Abstract
The mitogen-activated protein kinase (MAPK) pathways control diverse cellular functions in pathogenic fungi, including sexual differentiation, stress-response, and maintenance of cell wall integrity. Here we characterized a C. neoformans gene, which is homologous to the yeast Ste50 that is known to play an important role in mating pheromone response and stress response as an adaptor protein to the Ste11 MAPK kinase kinase in Saccharomyces cerevisiae. The C. neoformans Ste50 was not involved in any of the stress responses or virulence factor production (capsule and melanin) that are controlled by the HOG and Ras/cAMP signaling pathways. However, Ste50 was required for mating in both serotype A and serotype D C. neoformans strains. The ste50Δ mutant was completely defective in cell-cell fusion and mating pheromone production. Double mutation of the STE50 gene blocked increased production of pheromone and the hyper-filamentation phenotype of cells deleted of the CRG1 gene, which encodes the RGS protein that negatively regulates pheromone responsive G-protein signaling via the MAPK pathway. Regardless of the presence of the basidiomycota-specific SH3 domains of Ste50 that are known to be required for full virulence of Ustilago maydis, Ste50 was dispensable for virulence of C. neoformans in a murine model of cryptococcosis. In conclusion, the Ste50 adaptor protein controls sexual differentiation of C. neoformans via the pheromone-responsive MAPK pathway but is not required for virulence.
Keywords: Cryptococcus neoformans, MAPK, Ste50, Pheromone, Stress response
1. Introduction
The mitogen-activated protein kinase (MAPK) signaling pathways commonly exist in all eukaryotic organisms and play a wide variety of cellular roles in growth, differentiation, and stress response. All MAPK modules are comprised of three protein kinases: MAPK kinase kinase (MAPKKK), MAPK kinase (MAPKK), and MAPK. Humans contain three major MAPKs, ERK, JNK, and p38 MAPKs, which are involved in growth, differentiation, immunomodulation, stress response, and apoptosis (Johnson and Lapadat, 2002; Waskiewicz and Cooper, 1995). In fungi, several MAPK signaling pathways have been extensively characterized, including the pheromone response Kss1/Fus3-like MAPK pathway, the stress-activated Hog1-like MAPK pathway, and the cell wall integrity Mpk1-like MAPK pathway. Understanding of these MAPK pathways is particularly important for characterizing growth, differentiation, and virulence features of pathogenic fungi.
Three MAPK signaling pathways have been discovered in Cryptococcus neoformans, which causes pulmonary cryptococcosis and meningoencephalitis that is a life-threatening disease if not treated (Idnurm et al., 2005; Lin and Heitman, 2006). One MAPK pathway is mediated by the Cpk1 protein kinase. Disruption of CPK1 results in defective pheromone production, mating, and monokaryotic fruiting, which are essential for production of infectious spores (Davidson et al., 2000; Davidson et al., 2003; Wang et al., 2004). The Cpk1 MAPK is highly homologous to the Kss1 and Fus3 dual MAPK system that modulates mating and filamentous growth of S. cerevisiae. Similar to S. cerevisiae and other fungi, Ste11 MAPKKK and Ste7 MAPKK work upstream of the Cpk1 MAPK (Davidson et al., 2003). Upstream of the Cpk1 MAPK module are a pheromone-sensing G-protein coupled receptor Cpr1 (also known as Ste3) and its associated α-subunits (Gpa2 and Gpa3). These two α subunits (Gpa2 and Gpa3) and the β subunit Gpb1 of heterotrimeric GTP-binding protein (G protein) complex are known to be involved in the mating process (Hsueh et al., 2007). Unlike S. cerevisiae, however, Ste12 is not a major transcription factor for Cpk1 in C. neoformans since it plays only a minor role in mating (Alspaugh et al., 1998; Wickes et al., 1997; Yue et al., 1999).
The second MAPK in C. neoformans is an Mpk1 protein kinase, which is required for maintenance of cell wall integrity and growth at high temperature that is critical for C. neoformans to survive in the host (Kraus et al., 2003). Mkk1 (also known as Mkk2) MAPKKK and Bck1 MAPKK are two upstream protein kinases for the Mpk1 MAPK (Gerik et al., 2005; Kojima et al., 2006). The Mkk1-Bck1-Mpk1 pathway is activated by protein kinase C (PKC) that responds to cell wall perturbing agents and high temperature (Gerik et al., 2008; Gerik et al., 2005; Kraus et al., 2003). PKC plays an additional role in controlling melanin biosynthesis (Heung et al., 2004).
The third MAPK in C. neoformans is the Hog1 stress-activated protein kinase that responds to a variety of environmental stresses, including osmotic changes, oxidative and genotoxic damages, UV irradiation, high temperature, antifungal drugs, toxic metabolites, and heavy metal exposure (Bahn, 2008; Bahn et al., 2007a; Bahn et al., 2005b; Bahn et al., 2006). Hog1 is activated by Pbs2 MAPKK and Ssk2 MAPKKK (Bahn et al., 2007a; Bahn et al., 2005b). Upstream of the Hog1 MAPK module, the two-component-like phosphorelay system, composed of two hybrid sensor kinases (Tco1 and Tco2), a histidine-containing phosphotransfer protein (Ypd1), and two response regulators (Ssk1 and Skn7), relays its signal to Ssk2 (Bahn et al., 2006; Ko et al., 2009). Interestingly, the HOG (High Osmolarity Glycerol response) pathway, consisting of the Hog1 MAPK module and the phosphorelay system, also controls production of two virulence factors, capsule and melanin as well as sexual differentiation, indicating that the Hog1 MAPK pathway cross-talks with other signaling pathways (Bahn, 2008; Bahn et al., 2007a; Bahn et al., 2005b; Bahn et al., 2006). In addition, recent transcriptome analysis revealed that the HOG pathway negatively controls ergosterol biosynthesis, which affects susceptibility to the polyene and azole drugs (Ko et al., 2009).
Although the basic signaling components and regulatory mechanism of each MAPK pathway have been characterized in C. neoformans, the potential cross-talk between the MAPK pathways has not been well characterized. Therefore, we aimed to investigate a signaling component that has been reported to link multiple MAPK pathways to other signaling pathways in fungi. The Ste50 adaptor protein is a protein kinase regulator that contains two evolutionarily conserved domains, SAM (Sterile Alpha Motif) and RA (Ras Association), at the amino and carboxy terminals, respectively (O'Rourke and Herskowitz, 1998; Posas et al., 1998). The SAM domain is a protein-binding domain playing a role in signal transduction and transcriptional regulation in eukaryotes (Bhunia et al., 2009; Slaughter et al., 2008). The RA domain is required for proper localization of the cargo proteins that Ste50 delivers (Truckses et al., 2006; Wu et al., 2006). The RA domain of Ste50 in S. cerevisiae controls HOG pathway activation for osmoregulation through interaction with the cytoplasmic single-transmembrane protein Opy2 (Ekiel et al., 2009). In S. cerevisiae, Ste50 is involved in multiple MAPK signaling pathways, governing mating, filamentous growth, and stress response (Posas et al., 1998; Rad et al., 1992; Ramezani Rad et al., 1998). It acts as an adaptor protein between the G-protein associated Cdc42-Ste20 protein complex and its effector protein Ste11 MAPKKK (O'Rourke and Herskowitz, 1998; Posas et al., 1998). Ste50-binding inhibits the interaction between the regulatory N-terminus and catalytic C-terminus of Ste11, which allows Ste11 to autophosphorylate (Wu et al., 1999). In S. cerevisiae, Ste11 MAPKKK regulates multiple MAPK pathways, including the Fus3 MAPK for the mating process, the Kss1 MAPK for filamentous growth, and the Hog1 MAPK for stress response (Lee and Elion, 1999; O'Rourke and Herskowitz, 1998; Posas and Saito, 1997). In addition to its role as an adaptor for Ste11, Ste50 has also been implicated in the Ras-cAMP signaling pathway, possibly due to the presence of the RA domain at the C-terminus (Poplinski et al., 2007; Ramezani-Rad, 2003). Indeed, Ste50 also interacts with Ras1 and Ras2 via the RA domain (Ramezani-Rad, 2003). In contrast to the Ste50 in ascomycota, the Ste50 in basidiomycota contains two additional SH3 domains at the C-terminus (Klosterman et al., 2008). Expectedly, the N-terminal SAM and RA domains are required for filamentous growth of U. maydis (Klosterman et al., 2008). Interestingly, the SH3 domains are dispensable for filamentation in U. maydis but are required for virulence (Klosterman et al., 2008), indicating that the SH3 domains of Ste50 may play an essential role in pathogenicity of basidiomycete fungal pathogens.
To investigate the role of Ste50 in the C. neoformans Cpk1-, Hog1-, and Ras-signaling pathways, we deleted the STE50 gene and analyzed the mutant for changes in signaling through the MAPK and Ras pathways. Ste50 is dispensable for stress response and production of two major virulence factors, capsule and melanin, indicating that Ste50 is not involved in regulation of the Hog1 and Ras-signaling pathways. However, Ste50 was required for sexual reproduction via the Cpk1 MAPK pathway. In contrast to other ascomycetes and basidiomycetes where Ste50 is involved in multiple signaling pathways, our study demonstrates that the Ste50 in C. neoformans is dedicated to control of the pheromone-responsive MAPK pathway.
2. Materials and Methods
2.1. Strains, plasmid, and media
The C. neoformans strains used in this study are listed in Table 1 and were cultured in YPD (yeast extract-peptone-dextrose) medium. V8 medium (pH 5.0) (Cambell, Camden, NJ) for mating, agar-based DME (Dulbecco’s modified Eagle’s) medium (Invitrogen, Carlsbad, CA) for capsule production, and Niger seed or L-DOPA medium for melanin production were prepared as previously described (Bahn et al., 2004).
Table 1.
Strains used in this study.
| Strain | Genotype | Parent | Reference |
|---|---|---|---|
| C. neoformans | |||
| H99 | MATα | (Perfect et al., 1993) | |
| KN99a | MATa | (Nielsen et al., 2003) | |
| JEC21 | MATα | (Moore and Edman, 1993) | |
| JEC20 | MATa | (Moore and Edman, 1993) | |
| YSB42 | MATα cac1::NAT-STM#159 | H99 | (Bahn et al., 2004) |
| YSB64 | MATα hog1::NAT-STM#177 | H99 | (Bahn et al., 2005b) |
| YSB81 | MATa hog1::NEO | KN99a | (Bahn et al., 2005b) |
| YSB313 | MATα ste11::NAT-STM#242 | H99 | (Bahn et al., 2007a) |
| YSB96 | MATa ura5 crg1::URA5 aca1::NEO | YSB58 × JKH43 | (Bahn et al., 2004) |
| YSB119 | MATα aca1::NAT-STM#43 ura5 ACA1-URA5 | YSB108 | (Bahn et al., 2004) |
| YSB121 | MATa aca1::NEO ura5 ACA1-URA5 | YSB109 | (Bahn et al., 2004) |
| YSB317 | MATα ste50::NAT-STM#296 | H99 | This study |
| YSB318 | MATα ste50::NAT-STM#296 | H99 | This study |
| YSB319 | MATα ste50::NAT-STM#296 | H99 | This study |
| H99 crg1 | M A Tα ura5 crg1::URA 5 | F99 | (Wang et al., 2004) |
| PPW196 | M A Ta ura5 crg1::URA5 | F99a | (Wang et al., 2004) |
| YSB523 | MATa ste50::NEO | KN99a | This study |
| YSB564 | MATα ste50::NAT-STM#296 STE50-NEO- pJAF12 | YSB317 | This study |
| YSB593 | MATα ste50::NAT-STM #224 | JEC21 | This study |
| YSB632 | MATα ste50::NAT-STM #122 ura5 crg1::URA 5 | H99 crg1 | This study |
| YSB637 | MATα ste50::NAT-STM #122 ura5 crg1::URA 5 | PPW196 | This study |
Each NAT-STM# indicates the Nat marker with a unique signature tag.
2.2. cDNA analysis and two-hybrid assay
To perform two-hybrid assay between C. neoformans Ste11 and Ste50, first we cloned cDNAs for STE11 and STE50 genes. Each cDNA was amplified by RT-PCR using total RNA prepared from matings between JEC21 and JEC20 strains (serotype D C. neoformans MATα and MATa strains, respectively) as a template and primers JOHE11932/JOHE11933 for STE50 and B2093/B2094 for STE11 (Table S1). The RT-PCR products were cloned into a pCR2.1-TOPO vector, generating pCR-STE50c and pCR-STE11c, respectively. The cloned cDNA of STE50 and STE11 were sequenced and deposited to GenBank (accession number HQ113107 and HQ113108, respectively).
The cDNA fragment of the STE50 gene was subcloned into plasmid pGBT9 with in-frame fusion to the Gal4 binding domain, generating pGBT-STE50c. The cDNA insert of the STE11 gene was subcloned into pGAD424 with in-frame fusion to the Gal4 activation domain, generating pGAD-STE11c. The two plasmids, pGBT-STE50c and pGAD-STE11c, were cotransformed into the reporter yeast strain PJ69-4A and at least three independent Leu+ Trp+ transformants were selected on SD medium lacking leucine and tryptophan (SD-Leu-Trp). To qualitatively examine protein- interaction, we monitored the growth ability of the transformants on SD-Leu-Trp-His and SD-Leu-Trp-His-Ade media. For quantitative examination, β-galactosidase activity was measured as previously described (Rose and Botstein, 1983). As control plasmids, pGAD-ACA1 and pGBT-CAC1(2126–2260) were used for transformation and yeast two-hybrid assays (data not shown).
2.3. Construction of the ste50Δ mutants
The STE50 gene (CNAG_07507) were disrupted in the serotype A MATα strain H99, the congenic MATa strain KN99a, and the serotype D MATα strain JEC21 genetic backgrounds with a disruption cassette generated by overlap PCR followed by biolistic transformation, as previously described (Bahn et al., 2005a; Davidson et al., 2002). Primers for amplification of the 5′ and 3′ flanking regions of the serotype A and D STE50 genes are listed in Supplementary Table S1. M13Re and M13Fe primers were used to amplify the Natr or Neor dominant selectable marker. Each gel-extracted gene disruption cassette was biolistically transformed into the serotype A strains H99 or KN99a, or the serotype D strain JEC21 (Davidson et al., 2000). NATr or NEOr positive stable transformants were selected on YPD medium containing nourseothricin or G418, respectively. To confirm genotype of the ste50Δ mutants, both diagnostic PCR and Southern blot analysis were performed (Supplementary Fig. S1 and S2). All PCR amplifications were performed using the ExTaq polymerase (Takara, Shiga, Japan).
To construct the ste50Δ crg1Δ double mutant, the ste50 disruption allele constructed by double joint PCR (DJ-PCR) was biolistically transformed in the MATα crg1Δ mutant (H99 crg1) and MATa crg1Δ mutant (PPW 196) strains (Kim et al., 2009). The correct genotype of the ste50Δ crg1Δ double mutants was confirmed by diagnostic PCR and Southern blot analysis (Supplementary Fig. S3).
2.4. Construction of the ste50Δ+STE50 complemented strain
To verify the phenotypes of the ste50Δ mutants we constructed, the ste50Δ+STE50 complemented strains were generated as follows. First, the full-length STE50 gene, which contains 895 bp of the 5′-untranslated region (UTR), 2699 bp of the STE50 open reading frame (ORF), and 168 bp of the 3′-UTR, was amplified by PCR with H99 genomic DNAs as templates and primers B1360 and B1361 containing a NotI recognition site and directly cloned into the TOPO vector (Invitrogen) to generate plasmid pCR-STE50. After confirming the DNA sequence, the STE50 insert was subcloned into the plasmid pJAF12 (NEOr) to produce the plasmid pJAF12-STE50. For the targeted re-integration of the wild-type STE50 allele into its native locus, pJAF12-STE50 was linearized by NsiI (New England Biolabs) digestion and biolistically transformed into the ste50Δ mutant (YSB317).
2.5. Southern blot analysis
Genomic DNAs were isolated from each strain as follows. Cells were grown in YPD medium overnight (50 ml culture), spun-down, and lyophilized. The lyophilized cell pellet was vigorously vortexed with 3 mm glassbeads (Sigmund Lindner) in 10 ml of CTAB (Cetyl trimethylammonium bromide, Sigma) extraction buffer (100 mM Tris-HCl pH7.5, 0.7 M NaCl, 10 mM EDTA, 1% CTAB, 1% β-mercaptoethanol), incubated at 65°C for 30 min, and cooled to room temperature. Then the cell extracts were mixed with 10 ml chloroform and spun down for phase separation. Genomic DNA was precipitated from the aqueous phase with an equal volume of isopropanol, washed with 70% ethanol, and resuspended in TE buffer (10mM Tris and 1mM EDTA) containing RNase H. Isolated genomic DNA was digested with the indicated restriction enzymes (i.e. HindIII for checking deletion of the STE50 gene in both serotype A and D). The digested genomic DNAs were separated by 1% agarose gel-electrophoresis, denatured in 0.4 N NaOH, transferred to the nylon membrane (GE) in 0.4 N NaOH and 1 M NaCl, and fixed by 1200 J/m2 UV exposure. The membrane was hybridized with modified church hybridization solution (1mM EDTA, 0.25M Na2HPO4, 1% hydrolysated casein, 7% SDS, 6% H3PO4) containing STE50-specific probes that were PCR-amplified with primers listed in Supplementary Table 1 and radiolabelled by Amersham Rediprime II Random Prime Labelling System (GE), washed for 10 min with each washing solution I (2×SSC buffer and 0.1% SDS), solution II (2×SSC buffer and 0.1% SDS), and solution III (0.5× SSC buffer and 0.1% SDS), then the membrane was exposed to autoradiography film, overnight.
2.6. Northern blot analysis for monitoring pheromone gene expression during mating
The MATα and MATa C. neoformans strains were grown in YPD medium for 16 hrs at 30°C and equal concentration of cells (107 cells/ml) were resuspended in water. The α and a mixtures were spread onto V8 medium and incubated in the dark at room temperature for 1 days. Cells were pelleted at 4°C, frozen at −80°C, and lyophilized. Total RNA was isolated using Trizol (Invitrogen). Ten μg of RNA was separated in a 1% agarose gel made with DEPC (diethyl pyrocarbonate)-treated water and 1× MOPS running buffer. The gel was washed three times with distilled water, transferred to a nylon membrane using 20x SSC transfer buffer. The membrane were hybridized with pheromone gene-specific probes that were PCR-amplified with the primers listed in Supplementary Table 1 and radioactively labeled with Rediprime kit (GE), washed and developed as described in for the Southern blot analysis.
2.7. Assays for capsule and melanin production
For capsule induction, each C. neoformans strain was incubated for 16 hrs at 30°C in YPD medium, spotted onto agar-based DME medium, and further incubated for 2 days at 37°C. After incubation, capsule production levels were measured qualitatively using India ink staining and quantitatively by comparing the size of capsule for each mutant by microscopically measuring diameters of the capsule and the cell using an Olympus BX51 microscope equipped with a SPOT Insight digital camera (Diagnostic Instrument Inc.) as described previously (Bahn et al., 2004). For additional quantitative measurement of capsule size, packed cell volume was also measured by using hematocrit capillary tubes as described previously (Alspaugh et al., 2002). Briefly, cells grown on DME medium for 2 days were scraped from the medium, washed with PBS (Phosphate buffer saline) to remove released polysaccharide, and fixed with 10% formalin. Cell concentration was determined by using hemocytometer and adjusted to 1×109 cells/ml with PBS buffer. Forty microliters of the cell suspension was loaded into Microhematocrit capillary tubes (HIRSCHMANN LABOGERÄTE No. 9100275 Germany) with clay and parafilm sealed tips to prevent evaporation of the medium during incubation. The capillary tubes were placed vertically overnight at room temperature to allow cell packing by gravity. The packed volume of cells was measured by calculating the ratio of the length of packed cell volume phase/length of total volume phase. Two or three independent triplicate experiments were performed. Statistical difference in relative capsule size between strains was determined by Bonferroni’s multiple comparison test by using Prism 4 software (GraphPad Software). For melanin production, cells were spotted onto Niger seed medium or L-DOPA medium containing 0.1% glucose and incubated for up to 7 days at 30°C or 37°C. Melanin production was monitored and photographed daily.
2.8. Mating, cell fusion, and confrontation assays
All C. neoformans strains tested for mating, cell fusion, and confrontation assays were initially grown in YPD medium for 16 hrs at 30°C and resuspended in water. For mating, equal concentrations of MATα and MATa cells (107 cells/ml) were mixed, spotted (5 μl) onto V8 mating medium, and incubated in the dark at room temperature for 1 to 2 weeks. The spotted mating mixtures were monitored weekly for filamentation and photographed using an Olympus BX51 microscope equipped with a SPOT Insight digital camera (Diagnostic Instrument Inc.). Cell fusion efficiency was measured as previously described with minor modifications (Bahn et al., 2004). Briefly, 107 cells/ml of each MATα and MATa strain containing Natr or Neor markers, respectively, were mixed in an equal volume and 5 μl of this cell mixture was spotted onto V8 medium and incubated for 24 hrs at room temperature in the dark. The cells were scraped from the medium, resuspended in 1 ml dH2O, and 200 μl of cell suspension was plated onto YPD medium containing nourseothricin and G418. The number of colonies on each plate was determined after 4 days of incubation at room temperature. As control strains, YSB119 (Natr wild-type strain) and YSB121 (Neor wild-type strain) were used. For the confrontation assay to monitor pheromone production and response of cells, α cells were streaked in confrontation with a cells on V8 medium and incubated for 10 days at room temperature in the dark. Images of mating and confrontation assays were captured with an Olympus BX51 microscope equipped with a SPOT Insight digital camera (Diagnostic Instrument Inc.).
2.9. Sensitivity test for stress responses
Each strain was incubated overnight (about 16 hrs) at 30°C in YPD medium, serially diluted (1 to 104 dilutions) in dH2O, and spotted (4 μl) onto solid YP or YPD medium. To test osmosensitivity, cells were spotted on YP containing 0.5 M, 1 M, and 1.5 M of NaCl or KCl, or 1.5–2 M sorbitol. To test genotoxic DNA damaging stress, cells were spotted on solid YPD medium and exposed to 200, 300, or 400 J/m2 of UV by using a UV crosslinker (UVP), or spotted on YPD medium containing 10, 30, 50 mM hydroxyurea (HU) and 0.01, 0.02, 0.03% methylmethane sulfonate (MMS). To test temperature sensitivity, plates were incubated at 25, 30, 37, and 40°C. To examine oxidative stress, cells were spotted on YPD medium containing 2, 3, and 4 mM diamide and 2.5, 3, 3.5 mM hydrogen peroxide (H2O2) (Junsei, Tokyo, Japan). To test heavy metal stress and toxic metabolite sensitivity, cells were spotted on YPD medium containing 15, 22.5, and 30 μM cadmium sulfate (CdSO4) (Sigma, Saint Louis, MO) or methylglyoxal (MG). To test cell wall and membrane integrity, cells were spotted on YPD medium containing 0.01, 0.02, and 0.03% sodium dodecyl sulfate (SDS), dithiothreitol (DTT), and 0.2, 0.5, and 0.7% Congo red. To test antifungal drug susceptibility, the cells were spotted on YPD medium containing the indicated concentration of polyene (amphotericin B), azole (fluconazole, ketoconazole, Itraconazole), flucytosine, and phenylpyrrole (fludioxonil) drugs (Sigma). Each plate was incubated for 2–5 days, and photographed during the incubation period.
2.10. Virulence assays
All animal experiments were done at the University of Minnesota in strict accordance with good animal practice as defined by the National Institutes of Health Office of Animal Welfare (OLAW), and the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All experiments were reviewed and approved by the University of Minnesota Institutional Animal Care and Use Committee (IACUC) under protocol number 0712A22250.
The wild-type strain (H99), ste50Δ mutant (YSB317), and its complemented strain (YSB564) were cultured overnight in YPD broth. The resulting yeast cells were pelleted and resuspended in sterile PBS at a concentration of 1x106 cells/ml based on hemocytometer count. Mice were anesthetized by intraperitoneal injection. Animals were infected intranasally with 5 × 104 cells in 50 μl PBS. The concentration of cells in the inoculum was confirmed by plating serial dilutions and enumerating colony forming units (CFUs). Mice were monitored daily and those that showed signs of severe morbidity (weight loss, extension of the cerebral portion of the cranium, abnormal gait, paralysis, seizures, convulsion, or coma) were sacrificed by CO2 inhalation. Two independent experiments were preformed. In one experiment, groups of ten 6-week-old female C57/BL6 mice (Jackson Laboratories, Bar Harbor, ME) were infected as described above and monitored for time until morbidity. In a second experiment, groups of 7-week-old female A/J mice (Jackson Laboratories, Bar Harbor, ME) (8 mice for the wild-type strain, ten mice for the ste50Δ mutant, and six mice for the ste50Δ+STE50 strain) were infected as described above and monitored for time until morbidity. In this second experiment all mice were included in the survival data presented in Figure 7 as well as for statistical analysis. For statistical test, we used the Mann-Whitney U test (also called the Mann-Whitney-Wilcoxon, the Wilcoxon rank-sum, or the Wilcoxon-Mann-Whitney test), which is a non-parametric significance test. P-values <0.05 were considered significant.
Fig. 7.
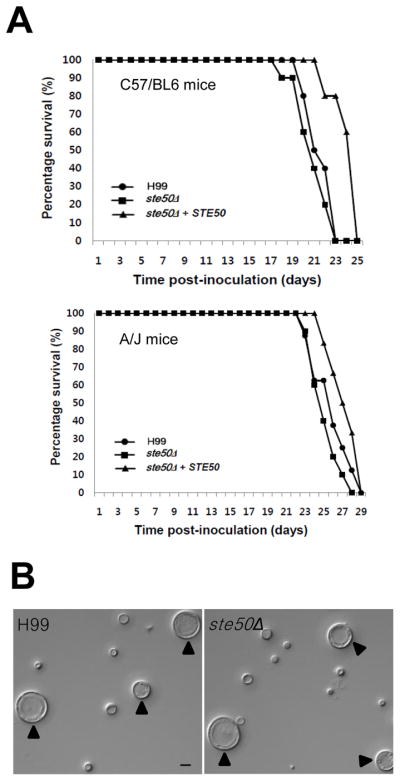
Ste50 is dispensable for virulence and titan cell formation of C. neoformans. (A) For virulence assays, groups of ten C57/BL6 or groups of six-ten A/J mice (see Materials and Methods) were infected with 5×104 cells of MATα WT (●: H99), ste50Δ (■: YSB317), and ste50Δ+STE50 complemented (▴: YSB564) strains by intranasal inhalation. Percent survival (%) was monitored daily until all mice were sacrificed. The ste50Δ mutant is as virulent as the WT strain. In both experiments, the P-values for the C57/BL6 experiment were: 0.35, 0.001, and 0.001 for H99/ste50Δ, ste50Δ/ste50Δ+STE50, and H99/ste50Δ+STE50, respectively. P-values for the A/J experiment were 0.57, 0.03, and 0.23 for H99/ste50Δ, ste50Δ/ste50Δ+STE50, and H99/ste50Δ+STE50, respectively. (B) For titan cell formation assay, A/J mice were infected with 1×106 cells of the wild-type (H99) and ste50Δ mutant (YSB317) strains and were sacrificed at 3 days post-infection. Cells in the lung lavage fluid were fixed with 3.7% formaldehyde and photographed. Bar = 10 μm, arrows denote titan cells.
To monitor titan cell formation in the wild-type (H99), ste50Δ mutant (YSB317), and complemented (YSB564) strains, the yeast cells were resuspended in sterile PBS at a concentration of 2×107 cells/ml based on hemocytometer count. Groups of two 7-week-old female A/J mice were anesthetized by intraperitoneal pentobarbital injection. Animals were infected intranasally with 1 × 106 cells in 50 μl PBS. Infected mice were sacrificed at 3 days post-infection by CO2 inhalation. Lungs were lavaged with 1.5 ml sterile PBS three times using a 20 gauge needle placed in the trachea. Cells in the lavage fluid were pelleted at 16,000g, resuspended in 3.7% formaldehyde, and washed with PBS. >500 cells per animal were analyzed for size by microscopy using an AxioImager microscope (Zeiss, Inc).
3. Results
3.1. Identification of the Ste50 homologue in C. neoformans
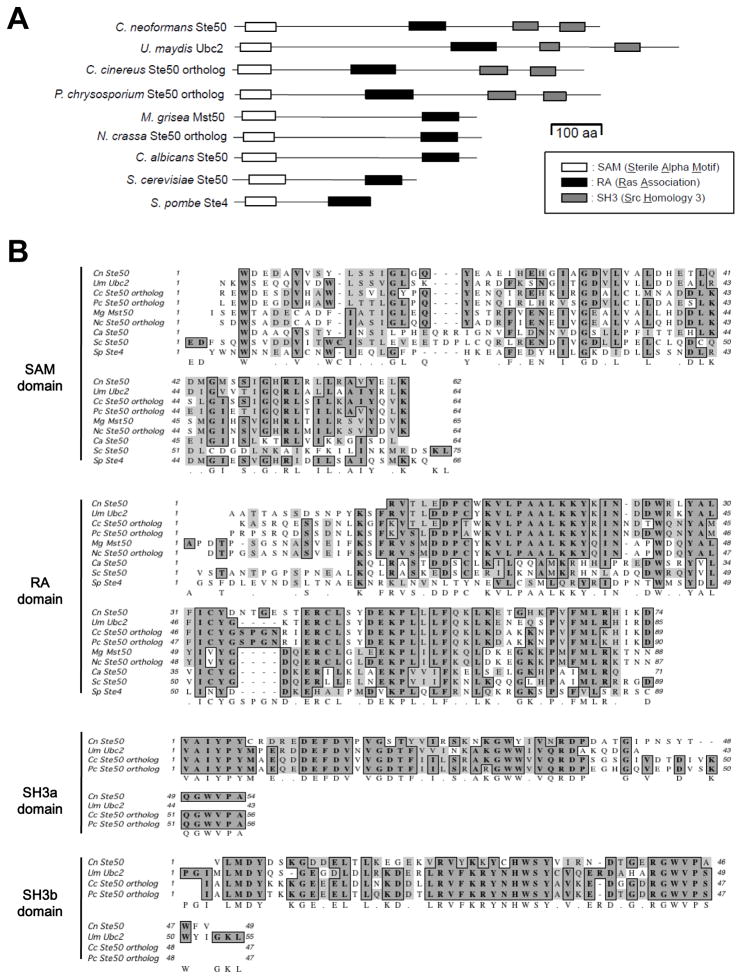
To identify the Ste50 ortholog in C. neoformans, we performed BLAST searches (tblastn) with the S. cerevisiae Ste50 protein sequence. In both serotype A (H99 strain) and D (JEC21 strain), a single Ste50 ortholog was discovered with a relatively low identity (score [Bits] 69.32; E-value, 1.12344e−12; identity 6.8%). The H99 (CNAG_07507.2) and JEC21 (CNC05800, 179.m00725) STE50 orthologous genes were predicted to encode proteins of 706 and 700 amino acids (aa), respectively. In order to investigate the detailed genomic structure of the STE50 gene, we performed cDNA analysis of the STE50 gene in the JEC21 strain. The cDNA analysis revealed that the open reading frame (ORF) of the STE50 gene consists of 12 exons and 11 introns and encodes a protein with 700 amino acids, corroborating the prediction from the JEC21 genome database. The predicted genomic structure of the STE50 gene in the H99 strain also consists of 12 exons and 11 introns (Broad Institute). Regardless of the low identity and similarity compared with Ste50 orthologs in other fungi, protein domain analysis by Pfam (Wellcome trust Sanger Institute, http://pfam.sanger.ac.uk/) revealed that the predicted C. neoformans Ste50 ortholog contains typical Ste50-functional domains, including a SAM (sterile alpha motif) domain at the N-terminal region, a RA (Ras-association) domain in the middle region, and two SH3 domains at the C-terminus (Fig. 1A and 1B). The C-terminal SH3 domains are thought to be basidiomycota-specific, has they have only been identified in Ste50 orthologs of basidiomycetous fungi (Klosterman et al., 2008).
Fig. 1.
Comparison of Ste50 orthologs between C. neoformans and other fungi. (A) Each Ste50 ortholog diagram shows functional protein domains, which were identified by the Pfam 24.0 database (http://pfam.sanger.ac.uk/). Each domain indicates the following: SAM, sterile alpha motif; RA, Ras- associated; SH3, Src homology 3. (B) Multiple sequence alignment of Ste50 orthologs is depicted by Clustal W alignment from MacVector software (versions 7.2.3, Accelrys). Protein sequences of Ste50 orthologs were retrieved from the following database: C. neoformans Ste50 - CNAG_07507 from the C. neoformans var. grubii H99 database of the Broad Institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html); Ustilago maydis Ubc2 - GenBank accession number AAK4932; Coprinus cinereus - locus CC1G_00975.3 from the C. cinereus Sequencing Project Database of the Broad Institute. (http://www.broadinstitute.org/annotation/genome/coprinus_cinereus/MultiHome.html); Phanerochaete chrysosporium Ste50 ortholog - protein ID 2321 at locus Phchr1/scaffold_3:1012644-101605 from the sequencing data produced by the United States Department of Energy, Joint Genome Institute (http://genome.jgi-psf.org/Phchr1/Phchr1.home.html); Neurospora crassa Ste50 ortholog - GenBank accession number XP_956774; Magnaporthe grisae Mst50 - GenBank accession number XP_359578; Candida albicans Ste50 - GenBank accession number XP_721713; Saccharomyces cerevisiae Ste50 - GenBank accession number NP_009898; Schizosaccharomyces pombe Ste4; GenBank accession number CAB38684.
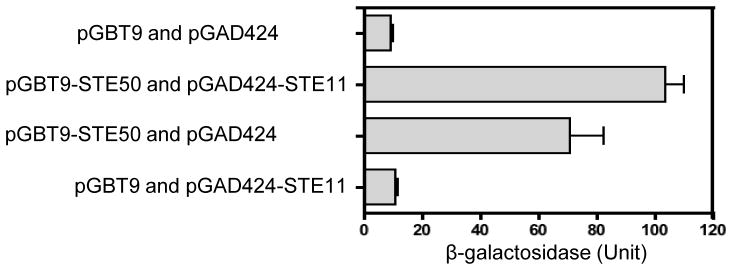
In S. cerevisiae, Ste50 regulates a diversity of responses including pheromone response, osmotolerance, and filamentous/invasive growth by interacting with components of the MAPK pathway, including the Ste11 MAPKKK (Posas et al., 1998; Rad et al., 1992; Ramezani Rad et al., 1998). The interaction of Ste50 with Ste11MAPKKK is also well conserved in filamentous fungi (Barr et al., 1996; Wu et al., 1999). Therefore, we examined the Ste50-Ste11 interaction to further confirm that the C. neoformans gene (CNC05800 in JEC21 strain or CNAG_07507.2 in H99 strain) is a Ste50 ortholog. Using the full-length cDNA of the STE50 and STE11 genes from serotype D JEC21 and JEC20 strains, we constructed pGBT-STE50 and pGAD-STE11 plasmids and performed a yeast two-hybrid assay as described in Materials and Methods. Quantitative measurement of protein interaction by β-galactosidase assay showed that Ste50 interacts with Ste11 in C. neoformans (Fig. 2). However, interaction between Ste50 and Ste11 appeared to be weak in C. neoformans since the reporter yeast strain PJ69-4A cotransformed with pGBT-STE50 and pGAD-STE11 grew in SD-Leu-Trp-His medium, but not in SD-Leu-Trp-His-Ade medium (data not shown). Interestingly, Ste50 itself has a weak transcriptional activation activity (Fig. 2). Therefore, based on sequence homology and its interaction with Ste11 MAPKKK, we named the C. neoformans gene as STE50.
Fig. 2.
Ste50 interacts with Ste11 in C. neoformans. Two-hybrid assay was performed with C. neoformans Ste50 and Ste11. β-galactosidase activity of a LacZ reporter gene in the PJ69-4A strain co-transformed with indicated vectors were measured with extracts of two independent Leu+ Trp+ transformants cultured in SD-Leu-Trp medium. Error bar indicates standard deviation.
3.2. Ste50 is not involved in stress response in C. neoformans
To characterize the function of Ste50 in C. neoformans, we constructed ste50Δ mutants in the serotype A H99 (MATα) and KN99a (MATa) strains, and the serotype D JEC21 (MATα) strain. The gene disruption cassette was generated by overlap PCR and introduced into each strain by biolistic transformation as described in Materials and Methods. Targeted, non-ectopic, integration of the STE50-disruption allele was confirmed by both diagnostic PCR and Southern hybridization (data not shown). To verify phenotypes observed in the ste50Δ mutants, we constructed multiple independent ste50Δ mutants in each strain background and also constructed ste50Δ+STE50 complemented strains, in which the wild-type STE50 gene was re-integrated into the native locus of the STE50 gene as described in Materials and Methods.
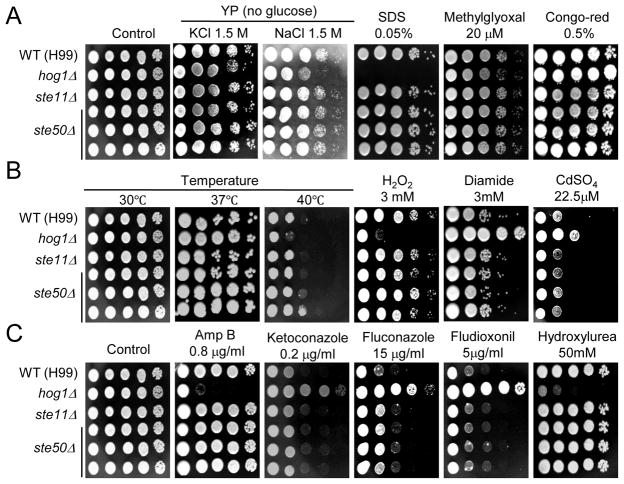
Since Ste50 is involved in stress response via the HOG pathway in S. cerevisiae, we monitored the capability of the ste50Δ mutant to resist a variety of external stresses that are controlled by the HOG pathway in C. neoformans (Bahn et al., 2007a; Bahn et al., 2005b; Bahn et al., 2006). Unlike the control HOG pathway mutants, the ste50Δ mutant was as resistant to osmotic stress as the wild-type strain (Fig. 3A). Furthermore, Ste50 appeared not to be involved in maintenance of cell wall and membrane integrity and detoxification of toxic metabolite since the ste50Δ mutant did not exhibit increased sensitivity to SDS, Congo red, and methylglyoxal (Fig. 3A). In test to monitor oxidative stress response, the ste50Δ mutant exhibited WT-levels of resistance to hydrogen peroxide and diamide (Fig. 3B). In contrast, the control hog1Δ mutant showed increased sensitivity and resistance to H2O2 and diamide, respectively, as reported previously (Fig. 3B). In addition, the ste50Δ mutant showed a wild-type response to heavy metal stress. In response to a variety of antifungal drugs, including polyene (amphotericin B), azoles (fluconazole, ketoconazole), and a phenylpyrrole-class fungicide (fludioxonil), the ste50Δ mutant exhibited wild-type levels of sensitivity, which was clearly distinguished from the control HOG pathway mutants (Ko et al., 2009) (Fig. 3C). Furthermore, the ste50Δ mutant was as resistant to hydroxyurea and methylmethane sulfonate as the wild-type strain (Fig. 3C and data not shown). Finally, the ste50Δ mutant did not exhibit any increased thermosensitivity, in contrast to the control hog1Δ mutant. Taken together, Ste50 is not involved in any of the known stress responses in C. neoformans.
Fig. 3.
Ste50 is not involved in the C. neoformans stress response. C. neoformans strains (the wild-type strain H99 and hog1Δ (YSB64), ste11Δ (YSB313), and ste50Δ (YSB317, YSB318, and YSB319) mutants) was grown overnight at 30°C in liquid YPD medium, 10-fold serially diluted (1–104 dilutions), and spotted (3 μl of dilution) on YPD or YP agar containing the indicated concentrations of KCl, NaCl, SDS, Methylglyoxal, Congo red (A), hydrogen peroxide (H2O2), diamide, CdSO4 (B), amphotericin B, ketoconazole, fluconazole, fludioxonil, and hydroxyurea (C). Cells were incubated at 30°C for 72 h and photographed. For the thermotolerance test (B), cells were spotted on YPD medium and incubated at 40°C for 4 days.
3.3. The role of Ste50 in melanin and capsule production in C. neoformans
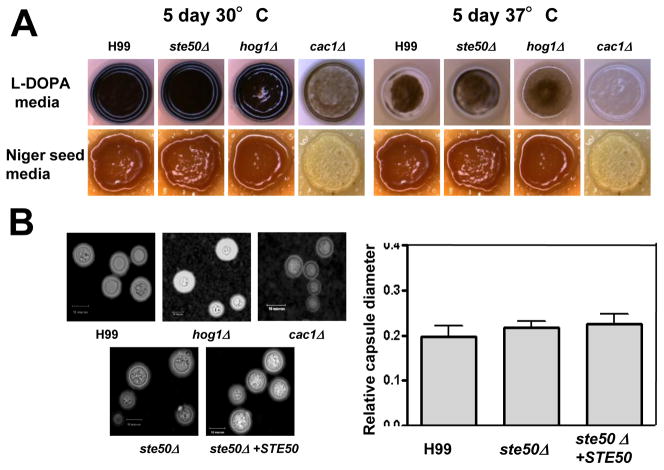
In C. neoformans, the HOG pathway mutants, such as hog1Δ, pbs2Δ, ssk2Δ, and ssk1Δ mutants, are enhanced in melanin and capsule production by controlling expression of LAC1 and CAP10, CAP59, CAP60, and CAP64 genes, respectively (Bahn et al., 2007a; Bahn et al., 2005b; Bahn et al., 2006; Ko et al., 2009). Therefore, we characterized the role of Ste50 for melanin and capsule production of C. neoformans. Unlike the control HOG pathway mutants, the ste50Δ mutant did not show any enhanced melanin and capsule production (Fig. 4).
Fig. 4.
Ste50 is not involved in capsule and melanin production of C. neoformans. (A) The following strains were spotted and grown on L-DOPA medium (glucose 0.1%) and Niger seed medium (glucose 0.1%) at 30°C or 37°C for 5 days: WT (H99) and ste50Δ (YSB317), hog1Δ (YSB64) and cac1Δ (YSB42) mutant strains. (B) The WT strain H99, hog1Δ (YSB64), cac1Δ (YSB42), ste50Δ (YSB317) and ste50Δ + STE50 (YSB564) strains were spotted and cultured on DME medium for capsule production at 37°C for 2 days. Cells were scraped, resuspended in distilled water, and visualized by India ink staining The packed volume of the cells (the wild-type strain [H99] and ste50Δ [YSB317] and ste50Δ + STE50 [YSB564] strains) was measured by calculating the ratio of the length of packed cell volume phase/length of total volume phase. Statistical differences in relative capsule size between strains was determined by Bonferroni’s multiple comparison test.
3.4. Ste50 is required for sexual reproduction of C. neoformans
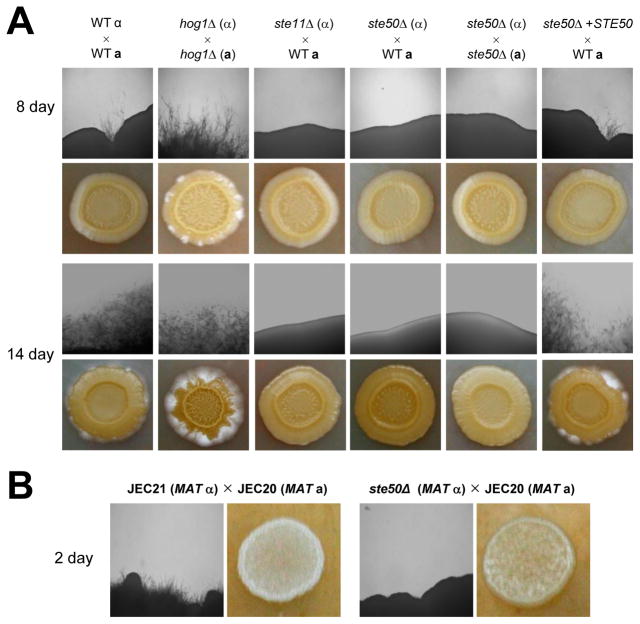
The finding that Ste50 is not involved in stress response, including thermotolerance, and virulence factor production, indicates that Ste50 functions independently from the HOG and Ras/cAMP pathways. We addressed whether Ste50 is involved in sexual differentiation of C. neoformans via the pheromone-response MAPK pathway since Ste50 is required for filamentous growth of both ascomycete and basidiomycete fungi as an adaptor protein for Ste11-like MAPKKK (Klosterman et al., 2008; Ramezani Rad et al., 1998). Our results clearly demonstrated that Ste50 was required for sexual differentiation of both serotype A and D C. neoformans. Even in unilateral crosses with the serotype A wild-type strains (H99 or KN99a strain), ste50Δ mutants exhibited severe mating defects on V8 mating medium (Fig. 5A). The ste50Δ mutant also exhibited a profound mating defect when it is mixed with the serotype D JEC20 (MATa) strain (data not shown). The mating defect observed in the ste50Δ mutant was restored to wild-type levels by introduction of the wild-type STE50 gene (Fig. 5A), further verifying the role of Ste50 in sexual differentiation.
Fig. 5.
Ste50 is required for sexual differentiation in both serotype A and serotype D C. neoformans strains. (A) Serotype A MATα and MATa strains were co-cultured on V8 medium (pH 5.0) for 2 weeks at room temperature in the dark: WT α × WT a (H99 and KN99), hog1Δ (α) × hog1Δ (a) (YSB64 and YSB81), ste11Δ (α)× WT (a) (YSB313 and KN99), ste50Δ (α) × WT (a) (YSB317 and KN99), ste50Δ (α) × ste50Δ (a) (YSB317 and YSB523), and ste50Δ + STE50 (α) × WT (a) (YSB564 and KN99). The images were photographed after 8 days and 14 days. (B) The following serotype D strains were co-cultured on V8 medium in the dark at room temperature up to 5 days and photographed after 2 days: JEC21 and JEC20 (α × a), YSB593 and JEC20 (ste50Δ × a)
We also investigated whether Ste50 is required for mating in the serotype D strain. Similar to the serotype A ste50Δ mutant, the serotype D ste50Δ mutant (JEC21 strain background) exhibited severe mating defects in unilateral crosses with the MATa JEC20 strain (Fig. 5B). These results strongly indicated that Ste50 is indispensable for sexual reproduction in C. neoformans serotype A and D strains.
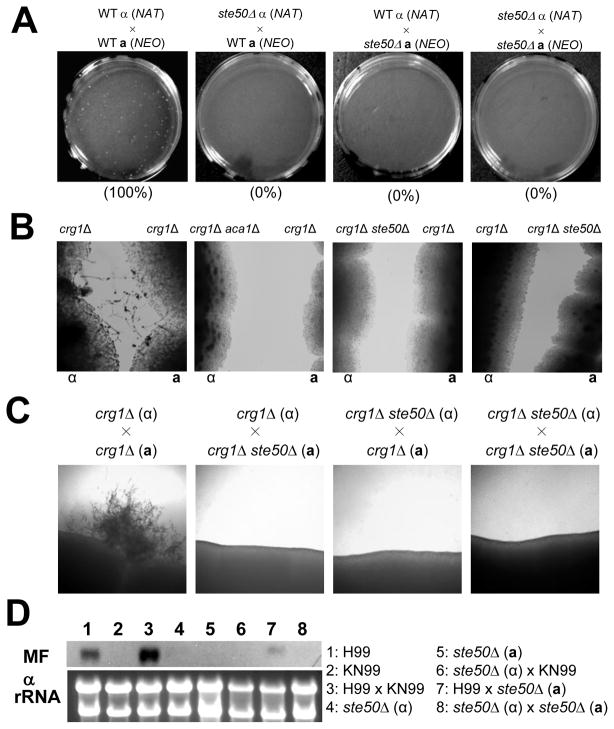
Next we investigated which mating step is impaired in the ste50Δ mutant. Mating in C. neoformans is achieved through the following steps. First, two α and a cells undergo cellular fusion in response to pheromone and initiate filamentous growth. The two dikaryotic nuclei (α and a ) progress along the filamentous structure using clamp connections until a basidium is formed and nuclear fusion (karyogamy) occurs. The diploid in the basidium undergoes meiosis to form four chains of basidiospores. As can be seen in Fig. 5, normal filamentous structures were not observed when the ste50Δ mutant is mated with wild-type strains, indicating that the ste50Δ mutant is impaired at early stages of mating. Therefore, we monitored the ability of the ste50Δ mutant to undergo cell fusion. YSB119 (MATα and Natr) and YSB121 (MATa and Neor) were used as control strains (Bahn et al., 2004). Both unilateral and bilateral mating crosses between the MATα and Natr and MATa and Neor ste50Δ mutants were performed (Table 1). Quantitative measurement of Natr Neor dikaryonic cell fusion products showed that ste50Δ mutants did not generate any cell fusion products, indicating that Ste50 is required for the initial cell-to-cell fusion process during mating (Fig. 6A).
Fig. 6.
Ste50 governs sexual differentiation via the pheromone-responsive Cpk1 MAPK pathway. (A) Cell-cell fusion assays were performed with the following strains: WT α (NAT) × WT a (NEO) (YSB119 and YSB121), ste50Δ α (NAT) × WT a (NEO) (YSB317 and YSB121), WT α (NAT) × ste50Δ a (NEO) (YSB119 and YSB522), ste50Δ α (NAT) × ste50Δ a (NEO) (YSB317 and YSB522). Cell fusion efficiency for each experimental set was calculated relative to the control strains (WT α (NAT) × WT a (NEO)). (B) Confrontation assays were performed with the following strains: MATα crg1Δ (H99 crg1), MATa crg1Δ (PPW 196), MATα crg1Δ aca1Δ (YSB96), MATα crg1Δ ste50Δ (YSB632), and MATa crg1Δ ste50Δ (YSB637). Indicated strains were streaked in confrontation with each other on V8 agar medium and incubated at room temperature in the dark. Images were photographed after 10 days. (C) Mating reactions were initiated for the following strains: The crg1Δ ste50Δ double mutants showed mating defect in both unilateral crossing and bilateral crossing. crg1Δ α × crg1Δ a (H99 crg1 and PPW 196), crg1Δ α × crg1Δ ste50Δ a (H99 crg1 and YSB637), crg1Δ ste50Δ α × crg1Δ a (YSB632 and PPW 196), crg1Δ ste50Δ α × crg1Δ ste50Δ a (YSB632 and YSB637). The images were photographed after 12 days. (D) Northern blot analysis for monitoring pheromone gene expression was performed with total RNA isolated from solo- or co-cultures of the indicated strain(s) grown for 24 hr under mating conditions: WTα (H99), WTa (KN99a), ste50Δα (YSB317), and ste50Δa (YSB522). The blot was probed with the MFα1 gene.
Next we addressed whether the defective cell-to-cell fusion results from abnormal cell fusion per se or defective pheromone sensing or production. Here we provide several lines of evidence indicating that the defective mating of the ste50Δ mutant results from inactivation of pheromone sensing and production. First, mutation of the STE50 gene blocked induction of pheromone-mediated conjugation tubes in crg1Δ mutants, which lack an RGS protein that plays a role in densensitizing the pheromone-responsive pathway (Fig. 6B). C. neoformans strains deleted for the CRG1 gene are hypersensitive to pheromone and form conjugation tubes when MATα and a crg1Δ mutants are confronted with each other (Nielsen et al., 2003; Wang et al., 2004). When the MATα ste50Δ crg1Δ double mutant was confronted by the MATa crg1Δ mutant, neither the ste50Δ crg1Δ double mutant nor the crg1Δ mutant strains formed conjugation tubes (Fig. 6B). Similarly, the MATα crg1Δ mutant did not form conjugation tubes when confronted with the MATa ste50Δ crg1Δ double mutant (Fig. 6B). Second, mutation of STE50 completely blocked sexual differentiation of the crg1Δ mutant in both unilateral and bilateral crosses (Fig. 6C). These data indicate that mutation of STE50 blocked pheromone production and sensing by the crg1Δ mutant, which suggests Ste50 acts downstream of Crg1 in the pheromone-responsive MAPK pathway. Third, expression of the mating pheromone gene (MFα1) was strongly repressed by mutation of the STE50 gene. Transcript levels of MFα1 in unilateral mating crosses with the ste50Δ mutant were dramatically decreased compared to wild-type crosses (Fig. 6D).
Crg1 negatively regulates melanin production under high glucose and temperature (37°C) conditions (Wang et al., 2004) in a manner independent of the pheromone-responsive Cpk1 MAPK pathway. In order to address whether Ste50 also works downstream of Crg1 in a Cpk1-independent manner, we tested whether mutation of STE50 could suppress the increase in melanin production observed in the crg1Δ mutant. We found that mutation of the STE50 gene was unable to suppress the increased melanin production of the crg1Δ mutant (data not shown), indicating that Ste50 acts downstream of Crg1 only for the mating process, but not for the melanin production.
3.5. Ste50 is not required for virulence and titan cell formation in C. neoformans
Ubc2, a Ste50 ortholog in Ustilago maydis, not only controls filamentous growth and mating, but also governs pathogenicity through the basidiomycete-specific carboxy terminal extension containing SH3 domains (Klosterman et al., 2008). The fact that C. neoformans Ste50 also contains the SH3 domains in the carboxy terminus prompted us to test its role in virulence. We used a murine inhalational model of cryptococosis as described in Materials and Methods. Unlike the U. maydis ubc2Δ mutant, the C. neoformans ste50Δ mutant was not attenuated in virulence compared to the wild type strain in a C57/BL6 mouse model (Fig. 7A, upper panel). When tested in another mouse genetic background (A/J), the ste50Δ mutant and ste50Δ+STE50 exhibited wild-type levels of virulence (Fig. 7A, lower panel). Therefore, we concluded that Ste50 is not required for virulence of C. neoformans regardless of the presence of the basidiomycete-specific SH3 domains in the carboxy terminus.
Pheromone sensing has also been implicated in formation of enlarged cells, known as titan cells, during the pulmonary infection (Okagaki et al., 2010; Zaragoza et al., 2010). Because Ste50 is required for mating and morphological differentiation, we tested the ste50Δ mutation for its affect on titan cell formation. The wild-type (H99) and ste50Δ mutant strains all showed equivalent levels of titan cell formation 3 days post-infection (Fig. 7B), which suggests Ste50 is not involved in titan cell formation and likely acts further downstream in the mating pathway.
4. Discussion
The present study aimed to identify and functionally characterize a key signaling mediator that contributes to cross-talk between signaling pathways in C. neoformans. For this purpose, we chose to investigate the role of the Ste50 adaptor protein in C. neoformans because Ste50 is known to be involved in multiple signaling pathways in the budding model yeast S. cerevisiae. In S. cerevisiae, Ste50 is involved in filamentous growth and cell wall integrity signaling via the Ste11-dependent pathway (Lee and Elion, 1999; Ramezani Rad et al., 1998), osmoadaptation signaling via the HOG pathway (O'Rourke and Herskowitz, 1998; Posas et al., 1998), and stress-tolerance signaling by interacting with the Ras-cAMP signaling pathway (Poplinski et al., 2007). These multiple functions of Ste50 are potentially due to the presence of SAM (Sterile-Alpha-Motif) and RA (Ras-Association) domains in the adaptor protein. Recently, Klosterman and co-workers reported that the Ste50 ortholog in basidiomycete fungi is further extended at the C-terminus and contains additional functional domains, such as SH3 domain (Klosterman et al., 2008). Notably, Ubc2, a Ste50 ortholog in U. maydis, also controls virulence of the plant fungus through the SH3 domains in the carboxy terminus, but in a manner independent from the SAM or RA domain in the amino terminus (Klosterman et al., 2008). At this point, however, a signaling cascade(s) governed by the SH3 domain of Ubc2 has not been elucidated.
Our study, however, demonstrates that the function of Ste50 in C. neoformans is restricted to mating/filamentous growth. In C. neoformans, Ste50 does not play a role in stress responses and virulence factor production that are known to be controlled by the HOG and Ras/cAMP signaling pathways (Alspaugh et al., 2000; Bahn, 2008; Bahn et al., 2007b; Ko et al., 2009; Pukkila-Worley and Alspaugh, 2004). The HOG pathway not only controls resistance of C. neoformans against diverse stresses, such as osmotic shock, oxidative and heavy metal stress, UV irradiation, DNA damages, and antifungal drug or toxic metabolite treatment, but also regulates sexual reproduction and production of virulence factors such as capsule and melanin (Bahn, 2008; Ko et al., 2009; Maeng et al., 2010). Besides mating and filamentous growth, however, none of phenotypes related to the HOG-signaling pathway were observed in the ste50Δ mutant. Supporting these data, our previous study demonstrates that the Ste11 MAPKKK, which is a Ste50-binding protein in S. cerevisiae, is not involved in activation of the HOG-signaling pathway in C. neoformans (Bahn et al., 2007a). In the model yeast, the Ste11 MAPKKK is activated by the Sho1 transmembrane protein with the help of Ste50 and subsequently activates the Pbs2-Hog1 MAPK pathway to counteract osmotic stress (Posas et al., 1998). However, C. neoformans does not have any proteins homologous to Sho1 in its genome. These observations combined with our previous study further indicate that the Ste50-Ste11 signaling pathway does not activate the Pbs2-Hog1 signaling pathway in C. neoformans. In C. neoformans, the Ras- and cAMP-signaling pathways also control cell survival under a plethora of environmental stresses, mating, and virulence factor production with some connection to the HOG pathway (Bahn, 2008; Maeng et al., 2010). However, none of the Ras- and cAMP-signaling related phenotypes, except mating, were observed in the ste50Δ mutant in C. neoformans.
The involvement of Ste50 in mating and filamentous growth of C. neoformans appears to be mainly mediated by the pheromone-responsive Ste11-Ste7-Cpk1 MAPK signaling pathway based on several findings made by the present study. First, Ste50 weakly interacts with Ste11 based on the results of a yeast two-hybrid assay. Second, similar to the ste11Δ mutant, the ste50Δ mutant did not exhibit any filamentation, even in a unilateral cross with the wild-type strain. The HOG pathway mutants, such as hog1Δ, pbs2Δ, ssk2Δ, or ssk1Δ mutants, show enhanced mating (Bahn et al., 2007a; Bahn et al., 2005b; Bahn et al., 2006). The cAMP mutants, such as aca1Δ, gpa1Δ, or cac1Δ mutants, are only partially impaired in the unilateral mating cross (Bahn et al., 2004). Third, the ste50Δ mutant was almost completely defective in producing mating pheromone and cell fusion in response to a mating partner, indicating that Ste50 is required for pheromone production. Fourth, mutation of STE50 can suppress phenotypes, such as enhanced pheromone production/sensing and mating, of the crg1Δ mutant. Taken together, these data suggest that Ste50 is one of the signaling components in the pheromone-responsive Cpk1 MAPK pathway.
The unique presence of the C-terminal SH3 domains in the Ste50 orthologs of basidiomycete fungi, including C. neoformans, suggests that Ste50 may have specialized functions in the basidiomycota, which have not been noticed in other fungi. Besides this study, the only functional study of Ste50 in the basidiomycota is done in U. maydis by Klosterman and co-workers (Klosterman et al., 2008). The Ste50 ortholog in U. maydis, Ubc2 adaptor protein, interacts with the Ubc4 MAPKKK, a Ste11 ortholog, and is involved in mating and filamentous growth through the SAM and RA domains. Surprisingly, while the two C-terminal SH3 domains are dispensable for mating, they are required for pathogenicity of U. maydis, strongly indicating that Ubc2 has a specialized function in virulence of this important fungal pathogen (Klosterman et al., 2008). In contrast, our study demonstrates that the role of the SH3 domains of Ste50 in fungal virulence is not a generalized phenomenon in the basidiomycota. In C. neoformans, the ste50Δ mutant exhibited wild-type levels of virulence in two independent mouse genetic backgrounds (A/J and C57/BL6 mice). Furthermore, Ste50 was not required for titan cell formation, which has been observed to occur during host infection of C. neoformans (Okagaki et al., 2010; Zaragoza et al., 2010). Taken together, these data indicate that Ste50 is not required for full virulence in this pathogen.
To the best of our knowledge, the function of Ste50 has never been reported in human fungal pathogens. For example, Candida albicans contains a single Ste50 ortholog (Orf19.1636), whose function is not known. However, the functional Ste50 binding partner, Ste11 MAPKKK, has been reported in C. albicans and C. glabrata. C. albicans Ste11 is not involved in stress responses but controls the cell wall damage stress response whereas C. glabrata Ste11 is required for hypotonic adaptation, mating response and virulence (Calcagno et al., 2005; Cheetham et al., 2007). Furthermore, ,functions of other signaling components, Cek1 (Cpk1 or Kss1/Fus3 MAPK ortholog), Hst7 (Ste7 MAPKK ortholog), and Cst20 (Ste20 ortholog), in the corresponding MAPK pathway have been characterized in C. albicans. The Cst20-Hst7-Cek1 MAPK pathway controls morphological transitions of C. albicans under certain filamentation inducing conditions via the Cph1 (Ste12 ortholog) transcription factor (Csank et al., 1998; Leberer et al., 1996). Interestingly, the role of each signaling components in pathogenicity of C. albicans is rather different. The hst7Δ and cph1Δ mutants exhibit WT-levels of virulence. In contrast, the cst20Δ or cek1Δ mutant show attenuated virulence (Csank et al., 1998; Leberer et al., 1996). These data suggest the role of Ste50 in virulence of C. albicans may reveal further differences in Ste50 activity in the pathogenic fungus. Similarly, A. fumigatus also contains a single Ste50 (Afu2g17130) and yet its role is not known. Therefore, the role of Ste50 in virulence and differentiation of human pathogenic fungi needs to be further studied.
In conclusion, the present study identified and functionally characterized the Ste50 ortholog in C. neoformans. Ste50 is involved in pheromone production/sensing and mating processes via the Cpk1 MAPK pathway, but not in diverse stress responses and virulence factor production.
Supplementary Material
Fig. S1. Construction of the serotype A MATα ste50Δ mutant. (A) Diagram for disruption of the STE50 gene in serotype A MATα H99 strain. Primers for the first-round and second-round PCR are indicated as bent arrows. Through recombination between 5′ and 3′ flanking region of STE50 gene, the intact STE50 gene is displaced with nourseothricin-resistant gene. (B) The correct genotype of the ste50 mutants were confirmed by Southern blot analysis using genomic DNA digested with the restriction enzyme EcoR and BamHI.
Fig. S2. Construction of the serotype A MATa ste50Δ mutant. (A) Diagram for disruption of the STE50 gene in serotype A MATa KN99a strain. Primers for the first-round and second-round PCR are indicated as bent arrows. Through recombination between 5′ and 3′ flanking region of the STE50 gene, the intact STE50 gene was displaced with the NEO dominant selectable marker. (B) The correct genotypes of the ste50Δ mutants were confirmed by Southern blot analysis using genomic DNAs digested with the restriction enzyme EcoR .
Fig. S3. Construction of the ste50Δ crg1Δ double mutant. (A) Diagram for disruption of the STE50 gene in H99 crg1 (crg1Δα) and PPW196 (crg1Δa) strains. Primers for the first-round PCR and double joint (DJ)-PCR are indicated as bent arrows. The native STE50 gene is specifically replaced with nourseothricin-resistant gene (NAT) through triple recombination between 5′ and 3′ flanking region of the STE50 gene. (B) The correct genotypes of the ste50Δ crg1Δ double mutants were confirmed by Southern blot analysis using genomic DNAs digested with the restriction enzyme Sac .
Acknowledgments
This work was supported by Pioneer Research Center Program through the National Research Foundation of Korea funded by Ministry of Education, Science and Technology (No. 2009-0081512), the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2009-0063344) (to YSB), and the National Institute of Allergy and Infectious Diseases (NIH) K22 grant AI070152 (to KN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alspaugh JA, et al. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36:352–65. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Alspaugh JA, et al. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol. 1998;25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- Alspaugh JA, et al. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS. Master and Commander in fungal pathogens: The two-component system and the HOG signaling pathway. Eukaryot Cell. 2008;7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, et al. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol. 2005a;15:2013–20. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Bahn YS, et al. Ssk2 mitogen-activated protein kinase kinase kinase governs divergent patterns of the stress-activated Hog1 signaling pathway in Cryptococcus neoformans. Eukaryot Cell. 2007a;6:2278–89. doi: 10.1128/EC.00349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, et al. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell. 2004;3:1476–91. doi: 10.1128/EC.3.6.1476-1491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, et al. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005b;16:2285–300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, et al. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17:3122–35. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, et al. Sensing the environment: lessons from fungi. Nat Rev Microbiol. 2007b;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- Barr MM, et al. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:5597–603. doi: 10.1128/mcb.16.10.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia A, et al. NMR structural studies of the Ste11 SAM domain in the dodecyl phosphocholine micelle. Proteins. 2009;74:328–43. doi: 10.1002/prot.22166. [DOI] [PubMed] [Google Scholar]
- Calcagno AM, et al. Candida glabrata Ste11 is involved in adaptation to hypertonic stress, maintenance of wild-type levels of filamentation and plays a role in virulence. Med Mycol. 2005;43:355–64. doi: 10.1080/13693780400006088. [DOI] [PubMed] [Google Scholar]
- Cheetham J, et al. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol Biol Cell. 2007;18:4603–14. doi: 10.1091/mbc.E07-06-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C, et al. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, et al. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–15. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Davidson RC, et al. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- Davidson RC, et al. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2003;49:469–85. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- Ekiel I, et al. Binding the atypical RA domain of Ste50p to the unfolded Opy2p cytoplasmic tail is essential for the high-osmolarity glycerol pathway. Mol Biol Cell. 2009;20:5117–26. doi: 10.1091/mbc.E09-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, et al. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2008;7:1685–98. doi: 10.1128/EC.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, et al. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol Microbiol. 2005;58:393–408. doi: 10.1111/j.1365-2958.2005.04843.x. [DOI] [PubMed] [Google Scholar]
- Heung LJ, et al. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. J Biol Chem. 2004;279:21144–53. doi: 10.1074/jbc.M312995200. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, et al. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol Biol Cell. 2007;18:3237–49. doi: 10.1091/mbc.E07-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, et al. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3:753–64. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–2. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Kim MS, et al. An efficient gene disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem Biophys Res Commun. 2009;390:983–988. doi: 10.1016/j.bbrc.2009.10.089. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, et al. Ubc2, an ortholog of the yeast Ste50p adaptor, possesses a basidiomycete-specific carboxy terminal extension essential for pathogenicity independent of pheromone response. Mol Plant Microbe Interact. 2008;21:110–21. doi: 10.1094/MPMI-21-1-0110. [DOI] [PubMed] [Google Scholar]
- Ko YJ, et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell. 2009;8:1197–1217. doi: 10.1128/EC.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, et al. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology. 2006;152:591–604. doi: 10.1099/mic.0.28571-0. [DOI] [PubMed] [Google Scholar]
- Kraus PR, et al. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol Microbiol. 2003;48:1377–87. doi: 10.1046/j.1365-2958.2003.03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, et al. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A. 1996;93:13217–22. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci U S A. 1999;96:12679–84. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- Maeng S, et al. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell. 2010;9:360–78. doi: 10.1128/EC.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TD, Edman JC. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–70. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun. 2003;71:4831–41. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–86. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, et al. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, et al. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–9. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplinski A, et al. Ste50 adaptor protein influences Ras/cAMP-driven stress-response and cell survival in Saccharomyces cerevisiae. Curr Genet. 2007;51:257–68. doi: 10.1007/s00294-007-0124-3. [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–5. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Posas F, et al. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol. 1998;18:5788–96. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Alspaugh JA. Cyclic AMP signaling in Cryptococcus neoformans. FEMS Yeast Res. 2004;4:361–7. doi: 10.1016/S1567-1356(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Rad MR, et al. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol Gen Genet. 1992;236:145–54. doi: 10.1007/BF00279653. [DOI] [PubMed] [Google Scholar]
- Ramezani-Rad M. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr Genet. 2003;43:161–70. doi: 10.1007/s00294-003-0383-6. [DOI] [PubMed] [Google Scholar]
- Ramezani Rad M, et al. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol Gen Genet. 1998;259:29–38. doi: 10.1007/s004380050785. [DOI] [PubMed] [Google Scholar]
- Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–80. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, et al. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS One. 2008;3:e1931. doi: 10.1371/journal.pone.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckses DM, et al. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:912–28. doi: 10.1128/MCB.26.3.912-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, et al. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot Cell. 2004;3:1028–35. doi: 10.1128/EC.3.4.1028-1035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Cooper JA. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- Wickes BL, et al. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–60. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- Wu C, et al. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 2006;20:734–46. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, et al. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2425–40. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C, et al. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–15. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, et al. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010;6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Construction of the serotype A MATα ste50Δ mutant. (A) Diagram for disruption of the STE50 gene in serotype A MATα H99 strain. Primers for the first-round and second-round PCR are indicated as bent arrows. Through recombination between 5′ and 3′ flanking region of STE50 gene, the intact STE50 gene is displaced with nourseothricin-resistant gene. (B) The correct genotype of the ste50 mutants were confirmed by Southern blot analysis using genomic DNA digested with the restriction enzyme EcoR and BamHI.
Fig. S2. Construction of the serotype A MATa ste50Δ mutant. (A) Diagram for disruption of the STE50 gene in serotype A MATa KN99a strain. Primers for the first-round and second-round PCR are indicated as bent arrows. Through recombination between 5′ and 3′ flanking region of the STE50 gene, the intact STE50 gene was displaced with the NEO dominant selectable marker. (B) The correct genotypes of the ste50Δ mutants were confirmed by Southern blot analysis using genomic DNAs digested with the restriction enzyme EcoR .
Fig. S3. Construction of the ste50Δ crg1Δ double mutant. (A) Diagram for disruption of the STE50 gene in H99 crg1 (crg1Δα) and PPW196 (crg1Δa) strains. Primers for the first-round PCR and double joint (DJ)-PCR are indicated as bent arrows. The native STE50 gene is specifically replaced with nourseothricin-resistant gene (NAT) through triple recombination between 5′ and 3′ flanking region of the STE50 gene. (B) The correct genotypes of the ste50Δ crg1Δ double mutants were confirmed by Southern blot analysis using genomic DNAs digested with the restriction enzyme Sac .