Abstract
Fetal hemoglobin (HbF) levels are generally inversely proportional to severity of sickle cell disease (SCD) for given sickle phenotypes. Molecular regulation of HbF occurs through complex interactions cis and trans to the beta globin gene locus. Novel insights made through population-based genetic epidemiologic studies of non-anemic populations were replicated in SCD groups, despite large differences in HbF levels. Identification of the lymphoid transcription factor BCL11A as a key suppressor of HbF expression validates approaches using population genetics to study HbF expression. We review these methods and findings, and speculate on applying pharmaco-genetics to optimize hydroxyurea therapy aimed at increasing HbF.
Keep words: sickle cell disease, fetal hemoglobin, genetic epidemiology, BCL11A, hydroxyurea
Introduction
Sickle cell disease (SCD) affects 90–100,000 people in the U.S.[1], including more than 1700 newborns annually [2]. While population screening, specialized care and services have largely eliminated early child mortality from SCD and prolonged average life expectancy, disease-related morbidity and mortality accumulate by early adulthood [1]. The drug hydroxurea (HU) holds expanding promise for improved clinical outcomes. However, clinical response to treatment varies between individuals such that use of HU remains empiric [3]. This review describes newly identified genetic mechanisms for regulating fetal hemoglobin (HbF) as a disease modifier and prospects for prospectively assessing efficacy of HU therapy.
Mechanisms regulating fetal hemoglobin
Severity of SCD varies widely between affected individuals. Within genotypes of HbSS, HbSC and HbS-Beta thalassemia, lower fetal hemoglobin (HbF) level is the strongest predictor of worse disease [4]. Polymerization of sickle hemoglobin, the underlying patho-physiology of SCD, is actively inhibited by HbF – but not by HbA. Plunging HbF levels at 9–18 months of life to below 20% of total hemoglobin is usually accompanied by initial overt disease manifestations, such as pain, dactylitis, anemia, and hemolysis [4]. In children with SCD, HbF levels stabilize to adult levels by 3–4 years of age [4]. Stable levels vary among those with SCD from 3–20% of total hemoglobin, compared to only 0.5–2% for non-anemic individuals. Twin studies in non-anemic adults [5] and sibling studies in SCD [6] have documented high heritability of HbF in both populations. Therefore, genetic analyses of HbF regulation are key to understanding the spectrum of SCD severity, variability in response to HU and design of novel therapies.
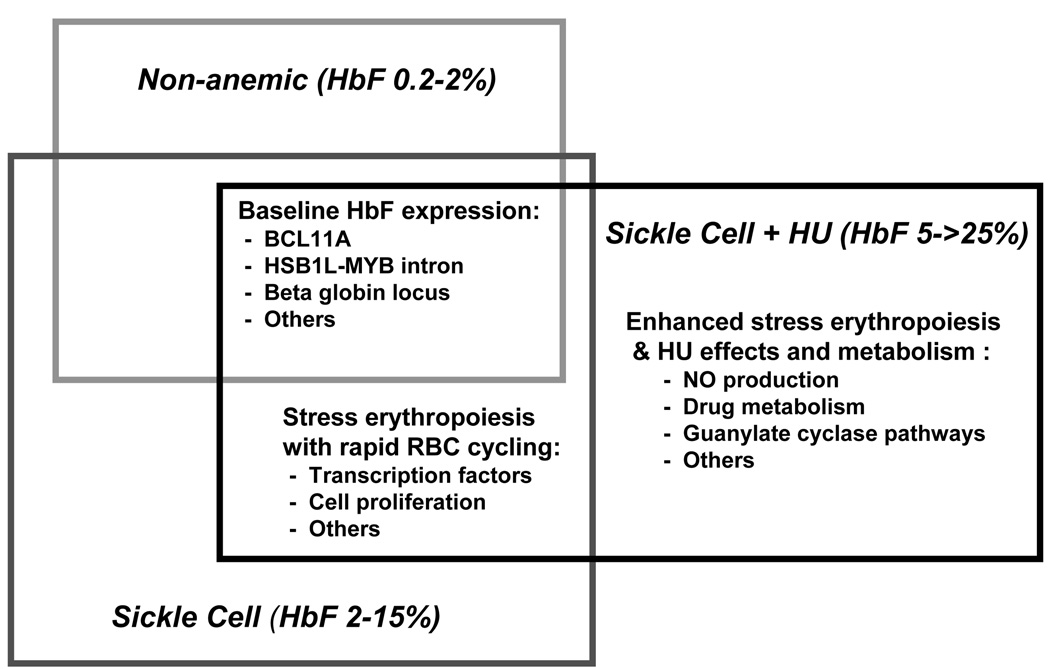
Current investigation takes advantage of mechanisms of HbF regulation shared by non-anemic and anemic populations, both SCD and beta thalassemia. Genetic mechanisms operate within, and external to, the beta globin locus on chromosome 11 (Figure 1). Regulation of the expression of gamma globin (for HbF) and suppression of beta globin (for HbS) occurs through combined transcriptional regulation, chromosomal remodeling and other epigenetic factors, and by the kinetics of red blood cell regeneration and differentiation [7,8]. Sickle carriers have normal ranges of HbF [9], implying that the sickle beta globin variant itself has limited impact on HbF elevation seen in SCD. Elevated HbF levels seen with disease are more likely secondary to the accelerated destruction and rapid regeneration of the RBC. This state of “stress erythropoiesis” leads to enhanced production of HbF and F cells [3,7]. Hence, HbF levels in SCD should be regulated by mechanisms controlling both basal HbF production and those resulting from sickle red cell homeostasis. This paradigm predicts that mechanisms regulating HbF production in non-anemic populations should overlap with those with SCD (Figure 2). Supporting this assertion is the recent identification of genetic determinants for HbF levels shared by those with or without SCD, described below.
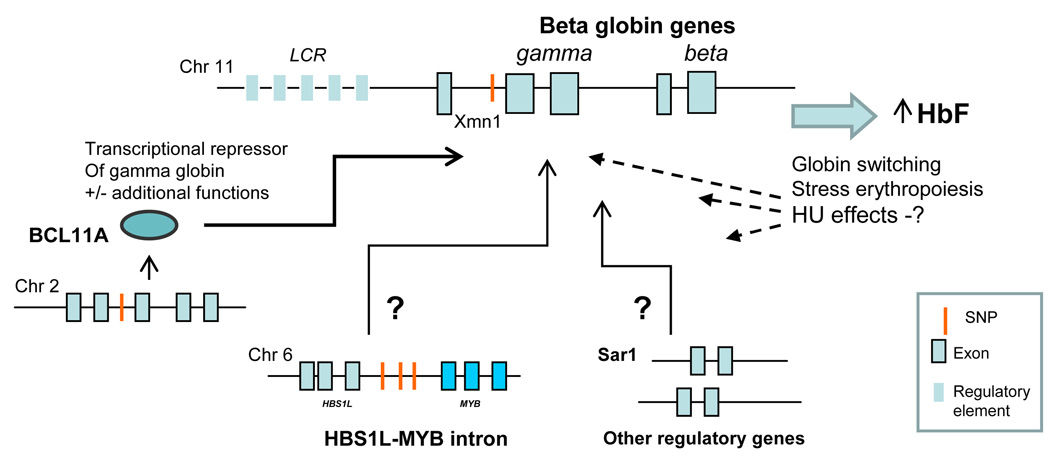
Figure 1.
The beta globin locus and interacting loci based on genetic epidemiologic data in normal and sickle cell adult populations. (Adapted from ref. 20).
Figure 2.
A model of overlapping mechanisms regulating HbF in normal and SCD populations, and those on hydroxyurea therapy.
Continuous distribution of HbF levels in populations with or without hemoglobinopathies implies that HbF is regulated as a complex genetic trait [9]. As such, HbF can be considered an endophenotype for SCD to detect underlying genetic risk factors. In general, an endophenotype, better characterizes the relationship between the gene and the disease locus since it lies within the biological pathway toward the disease of interest. Therefore, endophenotypes are presumably determined by fewer genes than the disease of interest, which reduces the complexity of the genetic analysis and enhances the statistical power to detect genetic associations [10,11]
Beta globin gene variants
Within the beta globin locus, cis acting elements controlling the postnatal switch from gamma globin to beta globin were initially identified through studies of naturally occurring variants with reduced switching away from gamma globin production, such that HbF predominates (HPFH) [7]. In the 1980’s, restriction length polymorphisms (RFLPs) variations within introns of the beta globin locus in SCD characterized different origins of the sickle mutation within Africa and Asia. Some markers track with differing HbF levels and disease severity in adults [12,13] and young children [14], especially the Xmn1 digestion site upstream of the beta globin locus (Figure 1), a polymorphism that largely defines the Senegalese SCD haplotype associated with higher HbF levels and milder disease. RFLPs generally constitute segments too large for fine mapping and have been largely succeeded by use of multiple single nucleotide polymorphisms (SNPs) across the genome, including the SNP for Xmn1.
Genome-wide approaches
To identify determinant of HbF regulation, genetic epidemiology use of thousands of SNPs to screen for regions of interest throughout the genome, so-called genome-wide association studies (GWAS). In contrast to, candidate gene studies select genes based on known or suspected disease mechanisms rather than permitting an unbiased scan to identify disease modifiers or susceptibility factors. Popularity of GWAS also stems from technologic advances for high throughput genetic analysis, with low SNP genotyping costs and low genotyping error rates. GWAS also accommodates analysis from family studies or from multiple unrelated affected individuals.
Despite these advantages, GWAS are limited by the large number of subjects needed to demonstrate modest effect sizes, as odd ratios of association between SNPs and causal variants typically are limited to less than 1.5, i.e., 50% of the increased risk of the outcome relative to the control population. Furthermore, to avoid false positives with the large number of statistical tests performed, putative associations must survive genome-wide multiple corrections and validation in one or more independent replication datasets. Replication sets for relatively rare disorders such as SCD and thalassemia may be sufficiently large for candidate gene studies but are rarely available for GWAS. As described below, recent studies have taken advantage of the overlap between HbF regulation in non-anemic European populations and in those with SCD by performing GWAS in the former and confirming by candidate gene approach in the latter.
Another complication of GWAS is that genetic architecture may differ between populations. Analysis of genome-wide polymorphisms from human populations demonstrate that some alleles at different polymorphic loci are found together more often than expected by chance based on their frequencies. This non-random pattern of allelic associations is called linkage disequilibrium (LD). If a SNP frequency varies among different populations, the LD pattern, or how the alleles from different SNPs are associated in haplotypes, will be also different, further complicating the statistical analysis of the genetic data. Since the extent of LD in a chromosomal region of genetic association can be different between the cases and controls, LD patterns are computed using the healthy control population as baseline to contrast to the LD found in the cases.
Overall, genotyping tools and statistical methods were developed for European and Asian populations. The high levels of haplotype diversity in African populations are a potential powerful tool in GWAS to identify the causal variants underlying the disease. This genetic diversity, however, leads to low levels of linkage disequilibrium. As allelic associations among SNPs are less frequent in comparison to European populations, African studies require denser SNP genotyping platforms to characterize genetic variability than those typically used in other population studies. Efforts are growing to gain deeper insights into genetics in African populations through GWAS from African populations with different human diseases phenotypes [15,16]. The International HapMap project has generated a catalogue of SNP allele and haplotype frequencies in Yoruba individuals from central Africa, who as a Bantu ethnic group are relevant to those with SCD. A more comprehensive effort to determine SNP allele frequencies among different populations of African origin for SCD would be useful to identify relevant genetic variants in complex traits such as HbF. Genetic studies of SCD in North and South Americans may also need to account for the largely admixed populations.
Major genetic findings and their implications
In Northern European adult populations, a GWAS comparing those on the extremes of HbF distribution revealed strong associations with SNPs in three different genes or gene loci as most strongly associated with the range of HbF expression: Xmn1-HBG2, HBS1L-MYB and BCL11A (Table 1, Figure 1) [9,17]. Subsequently, these same three genes were also found to be strongly associated with HbF levels in other non-anemic adult populations [9] and, potentially more medically significant, in two anemic disease populations: those with beta thalassemia [18] and in two distinct adult populations with SCD. In the latter, these same variants assessed by only five SNPs, accounted for a considerable proportion, i.e 20–30%, of the variation among adults with SCD [19; reviewed in ref. 20]. Notably, the same five SNP variants correlated with rates of SCD related pain crises [19], a finding that strongly reinforces the productivity of these genetic approaches.
Table I.
Candidate genes regulating fetal hemoglobin in anemic and non-anemic populations
| Reference | Chr | Gene | Population | * %var |
|---|---|---|---|---|
| Thein et al [17] | 6 | HBS1L-MYB | healthy North European | 17.6 |
| Menzel et al [34] | 2 | BCL11A | healthy Europeans | 15.1 |
| 6 | HBS1L-MYB | healthy Europeans | 19.4 | |
| 11 | β-globin | healthy Europeans | 10.2 | |
| Lettre et al [19] | 2 | BCL11A | SCD AfAm | 11.8–14.1 |
| SCD Brasil | 6.7–7.9 | |||
| 6 | HBS1L-MYB | SCD AfAm | 0.3–3.5 | |
| SCD Brasil | 3.4–7.4 | |||
| 11 | β-globin | SCD AfAm | 2.2 | |
| SCD Brasil | ||||
| Creary et al [35] | 6 | HBS1L-MYB | SCD AfCaribb | |
| healthy AfGerman | ||||
| healthy AfAm | ||||
| SCD WestAf | ||||
| Solovieff et al [25] | 2 | BCL11A | SCD AfAm | 10.3 |
| HbE/Thalass Thai/HKong | ||||
| 11 | OR51B5;OR51B6 | SCD AfAm | 5.6 | |
| HbE/Thalass Thai/HKong | ||||
Percentage of HbF levels variance explained by genetic polymorphisms
BCL11A
A single SNP within intron 2 of the oncogene BCL11A on chromosome 2p16 in healthy European populations accounts for 15% of the F-cell variability (Table 1) [9]. BCL11A is a lymphoid transcription factor that functions in early B cell development and whose disregulated expression is involved in leukemogeneisis [21]. Studies of cultured erythroid precursor cells and in vivo murine models of murine or humanized globin loci [8,22] demonstrated that BCL11A binds to the beta globin locus and plays a direct role in inhibiting the switch from gamma to beta globin, thereby dampening expression of HbF. Xu et al. recently reported that BCL11A binding to the beta globin locus affects chromosomal configuration and transcriptional silencing of gamma globin [8]. BCL11A may also provide signals for red cell proliferation and differentiation (Figure 1), [22] speculation based upon functions of its known interacting proteins in transcription, hemopoiesis and lymphoid differentiation. Intronic SNP variants within BCL11A associated with higher or lower HbF production may provide further clues to BCL11A’s role in HbF suppression.
In separate GWAS in healthy populations, BCL11A is also associated with red cell size (mean corpuscular volume, MCV) [23], suggesting that it may influence the regenerative kinetics of red cell generation and maturation. Overlapping functions regulating HbF and MCV may yield clues to its function in erythroid production and development. Identification of multiple roles for BCL11A’s regulatory function reinforces the concept that cell growth and differentiation in red cells, as in lymphocytes, are tightly linked.
Xmn1-HBG2
The Xmn1 genotype, now defined as a SNP, was also reported for non-anemic adults, accounting for 13–32% of the total variation of HbF (Table 1) in most population groups including Europeans, African and Asian Indians [12], and in those with SCD [19]. No delineated function for regulating HbF currently exists for is SNP or for the HBS1L-MYB intron or other sites described below.
HBS1L-MYB intron
A genome-wide linkage analysis of Asian-Indian kindred with probands showing mild SCD identified a major locus on chromosome 6q23 [9]. Higher resolution mapping on a sample of North European ancestry identified haplotype blocks in the intergenic region between HBS1L and MYB oncogene as strongly associated with F cell levels. In non-anemic Northern Europeans, the 6q locus accounts for approximately 19% of the population trait variance (Table 1), and also contributes to the HbF variation in SCD [19].
Other candidate genes
Two new GWAS reported other novel polymorphisms associated with SCD. Using two datasets totaling over 2000 African American adults with either severe or mild SCD, Steinberg and colleagues identified and replicated with varying degree of confidence 11 genes associated with disease severity, including a region on chromosome 11 that contains olfactory receptor genes affecting HbF levels [24,25]. How these regions or genes could affect HbF levels are not yet known.
Genetic influences on response to HU
HU, a ribonucleotide reductase inhibitor, is the sole FDA-approved pharmacologic treatment for SCD. HU likely increases levels of HbF by inducing stress erythropoiesis, likely through continued cytotoxicity of dividing erythroid precursors. Additional mechanisms of HU action have been posited [3,26]. Increased HbF improves red cell rheology, reduces red cell adhesion and lysis and increases hematocrit [3]. HU increases HbF and decreases symptoms in approximately half of adults with SCD [27,28] and in most pediatric patients, although with highly variable degrees of response [29]. While providing no cure, HU appears to convert more severely affected patients to a milder phenotype [30]. Despite HU’s unique and frequently dramatic impact on disease, the 2008 NHLBI consensus statement affirmed the under-utilization of HU in treating sickle cell disease [31].
Steinberg et al. demonstrated that the degree of HbF elevation from HU therapy is a heritable trait [6]. Likely overlap in the physiology of supra-normal HbF associated with both hemoglobinopathy and HU induction predicts a shared influence of the genetic polymorphisms described above. This prediction is supported by a reported correlation between levels of HbF at baseline and at maximum HU dose in children [29]. Only one published study broadly examines the genetics of HU response in SCD [28]. Using a retrospective cohort of adults from the seminal Multicenter Study of Hydroxyurea (MSH) trial and assessing over two dozen candidate genes, Ma et al. report significant associations between SNPs and HbF response to HU in or near genes involved in the metabolism of arginine to nitric oxide and in a transcription factor that induces DNA bending.
This report pre-dated the identification of BCL11A as a central regulator of HbF expression, leaving yet unknown the tantalizing question of whether it modifies HU’s effects. Additional mechanisms of HU could include effects on pathways involving BCL11A and the other polymorphisms described above, such as protein partners and/or upstream or downstream effectors and telomerase function [32].Other reports include associations between HU response in SCD and polymorphisms in the guanosine triphosphate (GTP)-binding protein gene sar1a [26], underscoring the reality that genetic pathways regulating the HbF response to HU are complex (Figures 1 and 2).
Of note, the majority of the 137 MSH subjects included in that report responded only modestly to HU, with less than a 5% change in HbF from baseline [28]. These data suggest that some adult cohorts may have limited physiologic capabilities to respond to HU and thus may not be the most informative group to study the genetics of HU response. Since the study population of is restricted to those with SCD on HU, sample sizes are modest and estimation of LD among markers relies on the cases only.
Children with SCD
To minimize influences on endophenotype from disease progression such as marrow infarction, investigation of the genetics of HbF regulation, RBC regeneration and genotype-phenotype in SCD populations arguably should focus on children. For example, in a GWAS analysis, Steinberg and colleagues describe age-dependent severity scores in the large CSSCD cohort that was based on the occurrence of disease complications [24]. In that cohort, over 80% of children less than 18 years of age had relatively uncomplicated disease. By age 18–40 years, approximately 80% of subjects had one or more major disease complication; virtually all of those over 40 had multiple complications. Differing response rates to HU in children (HUG study, ref. 29) and in some studies of adults (MSH study, ref. 27) underscore that clinical factors may complicate analysis in the latter group.
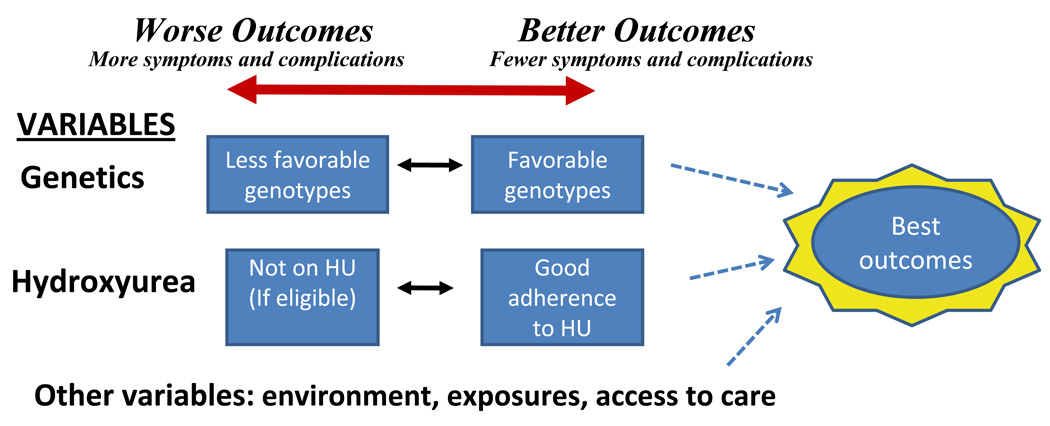
Prospectively identifying those children most likely to respond would provide persuasive rationale for initiating early HU. If clinical trials demonstrate that benefits of early HU outweigh potential harms (e.g. BABY-HUG, ref. 33), then HU use may shift from treating those most severely affected to generally applied prevention of SCD complications. For those with milder disease and/or likely to best respond to HU, aggressive HU therapy could dramatically alter long-term outcomes (Figure 3), as demonstrated in a Greek study of adults with SCD [30].
Figure 3.
A scheme of the range of clinical outcomes in SCD based on the genetics of disease modifiers such as HbF and response to hydroxyurea.
Future challenges
Recent population-based genetic research has led to discovery of novel genetic modifiers of HbF in SCD, beta thalassemia and in non-anemic populations. BLC11A and other yet unidentified players likely regulate HbF expression by modifying globin switching and by affecting the kinetics of RBC growth and differentiation through hitherto unknown mechanisms. Effects of HU predict that it regulates HbF via mechanisms that overlap with those controlling baseline HbF in normal and in globin disease populations rather than by entirely distinct pathways (Figure 2). Approaches used to date for understanding baseline HbF may also lead to pharmaco-genomic approaches to HU in SCD through improved prediction of HbF response in genetically definable sub-populations or individuals. Those patients less likely to have robust responses may be counseled towards other therapies such as hematopoietic transplantation. New insights into HbF regulation enhance its appeal as a model for the study of disease modifiers of single gene disorders and as a therapeutic target.
Acknowledgements
This work was supported by NIH grant number 3UL1RR024156-04S4. The authors thank Dr. Arthur Bank for his editorial support.
Footnotes
We have no conflicts of interest to disclose.
References
- 1.Hassell KL. Population estimates of sickle cell disease in the U.S. Am J Prev Med. 2010;38:S512–S521. doi: 10.1016/j.amepre.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 2. [Accessed 4/15/10];2006 http://genes-r-us.uthscsa.edu/ data.
- 3.Platt OS. Hydroxyurea for the treatment of sickle cell anemia. New Engl J Med. 2008;358:362–369. doi: 10.1056/NEJMct0708272. [DOI] [PubMed] [Google Scholar]
- 4.Powars DR, Weiss JN, Chan LS, Schroeder WA. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63:921–926. [PubMed] [Google Scholar]
- 5.Garner C, Tatu T, Reittie JE, Littlewood T, et al. Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood. 2000;95:342–346. [PubMed] [Google Scholar]
- 6.Steinberg MH, Voskaridou E, Kutlar A, Loukopoulos D, et al. Concordant fetal hemoglobin response to hydroxyurea in siblings with sickle cell disease. Am J Hematol. 2003;72:121–126. doi: 10.1002/ajh.10264. [DOI] [PubMed] [Google Scholar]
- 7.Bank A. Regulation of human fetal hemoglobin: new players, new complexities. Blood. 2006;107:435–443. doi: 10.1182/blood-2005-05-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thein SL, Menzel S, Lathrop M, Garner C. Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum Mol Genet. 2009;18:R216–R223. doi: 10.1093/hmg/ddp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan WH, Lynn KS, Chen CH, Wu YL, et al. Using endophenotypes for pathway clusters to map complex disease genes. Genet Epidemiol. 2006;30:143–154. doi: 10.1002/gepi.20136. [DOI] [PubMed] [Google Scholar]
- 11.Schliekelman P. Statistical power of expression quantitative trait loci for mapping of complex trait loci in natural populations. Genetics. 2008;178:2201–2216. doi: 10.1534/genetics.107.076687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilman JG, Huisman THJ. DNA sequence variation associated with elevated fetal Gg globin production. Blood. 1985;66:783–787. [PubMed] [Google Scholar]
- 13.Nagel RL, Erlingsson S, Fabry ME, Croizat H, Susuka SM, Lachman H, Sutton M, Driscoll C, Bouhassira E, Billett HH. The Senegal DNA haplotype is associated with the amelioration of anemia in African-American sickle cell anemia patients. Blood. 1991;77:1371–1375. [PubMed] [Google Scholar]
- 14.Green NS, Fabry ME, Kaptue-Noche L, Nagel RL. Senegal haplotype is associated with higher HbF than Benin and Cameroon haplotypes in African children with sickle cell anemia. Am J Hematol. 1993;44:145–146. doi: 10.1002/ajh.2830440214. [DOI] [PubMed] [Google Scholar]
- 15.Adeyemo A, Gerry N, Chen G, Herbert A, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000564. e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jallow M, Teo YY, Small KS, Rockett KA, et al. Wellcome Trust Case Control Consortium; Malaria Genomic Epidemiology Network. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thein SL, Menzel S, Peng X, Best S, et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci U S A. 2007;104:11346–11351. doi: 10.1073/pnas.0611393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uda M, Galanello R, Sanna S, Lettre G, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lettre G, Sankaran VG, Bezerra MA, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michelson AM. Developmental biology. From genetic association to genetic switch. Science. 2008;322:1803–1804. doi: 10.1126/science.1169216. [DOI] [PubMed] [Google Scholar]
- 21.Singh H, Pongubala JM, Medina KL. Gene regulatory networks that orchestrate the development of B lymphocyte precursors. Adv Exp Med Biol. 2007;596:57–62. doi: 10.1007/0-387-46530-8_5. [DOI] [PubMed] [Google Scholar]
- 22.Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, Tucker PW, Orkin SH. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebastiani P, Solovieff N, Hartley SW, Milton JN, et al. Genetic modifiers of the severity of sickle cell anemia identified through a genome-wide association study. Am J Hematol. 2010;85:29–35. doi: 10.1002/ajh.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solovieff N, Milton JN, Hartley SW, Sherva R, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5’ olfactory receptor gene cluster. Blood. 2010;115:1815–1822. doi: 10.1182/blood-2009-08-239517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumkhaek C, Taylor JG, 6th, Zhu J, Hoppe C, Kato GJ, Rodgers GP. Fetal haemoglobin response to hydroxycarbamide treatment and sar1a promoter polymorphisms in sickle cell anaemia. Br J Haematol. 2008;141:254–259. doi: 10.1111/j.1365-2141.2008.07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charache S, Terrin ML, Moore RD, Dover GJ, et al. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med. 1995;332:1317–1322. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 28.Ma Q, Wyszynski DF, Farrell JJ, et al. Fetal hemoglobin in sickle cell anemia: genetic determinants of response to hydroxyurea. Pharmacogenomics J. 2007;7:386–394. doi: 10.1038/sj.tpj.6500433. [DOI] [PubMed] [Google Scholar]
- 29.Ware RE, Eggleston B, Redding-Lallinger R, et al. Predictors of fetal hemoglobin response in children with sickle cell anemia receiving hydroxyurea therapy. Blood. 2002;99:10–14. doi: 10.1182/blood.v99.1.10. [DOI] [PubMed] [Google Scholar]
- 30.Voskaridou E, Christoulas D, Bilalis A, Plata E, et al. The effect of prolonged administration of hydroxyurea on morbidity and mortality in adult patients with sickle cell syndromes: results of a 17-year, single-center trial (LaSHS) Blood. 2010;115:2354–2363. doi: 10.1182/blood-2009-05-221333. [DOI] [PubMed] [Google Scholar]
- 31.Brawley OW, Cornelius LJ, Edwards LR, et al. National Institutes of Health Consensus Development Conference statement: hydroxyurea treatment for sickle cell disease. Ann Intern Med. 2008;148:932–938. doi: 10.7326/0003-4819-148-12-200806170-00220. [DOI] [PubMed] [Google Scholar]
- 32.Snyder AR, Zhou J, Deng Z, Lieberman PM. Therapeutic doses of hydroxyurea cause telomere dysfunction and reduce TRF2 binding to telomeres. Cancer Biol Ther. 2009;8:1136–1145. doi: 10.4161/cbt.8.12.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson BW, Miller ST, Rogers ZR, Rees RC, et al. The pediatric hydroxyurea phase III clinical trial (BABY HUG): challenges of study design. Pediatr Blood Cancer. 2010:250–255. doi: 10.1002/pbc.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]