Abstract
Cell adhesion to the extracellular matrix is mediated by adhesion receptors, mainly integrins, which upon interaction with the extracellular matrix, bind to the actin cytoskeleton via their cytoplasmic domains. This association is mediated by a variety of scaffold and signaling proteins, which control the mechanical and signaling activities of the adhesion site. Upon transformation of fibroblasts with active forms of Src (e.g., v-Src), focal adhesions are disrupted, and transformed into dot-like contacts known as podosomes, and consisting of a central actin core surrounded by an adhesion ring. To clarify the mechanism underlying Src-dependent modulation of the adhesive phenotype, and its influence on podosome organization, we screened for the effect of siRNA-mediated knockdown of tyrosine kinases, MAP kinases and phosphatases on the reorganization of the adhesion-cytoskeleton complex, induced by a constitutively active Src mutant (SrcY527F). In this screen, we discovered several genes that are involved in Src-induced remodeling of the actin cytoskeleton. We further showed that knockdown of Src in osteoclasts abolishes the formation of the podosome-based rings and impairs cell spreading, without inducing stress fiber development. Our work points to several genes that are involved in this process, and sheds new light on the molecular plasticity of integrin adhesions.
Keywords: Src, podosomes, actin, cytoskeleton, siRNA, screening, cell adhesion, microscopy
Introduction
Cell adhesion to the extracellular matrix occurs at specialized sites where adhesion receptors, mainly integrins, bridge between the extracellular matrix and the actin cytoskeleton, via a network of scaffold and signaling proteins (Berrier and Yamada, 2007; Campbell, 2008; Zamir and Geiger, 2001). There are several types of integrin-based adhesions that share many molecular components, but differ in their overall structure, function and actin machinery (Albiges-Rizo et al., 2009). Focal adhesions, the best-characterized integrin adhesions, are elongated structures that contain clusters of transmembrane integrin receptors bound to the ECM at one end, and to actin stress fibers at the other (Geiger et al., 2001; Block et al., 2008). The organization and dynamics of these structures are tightly regulated by associated signaling components, as evidenced by the fact that an active form of pp60src (Src) such as v-Src, induces deterioration of stress fibers and formation of dot-like contact sites, called podosomes (Tarone et al., 1985; Nitsch et al., 1989).
Podosomes are small (~1μm in diameter), actin-rich and highly dynamic adhesion-related cytoskeletal structures that are often associated with matrix degradation (Linder and Aepfelbacher, 2003; Linder and Kopp, 2005; Gimona and Buccione, 2006). Podosome architecture is quite different from that of focal adhesions, consisting of a central core of actin filaments surrounded by a peripheral adhesion-mediating ring domain. The adhesion ring is composed of integrins and many other adhesion-associated proteins such as paxillin, talin, tensin, p130cas and vinculin (Hiura et al., 1995; Lakkakorpi et al., 1993; Zambonin Zallone et al., 1989), while the core domain contains, in addition to F-actin, actin-associated proteins such as WASP or N-WASP (Calle et al., 2004), Arp 2/3 (Hurst et al., 2004), fimbrin (Marchisio et al., 1987), gelsolin (Chellaiah et al., 2000), α-actinin (Nermut et al., 1991), and cortactin (Hiura et al., 1995). Podosomes also contain and secrete matrix metalloproteases [MMPs; (Sato et al., 1997)].
Podosomes are the primary adhesion structures of monocyte-derived cells such as macrophages, osteoclasts (Marchisio et al., 1984) and dendritic cells (Burns et al., 2001). They can also be found in smooth muscle (Hai et al., 2002) and endothelial cells (Moreau et al., 2003). Because podosomes are found mainly in motile cells and control the activity of matrix metalloproteases, they are thought to contribute to tissue invasion and matrix remodeling.
Individual podosomes commonly assemble into higher-order structures. In v-Src transformed fibroblasts, for example, podosomes mostly group together to form ring-like structures called rosettes (Gavazzi et al., 1989), while in osteoclasts, podosomes undergo major reorganization during their maturation, ranging from individual structures, scattered throughout the cell’s ventral membrane via ordered clusters, to well-developed rings that extend toward the cell periphery, merge with neighboring rings, and eventually stabilize, forming a peripheral belt structure (Luxenburg et al., 2006a; Luxenburg et al., 2007).
The molecular infrastructure underlying all integrin adhesions is the so-called “integrin adhesome”, a complex, highly-interconnected network of more than 150 proteins that control the mechanical and signaling activities associated with these structures. When integrins interact with the extracellular matrix, there is a local increase in protein tyrosine phosphorylation, due to the activation of several tyrosine kinases (Clark and Brugge, 1995). The integrin adhesome is enriched in tyrosine kinases and phosphatases (Zaidel-Bar, 2010), and many of the protein-protein interactions essential for building the adhesion scaffold are mediated by the binding of SH2 domain-containing proteins to phosphorylated tyrosines of partner molecules (Zaidel-Bar et al., 2007a).
The Src tyrosine kinase is a major regulator of integrin-mediated adhesions, required for the efficient adhesion and spreading of a variety of cell types. Src−/− fibroblasts exhibit reduced adhesion and delayed spreading, compared to wild-type cells (Felsenfeld et al., 1999; Kaplan et al., 1995). Moreover, Src tyrosine kinase was shown to be involved in the regulation of podosome dynamics. Src scaffolding activity supports the assembly of immature, stationary podosomes, while Src catalytic activity is essential for podosome maturation and turnover. Furthermore, osteoclasts expressing constitutively active Src form ectopic podosome rings with high turnover (Luxenburg et al., 2006b).
Thus, Src and its substrates seem to be essential for podosome formation. For example, expression of activated Src in p130Cas-deficient fibroblasts results in impaired podosome formation (Honda et al., 1998), similar to knockdown of Pyk2 in osteoclasts (Duong et al., 2001). Src is also essential for osteoclast activity in vivo, since Src knockout mice suffer from severe osteopetrosis caused by deficient osteoclast activity (Soriano et al., 1991). Osteoclasts derived from such mice do not develop normal podosomes, cannot spread properly, and fail to give rise to mature super-structures (Lakkakorpi et al., 2001).
Tyrosine dephosphorylation is also involved in podosome turnover. The tyrosine phosphatase Shp-2 is necessary for Src-induced podosome formation (Hakak et al., 2000), and PTP-PEST, a tyrosine phosphatase for p130Cas, is a podosome component in osteoclasts (Linder and Aepfelbacher, 2003). Interestingly, despite intensive investigations into the regulation of integrin adhesions, the mechanisms and signaling pathways governing these processes, upstream or downstream of Src, are still poorly understood.
In this study, we investigated the role of Src in the assembly of various forms of integrin adhesions and, in particular, podosomes. Toward that end, we over-expressed a constitutively active Src mutant (Src-Y527F) that cannot be downregulated by inhibitory phosphorylation of tyrosine 527 (amino acid numbering is based on the sequence of chicken Src) in mouse embryonic fibroblasts (MEF). Microscopy-based examination of the transformed cells revealed major changes in actin organization – manifested by the loss of actin stress fibers and formation of podosome-based rings of different shapes.
To explore the signaling pathways downstream of Src that are involved in this phenotypic transformation, an siRNA-screen was performed, in which we knocked down tyrosine kinases, MAP kinases and phosphatases in SrcY527F-expressing fibroblasts, and analyzed their effects on actin organization. In this screen, several candidate genes were found to affect the Src-induced changes. Many of the siRNAs led to an increase in stress fiber formation, without affecting podosome ring formation. These findings indicate the existence of two different pathways regulated by Src, one leading to podosome assembly, and the other leading to stress fiber disassembly.
We further examined whether the molecules affecting Src-induced podosome formation in fibroblasts also affect sealing zone assembly in osteoclasts. The hits identified in the fibroblast siRNA screen had diverse effects on podosome formation in osteoclasts: For example, knocking down Src expression in osteoclasts dramatically reduced sealing zone formation, yet this treatment did not induce stress fiber formation in these cells. Our work sheds light on the molecules involved in the dynamic and structural effects of Src on the modulation of integrin adhesions.
Results
Constitutively active Src induces remodeling of cell adhesions and the associated actin cytoskeleton
To explore the mechanism underlying remodeling of focal adhesions and the actin cytoskeleton by active Src, we created a stable MEF cell line that expresses constitutively active Src. MEFs were infected with retroviruses encoding SrcY527F, or with YFP as a control. Control fibroblasts (MEF), labeled for paxillin, displayed 'classical' focal adhesions, located mainly at the cell periphery, and associated with the termini of actin stress fibers (Fig. 1A).
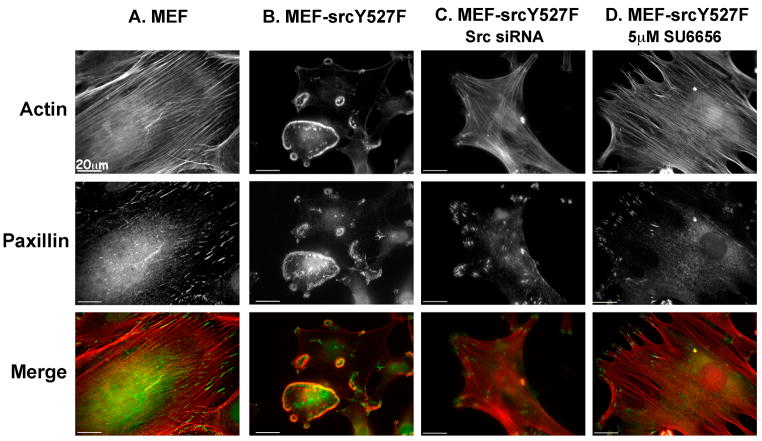
Figure 1. The SrcY527F constitutively active mutant causes extensive morphological changes in MEF cells that can be reversed by an Src inhibitor or by siRNA specific to Src.
(A) Control MEF and (B) MEF-SrcY527F cells were permeabilized, fixed and stained for actin (red) and paxillin (green). (C) MEF-SrcY527F cells were transiently transfected with Src siRNA and maintained in culture for 72 h before permeabilization, fixation and staining. (D) MEF-SrcY527F cells were treated with the Src family kinase inhibitor SU6656 (5μM) for 2 h and were then permeabilized, fixed, and stained for actin and paxillin.
Expression of deregulated Src had a dramatic effect on the cells, manifested by complete loss of actin stress fibers and paxillin-rich focal adhesions, coupled with the formation of highly organized, ring-shaped podosome arrays (Fig. 1B). Western blot analysis of the MEF-SrcY527F cells revealed a marked increase in the levels of Src tyrosine phosphorylation as well as active Src, compared with control MEFs (Supplementary Fig. S1). Transfection of the cells with siRNA targeting Src, or treatment with the Src inhibitor SU6656, induced restoration of focal adhesions and stress fibers and disappearance of the podosome rings, confirming the specificity and reversibility of the phenotype, and its dependence on Src enzymatic activity (Fig. 1C and D).
Staining of the MEF-SrcY527F cells for actin and paxillin displayed high cell-to-cell variability, which was not correlated with the levels of SrcY527F expression, and single-cell cloning failed to result in a morphologically uniform population (data not shown).
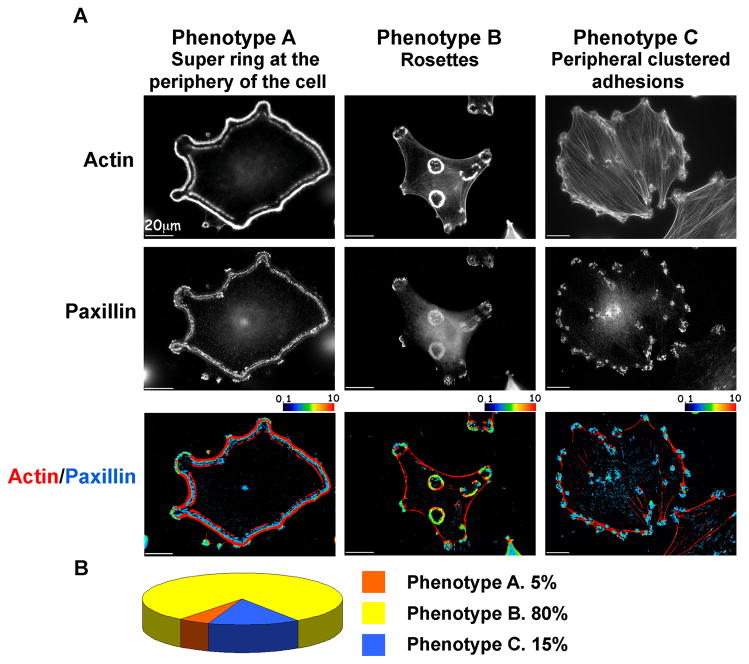
Based on the morphology and location of the actin rings, we identified three prominent “phenotypes”, which we refer to as “Phenotypes A, B and C” (Fig. 2A). While a few cells displayed a peripheral actin belt, similar to the sealing zone of osteoclasts (Luxenburg et al., 2006a), most of the cells completely lost their stress fibers and formed podosome rosettes. Moreover, some of the cells maintained poorly developed stress fibers and peripheral adhesions. The prominence of each phenotype in the total cell population is shown in Figure 2B.
Figure 2. The three major phenotypes of MEF-SrcY527F cells.
(A) MEF-SrcY527F cells were permeabilized, fixed and stained for actin and paxillin. Three groups of adhesion structures were defined: Phenotype A, displaying super rings at the periphery of the cell; Phenotype B with rosettes, and Phenotype C, with clusters of peripheral adhesions. Spatial ratio images are shown. Red indicates highly localized actin, whereas blue indicates high paxillin levels. Intermediate color levels, as shown in the color scale, indicate co-localization.
(B) Phenotype distribution in the total population of MEF-SrcY527F cells.
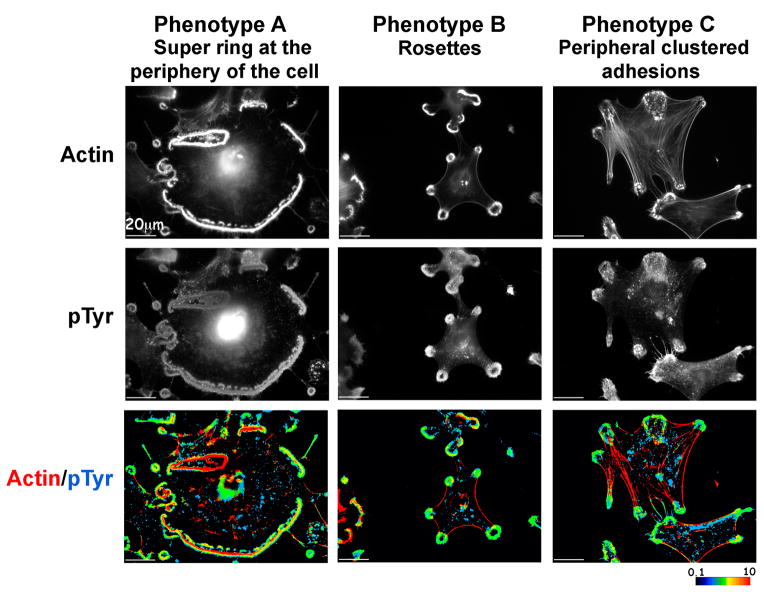
Staining of the cells (Fig. 3) reveals a clear segregation between actin and phosphotyrosine in Phenotype A as opposed to Phenotype B, and stress fibers anchored to large adhesions in Phenotype C. The structural domains of podosomes; namely, the F-actin-rich core surrounded by a paxillin-rich ring domain, were clearly resolved in scattered podosomes, but not in podosomes associated with rosettes, or with the peripheral ring (Supplementary Fig. S2).
Figure 3. Distribution of actin and tyrosine-phosphorylated proteins in the three major phenotypes of MEF-SrcY527F cells.
Representative MEF-SrcY527F cells from each phenotype were permeabilized, fixed, and stained for actin and general phospho-tyrosine. Spatial ratio images are shown. Red indicates highly localized actin, whereas blue indicates high phospho-tyrosine levels. Intermediate color levels, as shown in the color scale, indicate co-localization.
In order to assess the dynamics of the two domains, we prepared two color movies that reveal that the actin core is much more dynamic than plaque proteins such as paxillin (Supplementary Fig. S3, Supplementary Movie 1), as previously shown for podosomes in other cell types (Destaing et al., 2003; Luxenburg et al., 2006b).
The dynamic properties of the adhesions in the three phenotypes were compared by transfecting the cells with GFP-tagged α-actinin (or actin, for the dynamic B-type rings). The large ring or peripheral belt seen in Phenotype A expanded centrifugally; i.e., the cells retained the belt at the cell periphery, enabling it to expand as the cells spread (Fig. 4A, Supplementary Movie 2).
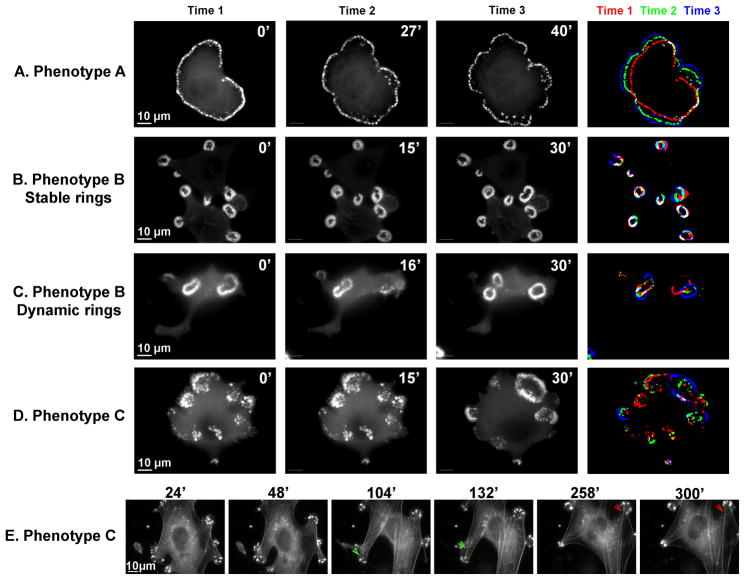
Figure 4. Dynamics of the adhesion structures in the three major phenotypes of MEF-SrcY527F cells.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin (A, B, D), mCherry actin (C) or co-transfected with GFP-cortactin and mCherry actin (E). Time-lapse movies were recorded 24 h later. Frames from the movies, and corresponding times, are shown. An image of the superimposed times is also shown, (A–D) in order to appreciate the dynamics. (E) Stress fibers seem to be anchored to podosomes, but can slide from one podosome to the other, as shown by the pairs of arrowheads.
In Phenotype B, rosette dynamics appears to be dependent on ring size and density. Thus, small and compact rings are relatively stable, maintaining their position and size for more than an hour (Fig. 4B, Supplementary Movie 3), whereas larger rings tend to collapse and reform frequently (Fig. 4C, Supplementary Movie 4). All in all, podosome rings, including those that appear more stable, have very fast internal actin dynamics, indicating that single podosomes are in a constant state of assembly and turnover.
Analysis of time-lapse movies indicates that cells can switch from one phenotype to another within short periods of time (5- to 20 minutes), so that podosome clusters can transform into larger rings (Fig. 4D, Supplementary Movie 5).
In order to analyze the behavior of the stress fibers co-existing with podosomes in “Phenotype C”, we transfected cells with GFP-cortactin and mCherry-actin. Careful examination of the time-lapse movies indicates that the stress fiber-like structures in these cells are anchored to the podosomes. These linkages are, however, unstable, and the anchored stress fibers may detach from one “anchoring podosome” and shift horizontally towards other podosomes (Fig. 4E and Supplementary Movie 6). It seems likely that these “shifts” are driven by contractile forces generated by the associated stress fibers.
In order to visualize the structural organization of the podosomal rings in MEF-SrcY527F, cells were cultured on glass slides, and the cell body was removed by brief exposure to a hypotonic solution, followed by mechanical shearing, leaving attached only the cell ventral membrane, with its adhesive apparatus and the associated cytoskeleton. The samples were then fixed, critical point-dried, coated with 2–4 nm of chromium, and examined by scanning electron microscopy (SEM).
In clusters, individual podosomes, structurally similar to those seen by Luxenburg et al. in osteoclasts (Luxenburg et al., 2007) could be visualized.
The core diameter of these podosomes is, however, smaller, about 200 nm (compared to ~300 nm in osteoclasts; Fig. 5A and B). The rosette structure is about 3–4 μm wide, and filled with dense actin arrays (Fig. 5C). Podosome cores and radial fibers can be observed within this structure (Fig. 5D).
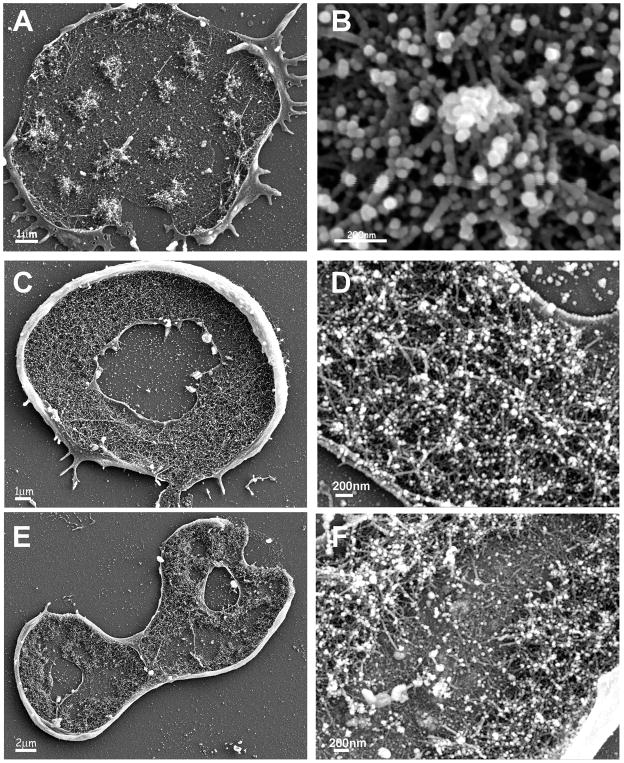
Figure 5. The structures of individual podosomes, podosomes in clusters, and podosome rings, as observed by scanning electron microscopy.
Cell bodies of MEF-SrcY527F cells were removed, leaving the cytoplasmic faces of the attached ventral membrane exposed. The samples were then fixed, and observed in a scanning electron microscope (SEM). (A) Podosome clusters. (B) An individual podosome. (C) Rosette structure (3–4μm wide). (D) Higher magnification reveals that the rosette is built from podosomes. (E) A structure of larger rings, built in part from two actin belts. Each of the actin belts is about 1–2μm wide, with an ~1–2μm gap between them. (F) Higher magnification reveals the double podosome belt.
Using live-cell video microscopy, we found that the larger rings tend to develop into super-structures similar to the osteoclast sealing-zone, and sometimes appear as a paxillin belt surrounded by dense inner and outer actin belts (Fig. 2A) that can be visualized by SEM (Fig. 5E). This structure is built of podosome subunits, and it is evident that the dense actin cores are anchored to the ventral membrane via radial actin fibers (Fig. 5F).
Screening for genes involved in Src-induced remodeling of the actin cytoskeleton and adhesion sites
To identify genes that participate in Src signaling to the cytoskeleton, we conducted an siRNA screen using automated, high-resolution light microscopy. For this purpose, MEF-SrcY527F cells were seeded in 96-well plates, and transfected 24 hours later with three mouse siRNA libraries (“SMARTpools”, Thermo Fisher Scientific; see Materials and Methods). These included libraries targeting 84 tyrosine kinases, 202 phosphatases, and 53 MAPK-related genes (Supplementary Table S1).
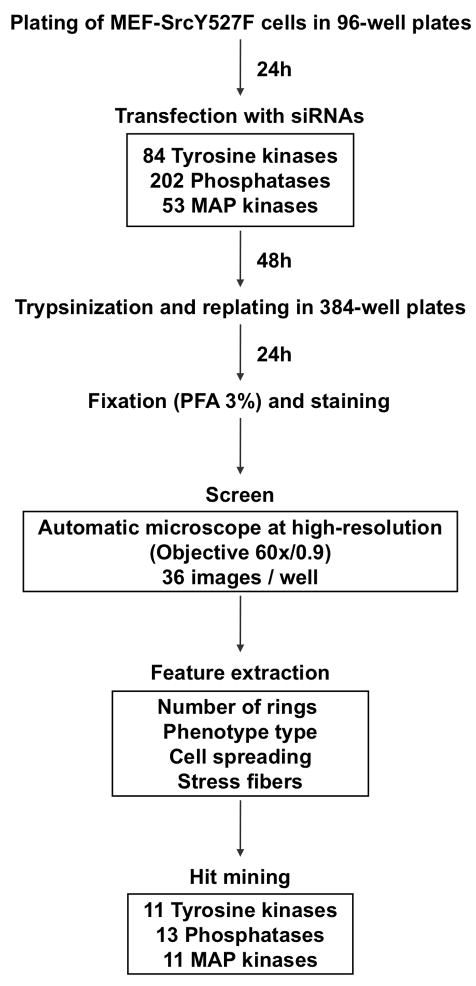
Following transfection, the cells were incubated for 48 hours, and then trypsinized and re-plated onto 384-well plates. Twenty-four hours later, the cells were fixed, stained for actin and DAPI, and screened using a WiScanTM (Idea BioMedical, Israel) automatic microscopy system, equipped with a high-resolution (60×, NA 0.9) objective. To obtain statistically sound datasets, we acquired 36 images per well, and analyzed them as montages to extract morphological information (see screen workflow, depicted in Fig. 6).
Figure 6. Workflow of the screen.
Schematic description of the screen, including the screening procedure, the number of screened siRNAs, the features extracted, and the number of hits.
In order to monitor the knockdown effects, we defined and scored 4 assessable morphological features: number of rings, prominence of stress fibers, dominant phenotype (namely, Phenotypes A–C, as specified above), and cell spreading. Nuclear staining was used to monitor the degree of toxicity, highlight changes in cell proliferation, and determine cell number.
The siRNAs with the most prominent effects were selected as hits (see Materials and Methods for a description of the scoring method).
The final list of hits includes 13 siRNAs from the phosphatase library, 11 from the tyrosine kinase library, and 11 from the MAPK library; all are presented in Table 1 and Figure 7. Searching biological databases for the involvement of our hits in focal adhesion or actin cytoskeleton regulation revealed that 17 of them were previously shown to be involved in these processes (Table 1).
Table 1.
List of the hits and their scores.
| SCORE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Alias | Official Full Name | Num.of rings | Phenotype Type | Spreading | Stress fibers | Phenotype | Focal adhesion | Regultion of actin cytoskeleton | ||
| A | B | C | |||||||||
| Mouse Phosphatases | |||||||||||
| Ppp1cb | 1200010B19 | Protein phosphatase 1, catalytic subunit, beta isoform | 0 | −1 | 0 | 1 | 1 | 2 | more SF, more spread, more C type cells | + | + |
| Ppp1r11 | Tctex5 | Protein phosphatase 1, regulatory (inhibitor) subunit 11 | 0 | −0.5 | 0 | 0.5 | 0.5 | 0.5 | more SF, more spread | ||
| Ppp1r2 | IPP-2 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 | 0 | −0.5 | 0 | 0.5 | 0.5 | 1.5 | more SF, more spread, more C type cells | ||
| Ppm1b | PP2CB | Protein phosphatase 1B, magnesium dependent, beta isoform | −0.5 | −0.5 | −0.5 | 0 | 0.5 | 1 | more SF, a little more spread, less rings | ||
| Ppp3cc | Calnc | Protein phosphatase 3, catalytic subunit, gamma isoform | −0.5 | −1 | −1 | 1 | 1 | 1 | more SF, more spread | ||
| Ptpn23 | PTP-TD14 | Protein tyrosine phosphatase, non-receptor type 23 | 1 | 0 | 1 | 0 | 0 | 0 | more rings | ||
| Ptpn11 | Shp2 | Protein tyrosine phosphatase, non-receptor type 11 (SHP2) | −1 | −2 | −1.5 | 1 | 1 | 1 | more SF, more spread, less rings | + | |
| Ptpru | PTP-lambdaProtein tyrosine phosphatase, receptor type, U | −1 | −1 | −1 | 0.5 | 0.5 | 1 | more SF, less rings | + | ||
| Psph | PSPase | Phosphoserine phosphatase | −1 | −2 | −1.5 | 1.5 | 1 | 1.5 | more SF, less rings, more C type cells | ||
| Inpp5d | SHIP-1 | Inositol polyphosphate-5-phosphatase D | 2 | 1.5 | 2 | −1 | 0 | 0 | more rings | + | |
| Pib5pa | Pib5pa | Phosphatidylinositol (4,5) bisphosphate 5-phosphatase, A | 1 | 1 | 1 | −1 | 0 | 0 | more rings | ||
| D9ertd660e | Gpd1l | Glycerol-3-phosphate dehydrogenase 1-like | 0 | 0 | 0 | 0 | 0 | 1 | more SF in parallel to B rings | ||
| Dusp8 | Nttp1 | dual specificity phosphatase 8 | −0.5 | 1 | −1 | −1 | 1 | 0 | Bigger rings (A type), more spread | ||
| Mouse Tyrosine Kinases | |||||||||||
| Musk | unp | Muscle, skeletal, receptor tyrosine kinase | 0 | 1 | −1.5 | 1.5 | 1 | 0.5 | more SF, more spread | ||
| Tyro3 | Etk-2 | TYRO3 protein tyrosine kinase 3 | 0 | 1 | −1 | 1.5 | 1 | 0.5 | more SF, more spread | + | |
| Flt4 | VEGFR-3 | FMS-like tyrosine kinase 4 | 0 | 1 | −1 | 1 | 0.5 | 0 | Bigger rings (A type) or small rings at the cell periphery | + | |
| Kdr | Flk-1 | kinase insert domain protein receptor | −0.5 | 0.5 | −1 | 0.5 | 0.5 | 0 | less rings , rings mainly in the periphery | + | |
| Ai326477 | Styk1 | serine/threonine/tyrosine kinase 1 | −1 | 1 | −1.5 | 0.5 | 0 | 0 | Bigger rings (A type), less rings | ||
| Fgfr3 | HBGFR | fibroblast growth factor receptor 3 | 0 | 1 | −1 | 0.5 | 0.5 | 0 | Bigger rings (A type), more spread | ||
| Insr | CD220 | insulin receptor | −0.5 | −1.5 | −1 | 1.5 | 1 | 1 | more SF, more spread | + | |
| Csk | AW212630 | c-Src tyrosine kinase | 1 | 0 | 1 | 0 | 0 | 0 | more rings | + | + |
| Ptk7 | CCK4 | PTK7 protein tyrosine kinase 7 | 0 | −1 | −1 | 1.5 | 1.5 | 0.5 | more spread | + | |
| Ros1 | c-ros | Ros1 proto-oncogene | 0 | −1 | −1 | 1.5 | 0.5 | 1 | more SF, more spread | ||
| Met | HGFR | met proto-oncogene | 1 | −1 | 1 | 1.5 | 0 | 1 | more SF, more rings | + | |
| Mouse Mitogen-Activated Protein Kinases (MAPK) | |||||||||||
| Mapk9 | JNK2 | Mitogen activated protein kinase 9 | −0.5 | 1.5 | −1.5 | −1 | 1 | 0 | more spread, bigger rings (A type) | + | |
| Mapk3 | ERK1 | Mitogen activated protein kinase 3 | 0 | −1.5 | 0 | 0.5 | 1 | 1 | more SF, more spread | + | + |
| Map2k1 | MEK1 | Mitogen activated protein kinase kinase 1 | 0 | 1.5 | −1 | −1 | 0 | 0 | Bigger rings (A type), double rings | + | + |
| Map2k2 | MEK2 | Mitogen activated protein kinase kinase 2 | 0.5 | 1 | −1 | −1 | 0 | 0 | Bigger rings (A type), more rings | + | |
| Map2k3 | MEK3 | Mitogen activated protein kinase kinase 3 | 0 | −1.5 | −1.5 | 2 | 0 | 0.5 | smaller rings at the cell periphery (C type) | ||
| Map2k4 | JNKK | Mitogen activated protein kinase kinase 4 | 0.5 | 1.5 | −1 | −1.5 | 1 | 0 | Bigger rings (A type), double rings, more spread | ||
| Map3k7 | TAK1 | Mitogen activated protein kinase kinase kinase 7 | 0.5 | −2 | −1.5 | 2 | 0.5 | 1 | more rings, small rings, more SF | ||
| Map3k10 | Mlk2 | mitogen activated protein kinase kinase kinase 10 | 0 | 1 | −0.5 | −1 | 0.5 | 0 | Bigger rings (A type), more spread | ||
| Map3k14 | Nik | mitogen-activated protein kinase kinase kinase 14 | −1 | 1.5 | −1.5 | −2 | 1.5 | 0 | Bigger rings (A type), more spread | ||
| Pak1 | Paka | p21 (CDKN1A)-activated kinase 1 | −0.5 | 1.5 | −1.5 | −2 | 2 | 0 | very big rings (A type), very spread | + | + |
| Pak2 | PAK-2 | p21 (CDKN1A)-activated kinase 2 | 0.5 | −2 | −1.5 | 1.5 | 0 | 0 | smaller rings in the periphery of the cell (C type) | + | + |
The list includes all of the hits, and their scores, including number of rings, phenotype type, degree of spreading, and stress fibers. For a detailed explanation of the scoring, see “Materials and methods”. A verbal description of the phenotype is also included. The standard gene symbol is indicated alongside a commonly used alias (from the Entrez gene database, NCBI). A description of the involvement of the gene hits in focal adhesion and in the actin cytoskeleton based on the KEGG database (Pathway)) (Kanehisa and Goto, 2000), Gene Ontology (biological process) and the Adhesome (Zaidel-Bar et al., 2007a) is also presented.
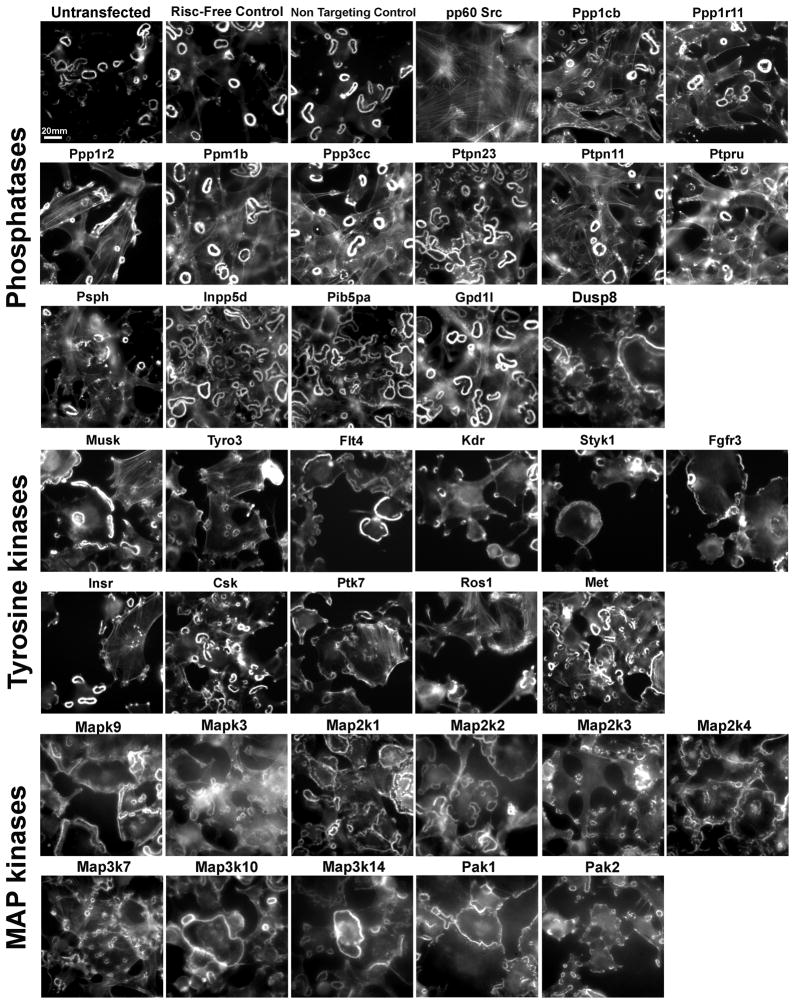
Figure 7. Images of the screen hits.
Single high–resolution, representative images of all the screen hits, stained for actin. The first four images were taken from controls (untransfected, Risc-Free, and non-targeting controls, and Src siRNA), and are shown for comparison.
Our findings reveal several prominent effects that can be divided into three major groups: 1. Restoration of stress fibers in addition to podosome rings (Fig. 7; e.g., Ppp1r2 and Ptpn11); 2. Induction of an increased number of rings per cell (Fig. 7; e.g., Inpp5d and Pib5pa); and 3. A shift in podosome organization characterized by one large ring at the cell periphery, resembling the sealing zone of osteoclasts (Fig. 7; e.g., Map2k4, Map2k14, Pak1).
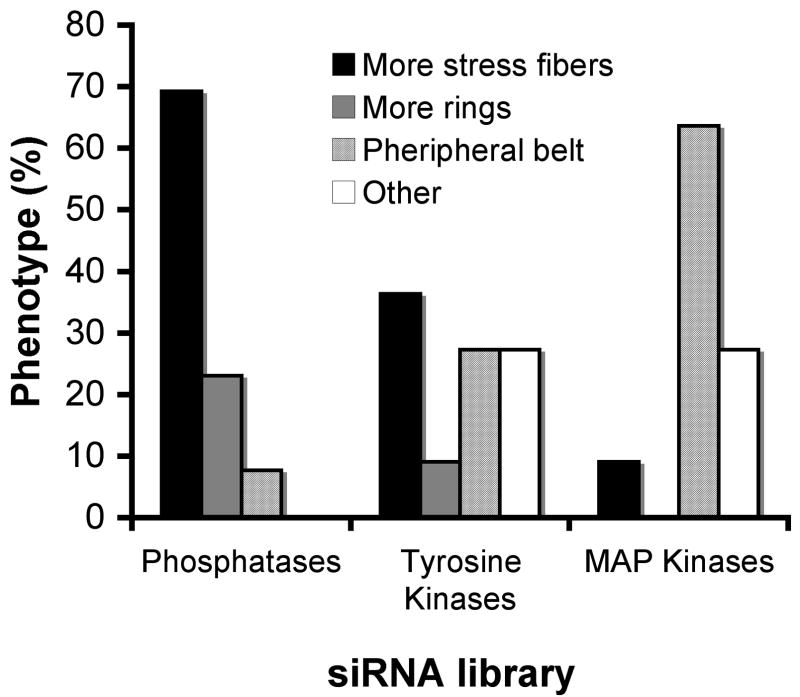
It is noteworthy that these major effects were not uniformly distributed among the three libraries. While most of the phosphatase hits (70%) caused restoration of stress fibers, most of the MAPK hits (64%) induced the peripheral belt phenotype, and only one MAPK, Mapk3, induced prominent reformation of stress fibers. The hits from the tyrosine kinase library induced variable, often conflicting, effects on the transfected cells. While knockdown of Musk, Tyro3, Insr, Ros1 and Met resulted in increased stress fiber formation, knockdown of Flt4, Styk1 and Fgfr3 resulted in increased peripheral belt formation (Fig. 8).
Figure 8. Distribution of the major effects in the three libraries.
The graph shows the percentage of the three major effects (more stress fibers, more rings, and a peripheral belt), of the siRNAs, in each of the three-siRNA libraries.
Src knockdown in osteoclasts impairs sealing zone formation, but does not result in the formation of stress fibers
To extend these findings and determine how modulation of the Src pathway affects podosome formation in a physiological setting, we carried out a preliminary screen for the effects of knockdown of selected hits on sealing zone formation in osteoclasts.
As one might expect, a most prominent phenotype was obtained following Src knockdown. Thus, RAW 264.7 cells infected with lentivirus, expressing Src shRNA (Fig. 9A), and induced to differentiate by treatment with RANK ligand and M-CSF, formed only small actin rings that were few in number, compared with the large rings seen in control RAW 264.7 cells. However, stress fibers did not appear in the knockdown cells, as was the case in fibroblasts.
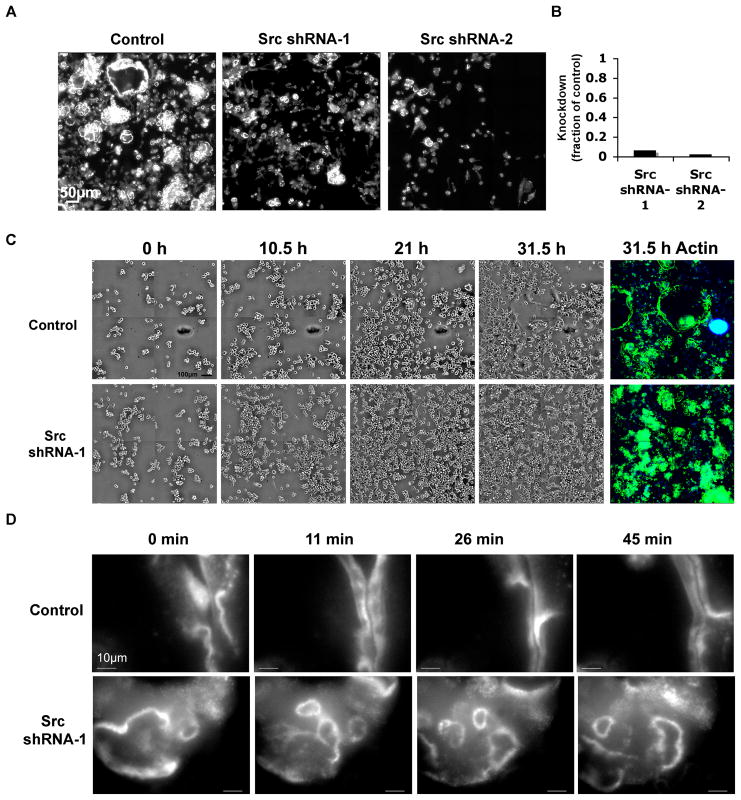
Figure 9. Effects of Src knockdown in RAW 264.7 osteoclasts.
(A) RAW 264.7 cells were infected with lentiviral vectors expressing shRNAs targeting Src, or treated with control media. The cells were allowed to differentiate for 72 h, and stained for actin. The image is a montage of 36 single images from the same well. (B) The graph shows the knockdown level of Src mRNA (the fraction of expression, compared to the non-targeted control), analyzed by RT-PCR in RAW 264.7 stable cell lines. The RNA amount was normalized to HPRT. Quantitative PCR was performed with the following primers: Src forward, 5’-GGTGCCTACTGCCTCTCTGTA-3’; Src reverse: 5’-GCGGGAGGTGATGTAGAAACC-3’; HPRT forward: 5′ CTGGTTAAGCAGTACAGCCCCAAA 3′;HPRT reverse 5′ TGGCCTGTATCCAACACTTCGAGA 3′. (C) Phase contrast, time-lapse movies of an Src shRNA-1 stable cell line, and of control cells, were produced 24 h after differentiation began. The figure shows 4 images of the 16-image montage taken at each time point. Four time points are shown, as indicated. At the end of the movie, the cells were stained with phalloidin-FITC (green) and DAPI (blue); the same field is shown for the stained image. (D) Fluorescent time-lapse movies of a differentiated Src shRNA-1 stable cell line, and of control cells, were produced at high magnification (100× /1.3). Four time points are shown; only a fraction of the cells is visible. Several unstable rings are seen in Src shRNA-1 cells; whereas two adjacent sealing zone-like rings are partially seen in control cells.
For an in-depth analysis of the effect of Src knockdown (KD) in osteoclasts, we established a stable RAW 264.7 cell line by selecting the infected cells with puromycin. Real-time PCR showed a drastic decrease (98%) in Src mRNA levels in stable cell lines derived from two distinct shRNA sequences, as compared to control cells (Fig. 9B).
Phase-contrast, time-lapse movies recording RAW 264.7 differentiation following induction with RANK ligand and M-CSF, showed that Src KD cells display reduced spreading compared to control cells, and fuse at considerably lower rates (Fig. 9C, Supplementary Movies 7 and 8). The cells were, however, very motile and protrusive before, during and after differentiation. Staining of the Src-KD cells with phalloidin-FITC at the end of the phase contrast movie (Fig. 9C: 35 hours, “actin”) indicated impaired peripheral ring formation following Src knockdown. Src knockdown effects may be due to effects in osteoclastogenesis that may lead to impaired cell fusion and spreading. Nevertheless Src also affects actin ring formation in the osteoclasts that were able to mature.
In order to analyze actin dynamics, Src KD and control cells were transfected with GFP-lifeact (Riedl et al., 2008), 48 hours after differentiation was induced. Time-lapse movies of the cells showed that even when Src KD cells were imaged at high magnification (100×/1.3), no stress fibers were seen. The cells did not form stable, actin-containing sealing zones; moreover, the small and unstable rings found in these cells generally did not fuse, but rather formed and expanded centrifugally, and then rapidly split into smaller rings, or disintegrated into scattered podosomes (Fig. 9D; Supplemental Movies 9 and 10).
These findings suggest that Src plays a central role in regulating podosome formation and reorganization in both types of cells: those that are physiologically programmed to form podosomes (e.g., osteoclasts), and those that normally organize their adhesion and cytoskeletal structures in the form of focal adhesions and stress fibers (e.g., fibroblasts). It is, however, notable that the formation of stress fibers is a much more cell-type restricted process. Thus, suppression of deregulated Src expression in fibroblasts restores stress fiber formation, while similar suppression in osteoclasts does not, though it does affect ring formation. Our ongoing characterization of the effects of other siRNAs hits, identified in our MEF-Src Y527F screen, on osteoclasts, is conducted along those lines; namely, distinguishing between regulators of podosome formation, and genes involved in the stress fiber/podosome switch.
Discussion
In this study, we addressed the role of Src in the regulation of the actin cytoskeleton in two different cellular systems, fibroblasts and osteoclasts.
The choice of these particular cells was motivated by the fact they display radically different forms of actin organization, and that Src is involved, somehow, in the transition from one form to the other. Specifically, MEFs display “classical” stress fibers and focal adhesions, which transform into podosomes upon overexpression of deregulated Src, and osteoclasts form podosomes as their standard cytoskeletal/adhesion structure, and lose these structures upon knockout of Src, or inhibition of its enzymatic activity (Destaing et al., 2008; Lakkakorpi et al., 2001). These well-documented observations left two major mechanistic questions unanswered: What are the proteins acting upstream or downstream of Src that are essential for the src-dependent cytoskeletal transformation? Moreover, are stress fibers and podosome readily inter-convertible actin structures, switched on and off by Src-mediated phosphorylation? The results described herein revealed several molecules that participate in Src-mediated reorganization of the cytoskeleton, and indicated that while excessive Src activity can induce podosome formation in fibroblasts, Src inhibition greatly affects podosomes in osteoclasts, but does not induce stress fiber formation.
Furthermore, we show that the podosome phenotype induced by over-expression of deregulated Src is not homogeneous, and conspicuous differences in ring size and location can be observed. As also shown in BHK cells (Badowski et al., 2008), podosomes induced by active Src behave very similarly to podosomes in osteoclasts (though with slower dynamics), suggesting that this system could be used to analyze the molecular pathways regulating podosome behavior in other cell types.
Although the molecular mechanisms by which active Src switches on podosome formation are only now being clarified (Oikawa et al., 2008), many open questions concerning Src regulation of the actin cytoskeleton remain to be addressed. Are the dynamic properties seen in Src-induced podosomes in transformed cells inherent to podosome structure and function? Are these properties shared by podosomes formed physiologically, in other cell types? What is the molecular mechanism that regulates stress fiber disassembly? Is it the change in the type of cell adhesion to the ECM that leads to stress fiber disassembly, or does some underlying mechanism actively regulate this process?
In this study, we show that the organization and dynamics of single podosomes and podosome rings and their ultrastructure, as observed by SEM, are very similar to corresponding structures in osteoclasts (Luxenburg et al., 2007). Clusters of peripheral adhesions, for example, can evolve into rosettes, including those that are very dynamic and fuse into larger rings that may then split again, and those that are quite stable. The belt structure was also seen to expand dynamically, thereby segregating actin and plaque proteins, a behavior also seen in osteoclasts (Luxenburg et al., 2006a).
In order to uncover new molecules and pathways involved in Src-induced actin remodeling, we performed an siRNA screen, targeting phosphatases, tyrosine kinases and MAPK. Thus far, we have identified 35 potential hits whose suppression leads to apparent changes in cell phenotype. Notably, targeting of phosphatases mainly led to the restoration of stress fibers, while targeting MAPK enhanced the Src-induced phenotype, resulting in enrichment of peripheral belts. Tyrosine kinase knockdown phenotypes were heterogeneous, and seemed to be dependent on the specific pathway regulated by the particular kinase. Of the 35 hits found, 18 of them were not previously shown to be involved in focal adhesion or actin cytoskeleton regulation (Table 1). Comparison of these hits with those of a previous screen, performed to clarify the effects of siRNA knockdown on focal adhesion formation (Winograd-Katz et al., 2009) revealed quite different sets of hits. This suggests that the pathways downstream of Src, leading to the induction of podosomes, differ from those regulating focal adhesions.
One interesting finding in our screen was the enrichment of stress fibers that co-exist with podosome rings, indicating that these two actin structures are not mutually exclusive. This phenotype was seen in Met and Map3k7 knockdowns, which resulted in the formation of additional stress fibers, together with more rings. Moreover, the co-existence of stress fibers with B type rings was also observed in many other gene knockdowns [e.g., Ppp1cb, Gpd1l, Ppp1r11, Ppp1r2, and Mapk3 (ERK1)].
These results suggest the existence of two different pathways downstream of Src, one regulating podosome assembly, organization, and turnover, and the other regulating stress fiber disassembly. Many of the hit phosphatases seem to inhibit this second pathway to a greater extent than their inhibition of Src-induced podosome ring formation. Indeed, Shp2 (PTPN11) was shown to mediate v-Src-induced morphological changes: thus, Shp2 −/− cells transformed with v-Src retain stress fibers and do not form excessive podosomes, as did Shp2 +/+ cells (Hakak et al., 2000). Shp2, known to be involved in ERK activation (Bennett et al., 1994), also led to the “more stress fibers” phenotype in our screen, indicating that ERK (ERK1) activation through Shp2 could be involved in the pathway leading to Src-mediated disassembly of stress fibers.
It has been suggested that active Src-induced disassembly of stress fibers is caused by inactivation of cytoplasmic Rho (Frame et al., 2002). This hypothesis has been supported by the observation that overexpression of constitutively active Rho in Src-transformed fibroblasts lead to the restoration of actin stress fibers (Mayer et al., 1999). Nevertheless, active Rho is localized to podosomes in Src-transformed cells, and is required for their assembly and function (Berdeaux et al., 2004). Therefore, the two different pathways we propose, one of stress fiber disassembly and the other of podosome assembly, could be aimed at the inactivation of cytoplasmic Rho on the one hand, and its recruitment and activation in podosomes, on the other.
The inhibition of Src in MEF-SrcY527F cells resulted in the disassembly of podosomes and assembly of stress fibers, indicating that Src is responsible for the two processes. Is Src responsible for these processes in all cells? In order to determine how modulation of the Src pathway affects cytoskeletal remodeling in other cell systems, we analyzed the effect effects of knockdown of a few of the siRNA screen hits in RAW 264.7 osteoclasts. Knockdown of some of the hits resulted in phenotypes, largely similar to those seen in MEF-SrcY527F cells, though the phenotypes differed in other ways; for example, no stress fibers were ever observed in osteoclasts. Further studies, aimed at analyzing the phenotypes obtained, are currently underway.
As a proof of principle, we first characterized the Src KD phenotype in osteoclasts. These KD cells displayed major effect on cell fusion, podosome dynamics and ring assembly, yet they did not form stress fibers. These findings suggest that the molecular composition of the adhesion machinery, specific to each cell type, determines its distinct organization. Nevertheless, the nature of the mechanisms regulating podosome formation in fibroblasts and osteoclasts may share some similarities.
Previous studies have demonstrated the effects of some of our hits on podosomes on Src-transformed fibroblasts; for example, inhibition of MEK in in RSV transformed BHK cells (by U0126 inhibitor) resulted in thicker podosome rings, due inhibition of podosome turnover (Badowski et al., 2008). Similarly, in our screen, knockdown of MEK1 and MEK2 resulted in larger rings, and even double rings (in the case of MEK1, see figure 7). It is interesting to note that the effects of MEK1 and MEK2 knockdown are very different from the effects of ERK1 knockdown, (which leads to more stress fibers) suggesting that other pathways, besides those involved in the phosphorylation of ERK1, are regulated by MEK1 and MEK2.
Another example is PAK1 that was shown to regulate caldesmon and podosome formation in Rous sarcoma virus- transformed fibroblasts (Morita et al., 2007). Caldesmon was also shown to be an integral component of podosomes in smooth muscle cells (Eves et al., 2006), providing further evidence that many regulators of podosomes in fibroblasts could be relevant for other podosome-forming cells.
Previous work showed that Src−/− osteoclasts cannot organize podosomes into rings (Destaing et al., 2008); moreover, Sanjay et al. (Sanjay et al., 2001) showed that these osteoclasts display focal adhesion-like adhesions. The apparent discrepancy between those results may be due to differences in the experimental systems. Very low amounts of Src are still expressed in our shRNA-treated cells, whereas Src −/− cells were obtained from Src knockout mice.
Another apparent discrepancy between our results and the effects of kinase- defective Src described previously (Luxenburg et al., 2006b; Destaing et al., 2008) could be explained by the fact that our cells express higher levels of other Src kinase family members contributing to the formation of podosome rings, while the effect of kinase-defective Src may include inhibition of any remaining activity of Src and Src-related proteins, thereby completely inhibiting both ring formation and podosome turnover.
Apart from those discrepancies, the Src KD phenotype in RAW 264.7 cells is similar to that described in previous reports involving Src anti-sense (Kumagai et al., 2004). In this work, Src anti-sense in RAW 264.7 cells led to little multi-nucleated cell formation, and weak mineral-hydrolyzing activity.
The inducible remodeling of the actin cytoskeleton from stress fibers into podosomes was also shown in smooth muscle cells after activation of PKC by phorbol esters (Hai et al., 2002; Lener et al., 2006) as well as in endothelial cells, following activation with diverse cytokines (Moreau et al., 2003; Billottet et al., 2008). In these cells, stress fiber disassembly seems to be coupled to podosome formation, similar to that seen in Src-transformed fibroblasts. It would be interesting to examine whether these functionally associated pathways could also be uncoupled in endothelial or smooth muscle cells, by means of siRNA perturbation. We cannot rule out the possibility that the unregulatable form of Src causes cells to be predisposed to pathway uncoupling; nevertheless, our system enabled us to reveal molecules involved in each of the aforementioned pathways, which may also play a role in other cells.
Taken together, the results of our study clarify the involvement of Src in the remodeling of the actin cytoskeleton, suggesting the existence of an Src-regulated pathway of stress fiber disassembly, and highlight candidate genes that may regulate actin remodeling downstream of Src.
Materials and Methods
Cell culture
Mouse embryonic fibroblasts (MEF) were retrovirally infected with SrcY527F in pBabe vector (a kind gift of A. Elson, Weizmann Institute of Science), with pBabeYFP used as a control. Cells expressing the constructs were selected with puromycin (Sigma-Aldrich, Rehovot, Israel). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal calf serum (Hyclone, Logan, UT, USA), 100 units/ml penicillin, and 100 μg/ml streptomycin, at 37° C in a 5% CO2 humidified incubator. SU6656 was obtained from Calbiochem (San Diego, CA, USA).
RAW 264.7 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). To induce osteoclast differentiation, 100 cells/mm2 were grown at 37°C in a 5% CO2 humidified atmosphere for 3 days in alpha MEM with Earle’s salts, L-glutamine and NaHCO3 (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and antibiotics (Biological Industries, Beit Haemek, Israel), 20ng/ml recombinant soluble receptor activator of NF kappa B ligand (RANK-L), and 20ng/ml macrophage colony-stimulating factor (mCSF) (R&D, Minneapolis, MN, USA).
Immunofluorescence staining and imaging
For staining, cells were seeded on non-coated cover slips. After 24 h, the cells were fixed and permeabilized for 2 min in 3% warm paraformaldehyde (PFA) (Merck, Darmstadt, Germany) + 0.5% Triton ×100 (Sigma-Aldrich), and with 3% PFA alone for an additional 30 min. After fixation, cells were washed 3 times with PBS (pH 7.4) and incubated with primary antibodies for 45 min. Cells were then washed with PBS and incubated for an additional 45 min with secondary antibodies. Primary antibodies in this study included: mAb anti-phospho-tyrosine (4G10; Upstate Biotechnology, Charlottesville, VA, USA) and mAb anti-paxillin, (Transduction Laboratories, Lexington, KY, USA). Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). Phalloidin-coumarin, phalloidin-FITC and DAPI were supplied by Sigma-Aldrich.
Immunofluorescent images were taken with a DeltaVision system (Applied Precision, Inc., Issaquah, WA, USA), consisting of an inverted microscope IX70 (Olympus, Tokyo, Japan) equipped with a temperature-controlled box (Life Imaging Services, Basel, Switzerland; www.lis.ch). Image processing and analysis were performed using the Priism software package (Applied Precision, Inc.). Fluorescent images were high pass-filtered (by subtracting from each pixel the average intensity in a 20×20 pixel box around it). Filtered images were used to produce pixel-by-pixel ratios, as previously described (Zamir et al., 1999).
DNA constructs, transfection, and time-lapse movies
The plasmids used for time-lapse movies included: GFP-cortactin (Kaksonen et al., 2000), GFP-paxillin (Zamir et al., 2000), GFP-α-actinin, kindly provided by Carol A. Otey (University of North Carolina, Chapel Hill, NC, USA), mCherry-actin, a generous gift from J. V. Small (Institute of Molecular Biology, Austrian Academy of Sciences, Salzburg, Austria), and Lifeact (Riedl et al., 2008), kindly provided by R. Fässler (Max Planck Institute of Biochemistry, Munich, Germany).
For the fluorescence dynamics movies, MEF-SrcY527F cells were seeded one day before transfection. The next day, transient transfection was performed using Lipofectamine 2000 (Invitrogen); 5 h later, cells were trypsinized and seeded on 35 mm glass-bottomed dishes. Cells were allowed to grow and express the plasmids and, 24 h after transfection, the medium was replaced with DMEM containing HEPES (25mM) without Phenol Red, and riboflavin solution (Biological Industries) supplemented with 10% FCS. Fluorescent time-lapse movies were acquired with a 100 X/1.3 objective. Cells were maintained on the microscope stage in a 37°C-heated chamber.
For time-lapse movies of RAW 264.7cells, cells were seeded in 384-well plates (Greiner Bio One GmbH, Fricjkenhausen, Germany; F-bottomed, mClear, black, tissue-culture-treated), and induced to differentiate for 48 h. Cells were then transfected with Fugene HD (Roche); 24 h after transfection, the medium was replaced by MEM alpha without Phenol Red (GIBCO) and supplemented with 10% FBS, 20ng/ml recombinant soluble receptor activator of NF kappa B ligand (RANK-L), and 20ng/ml macrophage colony-stimulating factor (mCSF) (R&D). Cells were kept at 37°C in a humidified atmosphere of 5% CO2, and fluorescent time-lapse movies were acquired with a 100 X/1.3 objective.
Fluorescent frames were high pass-filtered and used to produce pixel-by-pixel time ratio images (Zamir et al., 1999). Auto-correlation analyses were carried out as previously described (Zaidel-Bar et al., 2007b).
For phase-contrast time-lapse movies, RAW 264.7 cells were seeded in 12-well plates (MatTec Cultureware, Ashland, MA, USA) and induced to differentiate at 37°C in a humidified atmosphere of 5% CO2, for 24 h. Time-lapse movies were acquired with a 20×/0.45 objective in the automated microscope system described below.
Immunoblotting
Cell extracts from MEF and MEF-SrcY527F cells were subjected to BCA kit analysis for protein quantification (Pierce, Thermo Scientific), and equal amounts of protein from each sample were resolved by SDS-PAGE and analyzed by Western blot. Anti-GAPDH (Ambion, Inc) was used as a loading control. The following antibodies were used: mAb anti-phospho-tyrosine (4G10; Upstate Biotechnology), mAb anti-v-Src (Calbiochem), rabbit anti-Src pY418 (Biosource International, Camarillo, CA, USA).
Ventral membrane preparation for scanning electron microscopy (SEM)
The cell body was removed by brief exposure to a hypotonic solution followed by fluid shear flow, leaving the cytoplasmic face of the attached ventral membrane exposed (Luxenburg et al., 2007).
Following ventral membrane preparation, cells were immediately fixed with warm 2% gluteraldehyde (GA) (EMS, Hatfield, PA, USA), and rinsed with PBS for 30 min. Cells were then washed 3 times for 5 min each in PBS, and twice with cacodylate buffer (0.1M CaCO, 5Mm CaCl pH 7.3) (Merck), post-fixed with 1% OsO4 (EMS, Hatfield, PA, USA) for 45 min, and washed 3 times in cacodylate buffer, and then twice with H2O. The preparations were then incubated with 1% tannic acid (Merck) for 5 min, washed 3 times with H2O, incubated with 1% uranyl acetate (EMS) for 30 min, and washed 3 times with H2O. Dehydration in increasing concentrations of reagent-grade ethanol (2 × 5 min for 25%, 50%, 70%, and 95%, and 2 × 10 min for 100%) was followed by critical point drying using CPD30 (BAL TEC, Blazers, Lichtenstein), coated with 2 nm Cr using K575X (Emitech, Ltd., Kent, UK). The samples were visualized in a high-resolution SEM, Model Ultra 55 (Zeiss, Oberkochen, Germany).
SiRNA libraries
The screen was performed with Dharmacon SMARTpool technology, consisting of mixtures of 4 different siRNA sequences targeting the same gene. Three Mouse siARRAY® siRNA Libraries (Thermo Fisher Scientific, (http://www.dharmacon.com/HomePage.aspx) were screened: a phosphatase library containing 202 phosphatases, a tyrosine kinase library containing 84 kinases, and a MAPK library containing 53 genes involved in the MAPK signaling cascade.
Screening protocol
MEF-SrcY527F cells (2,500 cells/well) were plated in 100μl DMEM supplemented with 10% FCS (Hyclone Logan, UT, USA) in 96-well plates, and cultured for 24 h at 37ºC in a 5% CO2 humidified atmosphere. The next day, the medium was replaced by 80μl of fresh DMEM supplemented with 10% FCS. Cells were then transfected by direct addition of 20μl transfection mixture (5μl siRNA (1μM) in 5μl optiMEM with 0.7μl DharmaFECT4 in 9.3μl OptiMEM). Transfections were performed in duplicates (50nM final siRNA concentrations). The cells were incubated for 48 h and then trypsinized and replated in 384-well plates (Greiner Bio One GmbH; F-bottom, mClear, black, tissue culture-treated). Cells were fixed and stained for actin (phalloidin) and nuclei (DAPI) 24 h later.
Plates containing fixed cells in PBS were sealed with parafilm, and placed on the microscope stage for screening. Only plates with a transfection efficiency of better that 85% [as measured by siGLO (a fluorescent siRNA) and by mouse PLK1 siRNA [which induces cell death (LaPan et al., 2008)], were taken for analysis (Supplementary Fig. 4). The knockdown appeared to be very efficient, as demonstrated by MEF-SrcY527F cells that were transfected with Src siRNA, and regained the normal MEF phenotype (Supplementary Fig. 4).
The siRNA screen was performed in duplicates, at final SMARTpool concentrations of 50 nM. We compared the siRNA effects in treated cells to several controls (siCONTROL RISC-Free siRNA, siGLO Red Transfection Indicator, and siCONTROL Non-Targeting #2), all cultured in the same plate (Supplementary Fig. 4).
In order to define the knockdown effects, we visually scored 4 assessable morphological features: number of rings, prominence of stress fibers, dominant phenotype (namely, phenotypes A–C, as specified above) and cell spreading. Nuclear staining was used to monitor the degree of toxicity, highlight changes in cell proliferation, and determine cell number.
The score scale was from -2 to 2. A score of zero represents the control phenotype, including the typical distribution among the phenotypes: A- 5%, B- 80%, and C- 15% in unperturbed cells. A higher score (e.g., 1 or 2) indicates more cells with this phenotype (or more rings or spreading of the cells), and a lower score (e.g., -1 or -2) represents fewer cells with this phenotype (or less rings or spreading of the cells).
Automated microscopy for screening
The automated microscopy system used here was a prototype of the WiScanTM system (Idea Bio-Medical, Rehovot, Israel), based on an IX81 microscope (Olympus, Tokyo, Japan). Automation was provided by ProScan (Prior, Cambridge, UK), and included an XY stage, focus, shutters, and excitation and emission filter wheels. A fast laser AutoFocus attachment (Liron et al., 2006) was applied, to focus the objective before each image was acquired. The CCD camera is a Quantix 57 (Photometrics, Tucson, AZ, USA). The system is controlled by Resolve6D software running on the RedHat Linux operating system (Paran et al., 2006).
Lentivirus transduction and preparation of shRNA expressing stable cell lines
Mission shRNAs were purchased from Sigma-Aldrich, and the lentiviruses were produced, according to the manufacturer’s protocol. Stable cell lines constitutively expressing shRNAs were obtained by transducing RAW 264.7 cells with lentivirus expressing shRNAs, followed by selection with 2 μg/ml puromycin (Sigma-Aldrich). The Src1 sequence (clone NM_009271.1-476s1c1; Sigma) was:
CCGGGCAAGATCACTAGACGGGAATtroCTCGAGATTCCCGTCTAGTGAT CTTGCTTTTT, and the Src2 sequence (clone NM_009271.1-612s1c1; Sigma) was:
CCGGCCTAAATGTGAAACACTACAACTCGAGTTGTAGTGTTTCACATTTAGGTTTTT.
Supplementary Material
Cell extracts from MEF and MEF-SrcY527F cells were subjected to BCA kit analysis for protein quantification, and equal amounts of protein were analyzed by Western blot with anti-Src, anti-pY418-Src, and anti-general phospho-tyrosine antibody. GAPDH was used as a loading control.
MEF-SrcY527F cells were transiently co-transfected with GFP-paxillin and mCherry actin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 30 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Supplementary Figure 3. While the paxillin (green) is relatively static, there is a constant flow of actin (red); the rings position and size remain stable.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 40 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4. A stable, expanding ring at the periphery of the cell may be observed; the ring expands as the cell spreads.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 30 min. The rings maintain their size and location throughout the movie, but the internal dynamics is high. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were transiently transfected with GFP-actin, and 24 h later, recorded by time-lapse video microscopy at 2-min intervals for 30 min. The rings seen are extremely dynamic and constantly form, merge, split, and collapse. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 30 min. Clusters of peripheral, as well as central, podosomes, are seen. Podosomes can develop into an expanding ring. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were transiently co-transfected with GFP-cortactin and mCherry actin, and 24 h later, recorded by time-lapse video microscopy at 2-min intervals for 5 h. The actin (red) marks the stress fibers as well as the podosomes, while cortactin (green) marks only the podosomes. It appears that the stress fibers are anchored to the podosomes. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were permeabilized, fixed and stained for actin and paxillin. A spatial ratio image shows that the structure of a single podosome consists of two domains: an F-actin-rich core, surrounded by a paxillin-rich ring domain.
A time-lapse movie of MEF-SrcY527F cells transiently co-transfected with GFP-paxillin and mCherry actin was used to create temporal ratio images and auto-correlation analysis. The ratio between the two indicated time points is color-coded: new pixels are blue, pixels that disappeared are red, and unchanged pixels are represented by variable colors, depending on the specific local intensity ratio. Yellow indicates identical intensities in the two time points. Note the high predominance of blue and red in the actin labeling, as compared to the paxillin labeling. The auto-correlation graph was calculated by measuring the average correlation in intensity between frames taken at increasing intervals, from 0 sec to 300 sec. Actin decay occurs faster, indicating that actin is more dynamic than paxillin.
Representative montages of 6 × 6 images from untransfected MEF-SrcY527F cells, or MEF-SrcY527F cells transfected with control siRNAs, as indicated. (A) Montages of DAPI and actin (phalloidin) staining are shown, and a high-resolution image from the actin-stained montage is shown for each control. Bars: (montages) 50 μm; (high-resolution images) 10 μm.
For each siRNA, its library, target gene symbol, RefSeq (NCBI) accession number, gene name, Entrez gene ID, and a link to Gene (NCBI) are shown.
A phase-contrast, time-lapse movie of RAW 264.7 control cells was produced 24 h after differentiation began. The movie was recorded at 15 min intervals for 31.5 h. Each time, 16 images were taken from the well, and stitched together to form a montage. The display rate is 10 frames/sec. Bar: 20 μm. Images from this movie are shown in Figure 9.
A phase-contrast, time-lapse movie of RAW 264.7 Src shRNA-1 cells was produced, 24 h after differentiation began. The movie was recorded at 15 min intervals for 31.5 h. Each time, 16 images were taken from the well, and stitched together to form a montage. The display rate is 10 frames/sec. Bar: 20 μm. Images from this movie are shown in Figure 9.
RAW 264.7 control cells were transfected with lifeact, 48 h after differentiation began. The movie was recorded 30 h later, at 45 sec intervals, for 45 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 9.
RAW 264.7 Src shRNA-1 cells were transfected with lifeact, 48 h after differentiation began. The movie was recorded 30 h later, at 45 sec intervals, for 45 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 9.
Acknowledgments
We wish to thank Dafna Geblinger for help with the scanning electron microscopy, and Barbara Morgenstern for editorial assistance.
BG holds the Erwin Neter Professorial Chair in Cell and Tumor Biology. This study was supported by grants from the National Institutes of Health, Cell Migration Consortium (Grant U54 GM64346), the T3Net project of the EU-FP7, the Volkswagen Foundation, and the Jeanne and Joseph Nissim Foundation for Life Sciences Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci. 2009;122:3037–3049. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badowski C, Pawlak G, Grichine A, Chabadel A, Oddou C, Jurdic P, Pfaff M, Albiges-Rizo C, Block MR. Paxillin phosphorylation controls invadopodia/podosomes spatiotemporal organization. Mol Biol Cell. 2008;19:633–645. doi: 10.1091/mbc.E06-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci U S A. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdeaux RL, Diaz B, Kim L, Martin GS. Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J Cell Biol. 2004;166:317–323. doi: 10.1083/jcb.200312168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrier AL, Yamada KM. Cell-matrix adhesion. J Cell Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- Billottet C, Rottiers P, Tatin F, Varon C, Reuzeau E, Maitre JL, Saltel F, Moreau V, Genot E. Regulatory signals for endothelial podosome formation. Eur J Cell Biol. 2008;87:543–554. doi: 10.1016/j.ejcb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Block MR, Badowski C, Millon-Fremillon A, Bouvard D, Bouin AP, Faurobert E, Gerber-Scokaert D, Planus E, Albiges-Rizo C. Podosome-type adhesions and focal adhesions, so alike yet so different. Eur J Cell Biol. 2008;87:491–506. doi: 10.1016/j.ejcb.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Burns S, Thrasher AJ, Blundell MP, Machesky L, Jones GE. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood. 2001;98:1142–1149. doi: 10.1182/blood.v98.4.1142. [DOI] [PubMed] [Google Scholar]
- Calle Y, Jones GE, Jagger C, Fuller K, Blundell MP, Chow J, Chambers T, Thrasher AJ. WASp deficiency in mice results in failure to form osteoclast sealing zones and defects in bone resorption. Blood. 2004;103:3552–3561. doi: 10.1182/blood-2003-04-1259. [DOI] [PubMed] [Google Scholar]
- Campbell ID. Studies of focal adhesion assembly. Biochem Soc Trans. 2008;36:263–266. doi: 10.1042/BST0360263. [DOI] [PubMed] [Google Scholar]
- Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J Cell Biol. 2000;148:665–678. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Sanjay A, Itzstein C, Horne WC, Toomre D, De Camilli P, Baron R. The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol Biol Cell. 2008;19:394–404. doi: 10.1091/mbc.E07-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong LT, Nakamura I, Lakkakorpi PT, Lipfert L, Bett AJ, Rodan GA. Inhibition of osteoclast function by adenovirus expressing antisense protein-tyrosine kinase 2. J Biol Chem. 2001;276:7484–7492. doi: 10.1074/jbc.M008368200. [DOI] [PubMed] [Google Scholar]
- Eves R, Webb BA, Zhou S, Mak AS. Caldesmon is an integral component of podosomes in smooth muscle cells. J Cell Sci. 2006;119:1691–1702. doi: 10.1242/jcs.02881. [DOI] [PubMed] [Google Scholar]
- Felsenfeld DP, Schwartzberg PL, Venegas A, Tse R, Sheetz MP. Selective regulation of integrin--cytoskeleton interactions by the tyrosine kinase Src. Nat Cell Biol. 1999;1:200–206. doi: 10.1038/12021. [DOI] [PubMed] [Google Scholar]
- Frame MC, Fincham VJ, Carragher NO, Wyke JA. v-Src's hold over actin and cell adhesions. Nat Rev Mol Cell Biol. 2002;3:233–245. doi: 10.1038/nrm779. [DOI] [PubMed] [Google Scholar]
- Gavazzi I, Nermut MV, Marchisio PC. Ultrastructure and gold-immunolabelling of cell-substratum adhesions (podosomes) in RSV-transformed BHK cells. J Cell Sci. 1989;94(Pt 1):85–99. doi: 10.1242/jcs.94.1.85. [DOI] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- Gimona M, Buccione R. Adhesions that mediate invasion. Int J Biochem Cell Biol. 2006;38:1875–1892. doi: 10.1016/j.biocel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Hai CM, Hahne P, Harrington EO, Gimona M. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp Cell Res. 2002;280:64–74. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Hsu YS, Martin GS. Shp-2 mediates v-Src-induced morphological changes and activation of the anti-apoptotic protein kinase Akt. Oncogene. 2000;19:3164–3171. doi: 10.1038/sj.onc.1203655. [DOI] [PubMed] [Google Scholar]
- Hiura K, Lim SS, Little SP, Lin S, Sato M. Differentiation dependent expression of tensin and cortactin in chicken osteoclasts. Cell Motil Cytoskeleton. 1995;30:272–284. doi: 10.1002/cm.970300405. [DOI] [PubMed] [Google Scholar]
- Honda H, Oda H, Nakamoto T, Honda Z, Sakai R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, Katsuki M, Yazaki Y, Hirai H. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas. Nat Genet. 1998;19:361–365. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- Hurst IR, Zuo J, Jiang J, Holliday LS. Actin-related protein 2/3 complex is required for actin ring formation. J Bone Miner Res. 2004;19:499–506. doi: 10.1359/JBMR.0301238. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci. 2000;113(Pt 24):4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Swedlow JR, Morgan DO, Varmus HE. c-Src enhances the spreading of src−/− fibroblasts on fibronectin by a kinase-independent mechanism. Genes Dev. 1995;9:1505–1517. doi: 10.1101/gad.9.12.1505. [DOI] [PubMed] [Google Scholar]
- Kumagai N, Ohno K, Tameshige R, Hoshijima M, Yogo K, Ishida N, Takeya T. Induction of mouse c-src in RAW264 cells is dependent on AP-1 and NF-kappaB and important for progression to multinucleated cell formation. Biochem Biophys Res Commun. 2004;325:758–768. doi: 10.1016/j.bbrc.2004.10.094. [DOI] [PubMed] [Google Scholar]
- Lakkakorpi PT, Helfrich MH, Horton MA, Vaananen HK. Spatial organization of microfilaments and vitronectin receptor, alpha v beta 3, in osteoclasts. A study using confocal laser scanning microscopy. J Cell Sci. 1993;104(Pt 3):663–670. doi: 10.1242/jcs.104.3.663. [DOI] [PubMed] [Google Scholar]
- Lakkakorpi PT, Nakamura I, Young M, Lipfert L, Rodan GA, Duong LT. Abnormal localisation and hyperclustering of (alpha)(V)(beta)(3) integrins and associated proteins in Src-deficient or tyrphostin A9-treated osteoclasts. J Cell Sci. 2001;114:149–160. doi: 10.1242/jcs.114.1.149. [DOI] [PubMed] [Google Scholar]
- LaPan P, Zhang J, Pan J, Haney S. Quantitative optimization of reverse transfection conditions for 384-well siRNA library screening. Assay Drug Dev Technol. 2008;6:683–691. doi: 10.1089/adt.2008.142. [DOI] [PubMed] [Google Scholar]
- Lener T, Burgstaller G, Crimaldi L, Lach S, Gimona M. Matrix-degrading podosomes in smooth muscle cells. Eur J Cell Biol. 2006;85:183–189. doi: 10.1016/j.ejcb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Linder S, Kopp P. Podosomes at a glance. J Cell Sci. 2005;118:2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- Liron Y, Paran Y, Zatorsky NG, Geiger B, Kam Z. Laser autofocusing system for high-resolution cell biological imaging. J Microsc. 2006;221:145–151. doi: 10.1111/j.1365-2818.2006.01550.x. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Addadi L, Geiger B. The molecular dynamics of osteoclast adhesions. Eur J Cell Biol. 2006a;85:203–211. doi: 10.1016/j.ejcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, Addadi L. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2007;2:e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C, Parsons JT, Addadi L, Geiger B. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J Cell Sci. 2006b;119:4878–4888. doi: 10.1242/jcs.03271. [DOI] [PubMed] [Google Scholar]
- Marchisio PC, Cirillo D, Naldini L, Primavera MV, Teti A, Zambonin-Zallone A. Cell-substratum interaction of cultured avian osteoclasts is mediated by specific adhesion structures. J Cell Biol. 1984;99:1696–1705. doi: 10.1083/jcb.99.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchisio PC, Cirillo D, Teti A, Zambonin-Zallone A, Tarone G. Rous sarcoma virus-transformed fibroblasts and cells of monocytic origin display a peculiar dot-like organization of cytoskeletal proteins involved in microfilament-membrane interactions. Exp Cell Res. 1987;169:202–214. doi: 10.1016/0014-4827(87)90238-2. [DOI] [PubMed] [Google Scholar]
- Mayer T, Meyer M, Janning A, Schiedel AC, Barnekow A. A mutant form of the rho protein can restore stress fibers and adhesion plaques in v-src transformed fibroblasts. Oncogene. 1999;18:2117–2128. doi: 10.1038/sj.onc.1202537. [DOI] [PubMed] [Google Scholar]
- Moreau V, Tatin F, Varon C, Genot E. Actin can reorganize into podosomes in aortic endothelial cells, a process controlled by Cdc42 and RhoA. Mol Cell Biol. 2003;23:6809–6822. doi: 10.1128/MCB.23.19.6809-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mayanagi T, Yoshio T, Sobue K. Changes in the balance between caldesmon regulated by p21-activated kinases and the Arp2/3 complex govern podosome formation. J Biol Chem. 2007;282:8454–8463. doi: 10.1074/jbc.M609983200. [DOI] [PubMed] [Google Scholar]
- Nermut MV, Eason P, Hirst EM, Kellie S. Cell/substratum adhesions in RSV-transformed rat fibroblasts. Exp Cell Res. 1991;193:382–397. doi: 10.1016/0014-4827(91)90111-7. [DOI] [PubMed] [Google Scholar]
- Nitsch L, Gionti E, Cancedda R, Marchisio PC. The podosomes of Rous sarcoma virus transformed chondrocytes show a peculiar ultrastructural organization. Cell Biol Int Rep. 1989;13:919–926. doi: 10.1016/0309-1651(89)90074-x. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Itoh T, Takenawa T. Sequential signals toward podosome formation in NIH-src cells. J Cell Biol. 2008;182:157–169. doi: 10.1083/jcb.200801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran Y, Lavelin I, Naffar-Abu-Amara S, Winograd-Katz S, Liron Y, Geiger B, Kam Z. Development and application of automatic high-resolution light microscopy for cell-based screens. Methods Enzymol. 2006;414:228–247. doi: 10.1016/S0076-6879(06)14013-6. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne WC, Baron R. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, del Carmen Ovejero M, Hou P, Heegaard AM, Kumegawa M, Foged NT, Delaisse JM. Identification of the membrane-type matrix metalloproteinase MT1-MMP in osteoclasts. J Cell Sci. 1997;110(Pt 5):589–596. doi: 10.1242/jcs.110.5.589. [DOI] [PubMed] [Google Scholar]
- Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Tarone G, Cirillo D, Giancotti FG, Comoglio PM, Marchisio PC. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp Cell Res. 1985;159:141–157. doi: 10.1016/s0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- Winograd-Katz SE, Itzkovitz S, Kam Z, Geiger B. Multiparametric analysis of focal adhesion formation by RNAi-mediated gene knockdown. J Cell Biol. 2009;186:423–436. doi: 10.1083/jcb.200901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007a;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Benjamin G. Protein networks in integrin-mediated adhesions. Systems Biomedicine. 2010:139–151. [Google Scholar]
- Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007b;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- Zambonin Zallone A, Teti A, Gaboli M, Marchisio PC. Beta 3 subunit of vitronectin receptor is present in osteoclast adhesion structures and not in other monocyte-macrophage derived cells. Connect Tissue Res. 1989;20:143–149. doi: 10.3109/03008208909023882. [DOI] [PubMed] [Google Scholar]
- Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- Zamir E, Katz BZ, Aota S, Yamada KM, Geiger B, Kam Z. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112(Pt 11):1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- Zamir E, Katz M, Posen Y, Erez N, Yamada KM, Katz BZ, Lin S, Lin DC, Bershadsky A, Kam Z, Geiger B. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell extracts from MEF and MEF-SrcY527F cells were subjected to BCA kit analysis for protein quantification, and equal amounts of protein were analyzed by Western blot with anti-Src, anti-pY418-Src, and anti-general phospho-tyrosine antibody. GAPDH was used as a loading control.
MEF-SrcY527F cells were transiently co-transfected with GFP-paxillin and mCherry actin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 30 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Supplementary Figure 3. While the paxillin (green) is relatively static, there is a constant flow of actin (red); the rings position and size remain stable.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 40 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4. A stable, expanding ring at the periphery of the cell may be observed; the ring expands as the cell spreads.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 30 min. The rings maintain their size and location throughout the movie, but the internal dynamics is high. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were transiently transfected with GFP-actin, and 24 h later, recorded by time-lapse video microscopy at 2-min intervals for 30 min. The rings seen are extremely dynamic and constantly form, merge, split, and collapse. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were transiently transfected with GFP-α-actinin, and 24 h later, recorded by time-lapse video microscopy at 30-sec intervals for 30 min. Clusters of peripheral, as well as central, podosomes, are seen. Podosomes can develop into an expanding ring. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were transiently co-transfected with GFP-cortactin and mCherry actin, and 24 h later, recorded by time-lapse video microscopy at 2-min intervals for 5 h. The actin (red) marks the stress fibers as well as the podosomes, while cortactin (green) marks only the podosomes. It appears that the stress fibers are anchored to the podosomes. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 4.
MEF-SrcY527F cells were permeabilized, fixed and stained for actin and paxillin. A spatial ratio image shows that the structure of a single podosome consists of two domains: an F-actin-rich core, surrounded by a paxillin-rich ring domain.
A time-lapse movie of MEF-SrcY527F cells transiently co-transfected with GFP-paxillin and mCherry actin was used to create temporal ratio images and auto-correlation analysis. The ratio between the two indicated time points is color-coded: new pixels are blue, pixels that disappeared are red, and unchanged pixels are represented by variable colors, depending on the specific local intensity ratio. Yellow indicates identical intensities in the two time points. Note the high predominance of blue and red in the actin labeling, as compared to the paxillin labeling. The auto-correlation graph was calculated by measuring the average correlation in intensity between frames taken at increasing intervals, from 0 sec to 300 sec. Actin decay occurs faster, indicating that actin is more dynamic than paxillin.
Representative montages of 6 × 6 images from untransfected MEF-SrcY527F cells, or MEF-SrcY527F cells transfected with control siRNAs, as indicated. (A) Montages of DAPI and actin (phalloidin) staining are shown, and a high-resolution image from the actin-stained montage is shown for each control. Bars: (montages) 50 μm; (high-resolution images) 10 μm.
For each siRNA, its library, target gene symbol, RefSeq (NCBI) accession number, gene name, Entrez gene ID, and a link to Gene (NCBI) are shown.
A phase-contrast, time-lapse movie of RAW 264.7 control cells was produced 24 h after differentiation began. The movie was recorded at 15 min intervals for 31.5 h. Each time, 16 images were taken from the well, and stitched together to form a montage. The display rate is 10 frames/sec. Bar: 20 μm. Images from this movie are shown in Figure 9.
A phase-contrast, time-lapse movie of RAW 264.7 Src shRNA-1 cells was produced, 24 h after differentiation began. The movie was recorded at 15 min intervals for 31.5 h. Each time, 16 images were taken from the well, and stitched together to form a montage. The display rate is 10 frames/sec. Bar: 20 μm. Images from this movie are shown in Figure 9.
RAW 264.7 control cells were transfected with lifeact, 48 h after differentiation began. The movie was recorded 30 h later, at 45 sec intervals, for 45 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 9.
RAW 264.7 Src shRNA-1 cells were transfected with lifeact, 48 h after differentiation began. The movie was recorded 30 h later, at 45 sec intervals, for 45 min. The display rate is 10 frames/sec. Bar: 10 μm. Images from this movie are shown in Figure 9.