMC proteins.
Acknowledgments
We thank our NIDCD colleagues for advice and for critical review of the manuscript.
Abbreviations
- ER
endoplasmic reticulum
- TMC1
transmembrane channel-like gene 1 protein product
- TMD
transmembrane domain
- mTMC1
mouse TMC1
- KLH
keyhole limpet hemocyanin
- KDEL
amino acid sequence of an ER retention signal
- PVDF
polyvinylidene fluoride
Footnotes
This work was supported by National Institutes of Health intramural research fund Z01-DC-000060.
References
- 1.Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, Ghosh M, Menon PS, Deshmukh D, Oddoux C, Ostrer H, Khan S, Riazuddin S, Deininger PL, Hampton LL, Sullivan SL, Battey JF, Jr, Keats BJ, Wilcox ER, Friedman TB, Griffith AJ. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 2.Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, Hrabé de Angelis M, Avraham KB, Steel KP. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 3.Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J Physiol. 2006;574:677–698. doi: 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82:300–308. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 5.Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 2003;4:24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramoz N, Rueda LA, Bouadjar B, Montoya LS, Orth G, Favre M. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nature Genet. 2002;32:579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- 7.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 8.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 9.Choi BY, Stewart AK, Madeo AC, Pryor SP, Lenhard S, Kittles R, Eisenman D, Kim HJ, Niparko J, Thomsen J, Arnos KS, Nance WE, King KA, Zalewski CK, Brewer CC, Shawker T, Reynolds JC, Butman JA, Karniski LP, Alper SL, Griffith AJ. Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: genotype-phenotype correlation or coincidental polymorphisms. Hum Mutat. 2009;30:599–608. doi: 10.1002/humu.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 11.Margeta-Mitrovic M, Jan YN, Jan YL. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 12.Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plutner H, Davidson HW, Saraste J, Balch WE. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J Cell Biol. 1992;119:1097–1116. doi: 10.1083/jcb.119.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson R, Allen AJ, Oliver J, Brookman JL, High S, Bulleid NJ. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J. 1995;307:679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson PL, Flintoff WF. Topological and functional analysis of the human reduced folate carrier by hemagglutinin epitope insertion. J Biol Chem. 1999;274:16269–16278. doi: 10.1074/jbc.274.23.16269. [DOI] [PubMed] [Google Scholar]
- 16.Lin S, Lu X, Chang CC, Chang TY. Human acyl-coenzyme A: cholesterol acyltransferase expressed in Chinese hamster ovary cells: membrane topology and active site location. Mol Biol Cell. 2003;14:2447–2460. doi: 10.1091/mbc.E02-11-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 18.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 19.Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 20.Jensen LJ, Gupta R, Blom N, Devos D, Tamames J, Kesmir C, Nielsen H, Staerfeldt HH, Rapacki K, Workman C, Andersen CA, Knudsen S, Krogh A, Valencia A, Brunak S. Prediction of human protein function from post-translational modifications and localization features. J Mol Biol. 2002;319:1257–1265. doi: 10.1016/S0022-2836(02)00379-0. [DOI] [PubMed] [Google Scholar]
- 21.Jensen LJ, Gupta R, Staerfeldt HH, Brunak S. Prediction of human protein function according to Gene Ontology categories. Bioinformatics. 2003;19:635–642. doi: 10.1093/bioinformatics/btg036. [DOI] [PubMed] [Google Scholar]
- 22.Kota J, Ljungdahl PO. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J Cell Biol. 2005;168:79–88. doi: 10.1083/jcb.200408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannier B, Zhu X, Brown D, Birnbaumer L. The membrane topology of human transient receptor potential 3 as inferred from glycosylation-scanning mutagenesis and epitope immunocytochemistry. J Biol Chem. 1998;273:8675–8679. doi: 10.1074/jbc.273.15.8675. [DOI] [PubMed] [Google Scholar]
- 24.Yool AJ, Schwarz TL. Alteration of ionic selectivity of a K+ channel by mutation of the H5 region. Nature. 1991;349:700–704. doi: 10.1038/349700a0. [DOI] [PubMed] [Google Scholar]
- 25.Hartmann HA, Kirsch GE, Drewe JA, Taglialatela M, Joho RH, Brown AM. Exchange of conduction pathways between two related K+ channels. Science. 1991;251:942–944. doi: 10.1126/science.2000495. [DOI] [PubMed] [Google Scholar]
- 26.Dohke Y, Oh YS, Ambudkar IS, Turner RJ. Biogenesis and topology of the transient receptor potential Ca2+ channel TRPC1. J Biol Chem. 2004;279:12242–12248. doi: 10.1074/jbc.M312456200. [DOI] [PubMed] [Google Scholar]
- 27.Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 28.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 29.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]