Abstract
The promise of personalized medicine is now a clinical reality, with colorectal cancer genetics at the forefront of this next major advance in clinical medicine. This is no more evident than in the recent advances in testing of colorectal cancers for specific molecular alterations in order to guide treatment with the monoclonal antibody therapies cetuximab and panitumumab, which target the epidermal growth factor receptor (EGFR). In this review, we examine genetic mechanisms of colorectal cancer and how these alterations relate to emerging biomarkers for early detection and risk stratification (diagnostic markers), prognosis (prognostic markers), and the prediction of treatment responses (predictive markers).
Keywords: Colon Cancer, Biomarkers, EGFR, KRAS, K-Ras, BRAF, Microsatellite Instability, MSI, Chromosome Instability, Cetuximab, Panitumumab, Personalized Medicine
INTRODUCTION
The promise of personalized medicine is now a clinical reality, with colorectal cancer genetics at the forefront of this next major advance in clinical medicine. This is no more evident than in the testing of colorectal cancers for specific molecular alterations in order to guide treatment with the monoclonal antibody therapies cetuximab and panitumumab, which target the epidermal growth factor receptor (EGFR).1, 2, 3 Indeed, the discovery that acquired KRAS mutations are a robust predictive marker of resistance to cetuximab and panitumumab 4, 5 has led to clinically validated and cost-effective testing strategies to direct these drugs to appropriate patients. This discovery resulted from a detailed understanding of colorectal cancer genetics, including the role of KRAS mutations in colorectal carcinogenesis, as well as knowledge of the epidermal growth factor (EGFR) signaling pathways.6 The success of KRAS mutation testing in predicting treatment response is just the beginning of the use of genetic markers for directing the care of colorectal cancer patients. Many other genetic markers in colorectal cancer show promise for their use in treatment selection, prognosis, and early cancer detection. In this context, knowledge of the underlying genetic mechanisms of colorectal tumorigenesis and the potential of specific genetic lesions for clinical decision making is expected to become part of the working knowledge of care providers managing colon cancer patients. However, despite the promising advances in the molecular pathology of colorectal cancer that are highlighted in this review, it is important to emphasize that clinicopathological staging of tumor tissue is still the cornerstone of prognostication and treatment selection. The modern tumor-node-metastasis (TNM) classification system is recommended, although the original Dukes staging system is still used by some clinicians and is taught to pathologists in training.7 The pathologic features with greatest prognostic power are depth of tumor invasion, burden of lymphovascular invasion (estimated by the number of lymph nodes infiltrated by cancer), and presence of distant metastases. Efforts to correlate genetic alterations with histologic features have had limited success, although microsatellite instability is a molecular feature that shows modest correlation with certain histologic features such as cribriform architecture and medullary histology.8 Thus, molecular testing is usually required for accurate assessment of specific gene mutations or genomic instability that provide prognostic and predictive information beyond clinicopathologic features.
In this review, we examine genetic mechanisms of colorectal cancer and how these alterations relate to emerging biomarkers for early detection and risk stratification (diagnostic markers), prognosis (prognostic markers), and the prediction of treatment responses (predictive markers) (Table 1). The genetic features of colorectal cancer that are currently most clinically useful will be emphasized in this review, and a detailed description of the molecular genetics and molecular biology of the germane genetic and epigenetic alterations will be provided. We will conclude by reviewing the potential role for genetic markers in the selection of targeted colorectal cancer therapies that are in pre-clinical development or in Phase I and II trials.
Table 1.
Selected Biomarkers That Have Been Evaluated in Colorectal Cancer
| Biomarker | Molecular Lesion | Frequency in CRC |
Prediction | Prognosis | Diagnosis |
|---|---|---|---|---|---|
| KRAS | Codon 12/13 activating mutations; rarely codon 61, 117,146 |
40% | Yes | Possible | - |
| BRAF | V600E activating mutation |
10% | Probable | Probable | Lynch Syndrome |
| PIK3CA | Helical and kinase domain mutations |
20% | Possible | Possible | - |
| PTEN | Loss of protein by IHC | 30% | Possible | - | - |
| Microsatellite Instability (MSI) | Defined as >30% unstable loci in the NCI consensus panel or >40% unstable loci in a panel of mononucleotide microsatellite repeats9 |
15% | Probable | Yes | Lynch Syndrome |
| Chromosome Instability (CIN) | Aneuploidy | 70% | Probable | Yes | - |
| 18qLOH | Deletion of the long arm of chromosome 18 |
50% | Probable | Probable | - |
| CpG Island Methylator Phenotype (CIMP) |
Methylation of at least three loci from a selected panel of five markers |
15% | +/− | +/− | - |
| Vimentin (VIM) | Methylation | 75% | - | - | Early Detection |
| TGFBR2 | Inactivating Mutations | 30% | - | - | - |
| TP53 Mutations | Inactivating Mutations | 50% | - | - | - |
| APC Mutations | Inactivating Mutations | 70% | - | - | FAP |
| CTNNB1 (β-Catenin) | Activating Mutations | 2% | - | - | - |
| Mismatch Repair Genes | Loss of protein by IHC; methylation; inactivating mutations |
1–15% | - | - | Lynch Syndrome |
CRC- colorectal cancer; IHC- immunohistochemistry; FAP- Familial Adenomatous Polyposis
MOLECULAR MECHANISMS OF COLORECTAL CARCINOGENESIS
The adenoma/carcinoma progression sequence
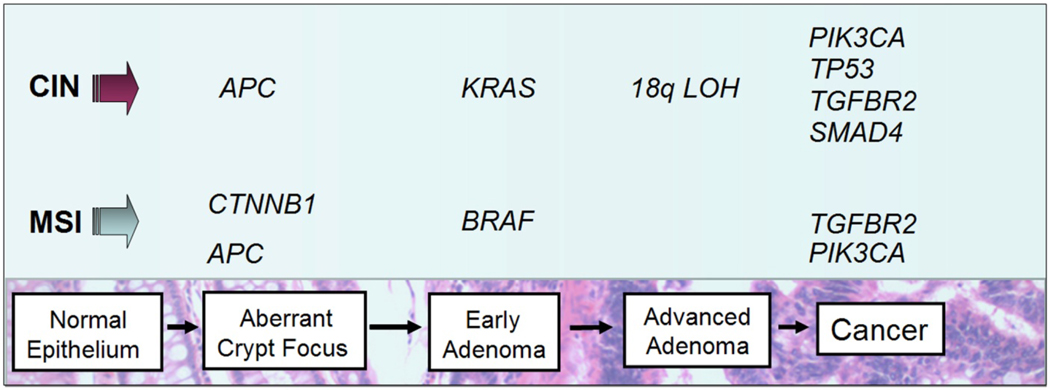
Colorectal cancer arises as the result of the accumulation of acquired genetic and epigenetic changes that transform normal glandular epithelial cells into invasive adenocarcinomas. Steps that transform normal epithelium to benign neoplasia (adenoma), followed by invasive carcinoma, and eventually metastatic cancer are described in the classic tumor progression model proposed by Fearon and Vogelstein (Figure 1).6 Since this model was originally proposed our understanding of the molecular pathogenesis has advanced considerably and led to numerous revisions of the Vogelstein and Fearon model. For instance, the original model proposed that only tubular and tubulovillous adenomas had the potential to progress to invasive adenocarcinoma. It is now recognized that serrated polyps including sessile serrated adenomas (SSA) and traditional serrated adenomas (TSA) also have the potential for malignant transformation.10, 11 These polyps are an alternative pathway to malignancy whereby a subset of hyperplastic polyps progress to serrated neoplasms (SSA or TSA) and a fraction of these serrated neoplasms progress to cancer. Premalignant serrated polyps more frequently arise in the proximal colon 12 and are associated with microsatellite instability and aberrant DNA methylation at CpG islands, whereas conventional tubular adenomas arise via biallelic inactivation of the APC tumor-suppressor gene and display chromosome instability.13 Furthermore, other molecular lesions, such as BRAF V600E mutations, are characteristically found more often in tumors arising via the serrated neoplasia pathway.13
Figure 1. The adenoma-to-carcinoma progression sequence.
Colorectal carcinogenesis progresses by at least two well-recognized pathways. The chromosome instability (CIN) pathway is characterized by classic tubular adenoma histology and the early acquisition of APC mutations that lead to deregulated WNT signaling, frequent activating mutations of the KRAS oncogene at the early adenoma stage, loss of heterozygosity at chromosome 18q (18qLOH) in late adenomas, and TP53 mutations that facilitate the transition to frank malignancy. By contrast, tumors that harbor microsatellite instability (MSI) frequently acquire BRAF mutations and are not associated with 18qLOH or TP53 mutations. Sporadic MSI cancers appear to commonly arise via the serrated neoplasia pathway, in which sessile serrated adenomas are the most frequently observed pre-cancerous lesions.
Genomic and Epigenomic Instability and Chromosomal Alterations
Genomic and epigenomic instability distinguishes neoplastic from normal colonic epithelium and is a hallmark feature of colorectal carcinogenesis.14, 15 At least four kinds of genomic or epigenetic instability have been described in colorectal cancers: (1) chromosomal instability (CIN), (2) microsatellite instability (MSI), (3) CpG island methylator phenotype (CIMP), and (4) global DNA hypomethylation. Overlap between these categories and imprecise use of these terms has led to confusion and confounds interpretation of the literature.16 Thus, in this section we will first define the different types of genomic and epigenetic instability in colorectal cancer and will delineate in general terms how these mechanisms are clinically relevant.
CIN
The most common form of genomic instability is chromosome instability, which is found in as many as 85% of colorectal cancers.17 Chromosome instability, which can be recognized by the presence of aneuploidy, is defined as the presence of numerical chromosome changes or multiple structural aberrations and is typically assessed by DNA flow cytometry.18 Despite the frequent occurrence of CIN in colorectal cancer, the mechanisms that give rise to this form of genomic stability and the role of aneuploidy in tumor progression remain poorly understood. However, there is some evidence that CIN promotes cancer progression by increasing clonal diversity 19, 20, 21. Importantly, from a clinical perspective, large meta-analyses have demonstrated that CIN is a marker of poor prognosis in colorectal cancers.18, 22
MSI
Microsatellite unstable tumors, which account for approximately 15% of colorectal cancers, are generally regarded as being mutually exclusive of CIN tumors because they display a normal karyotype and exhibit unique genetic features, although there does appear to be a subset of tumors that show both CIN and MSI.16 MSI colorectal cancer has been defined by the presence of at least 30% unstable loci in a panel of 5–10 loci consisting of mono- and di-nucleotide tracts selected at a National Cancer Institute consensus conference.23 Currently, many clinical laboratories assess MSI using a panel of 5 mononucleotide markers (BAT-25, BAT-26, NR-21, NR-24 and MONO-27) that were selected for high sensitivity and specificity.9 A subset of tumors with only 10–29% unstable loci has been designated as a form of microsatellite tumors designated “MSI-low”. Although there is evidence that MSI-low cancers have distinct features compared with MSI (also referred to as “MSI-High”, or “MSI-H”) and microsatellite stable tumors, there is considerable controversy regarding whether MSI-low is a unique molecular subclass of colorectal cancer.16, 17, 24 Colorectal cancer patients with MSI tumors have been shown to have a better prognosis compared to patients with CIN tumors18, 25, and probably respond differently to adjuvant chemotherapy compared to patients with microsatellite stable (MSS) cancer.26, 27, 28
In contrast to CIN, the mechanisms underlying MSI are relatively well understood and involve inactivation of genes in the DNA Mismatch Repair (MMR) family either by aberrant methylation or by somatic mutation.21 Furthermore, individuals with Lynch syndrome (hereditary non-polyposis colorectal cancer, HNPCC) almost exclusively develop MSI colorectal cancers because they have germline mutations in the MMR genes, which include MLH1, MSH2, MSH6, and PMS2. In contrast, sporadic MSI colorectal cancer most often have loss of MMR activity as the result of silencing of MLH1 by aberrant methylation.21, 29 It is also now recognized that sporadic MSI tumors are associated with the serrated neoplasia pathway and frequently carry BRAF V600E mutations, while cancers resulting from germline mutations in MMR genes (Lynch syndrome) do not have mutated BRAF.30, 31 Thus, the presence of a BRAF mutation in an MSI tumor effectively excludes the possibility that the tumor arose as the consequence of Lynch syndrome (Figure 2).
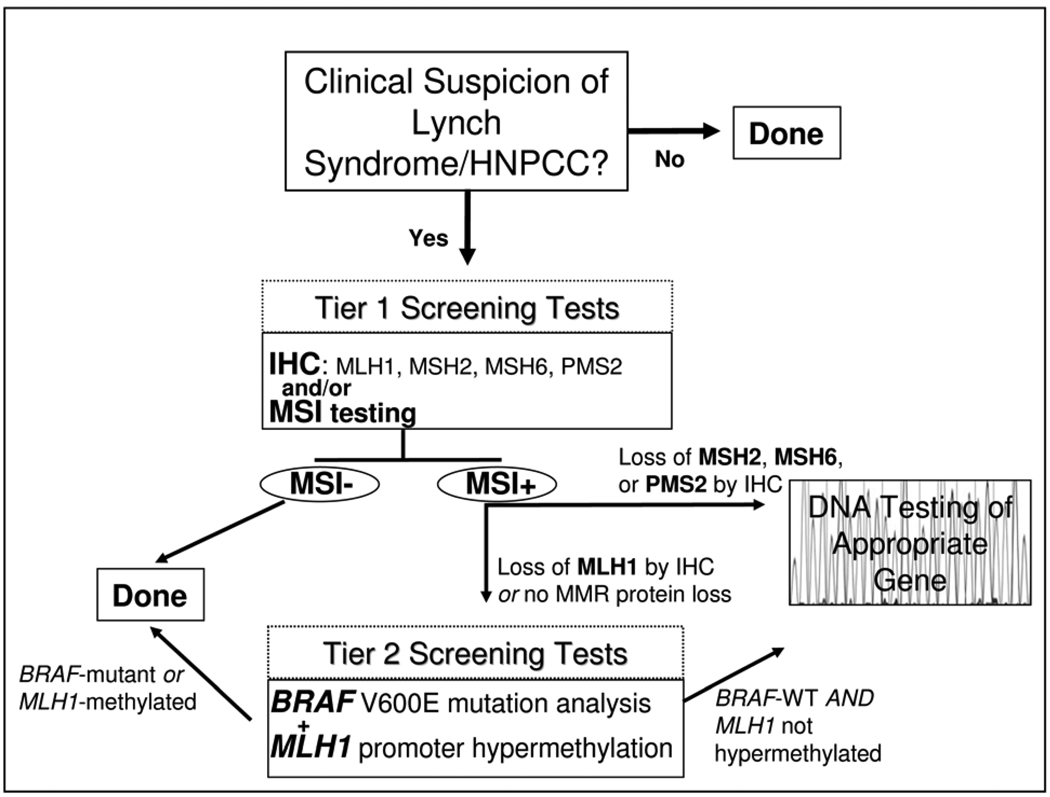
Figure 2. Testing strategies for Lynch Syndrome (HNPCC).
A multi-stage approach to facilitate the cost-effective diagnosis of Lynch Syndrome is outlined. Patients with a high clinical suspicion of Lynch Syndrome are first screened by immunohistochemistry (IHC) studies of the tumor tissue to assess for loss of Mismatch Repair proteins (MMR) expression and by MSI testing of the tumor DNA (Tier 1 Screening Tests). Patients with tumors that show microsatellite instability (MSI) with loss of MSH2, MSH6, or PMS2 by IHC undergo germline DNA mutation analysis of the gene corresponding to the missing protein. In contrast, patients with MSI tumors that lack MLH1 are further assessed with assessment of the tumor for MLH1 promoter methylation and mutant BRAF V600E (Tier 2 Screening Test) because most sporadic MSI colon cancers have methylated MLH1 and Lynch Syndrome MSI cancers rarely harbor BRAF mutations. When there is not evidence of MLH1 promoter methylation or BRAF mutation, mutation analysis of the MLH1 gene is performed to identify Lynch Syndrome patients with mutations in this gene.
CIMP
Epigenetic instability in colorectal cancer is manifested as both hypermethylation of gene promoters that contain CpG islands (the CpG Island methylator phenotype, CIMP), and global DNA hypomethylation. Mechanisms that give rise to CIMP are not yet clear, although the strong association between BRAF V600E mutations and CIMP colorectal cancer suggests a role for activated BRAF in the pathogenesis of the methylator phenotype and a link between sporadic MSI and CIMP.32, 33 However, in vitro studies of mutant BRAF in colorectal cancer cell lines have not demonstrated a direct cause and effect relationship between BRAF and CIMP.34 Furthermore, although CIMP tumors do appear to represent a distinct subset of colorectal cancer, the clinical utility of this designation is hindered by lack of a universally accepted definition of the methylator phenotype. CIMP is usually defined as methylation of at least three loci from a selected panel of five gene associated CpG islands. Because this panel is not always the same across studies, attempts are being made to facilitate standardization of CIMP markers for clinical use.33, 35 Some authors have proposed two classes of CIMP, CIMP-low, and CIMP-high, depending on the number of methylated marker loci detected.32 Another group suggested that CIMP colorectal cancers be divided into two distinct classes (called CIMP1 and CIMP2) based on the results of unsupervised cluster analysis of a large panel of methylation markers.36 Finally, considerable overlap between CIMP and sporadic MSI tumors adds to the challenge of incorporating CIMP-status into clinical trials and clinical decision making.33 Retrospective studies suggest CIMP will ultimately be shown to be a predictive marker for colorectal cancer, but the data is not adequate at this time to recommend its clinical use.36, 37 Thus, the discovery and classification of CIMP tumors has advanced our understanding of the molecular pathology of colorectal cancer but has not yet impacted clinical care.
In addition to aberrant gene methylation, a global decrease in methylation has also been identified in many colorectal cancers and is tightly associated with CIN tumors.38, 39 Further research is necessary to determine if measurement of global DNA hypomethylation in colorectal cancer has any role in the clinical setting.
Role of Specific Genetic Alterations and Signal Pathway Deregulation
Just as important as genomic and epigenomic instability for the pathogenesis of colorectal cancer is the accumulation of mutations in specific genes and the resulting deregulation of specific signaling pathways that control the hallmark behaviors of cancer: cell proliferation, differentiation, apoptosis, immortalization, angiogenesis, and invasion. The best-studied pathways that are deregulated in colorectal cancer are the WNT-β-catenin signaling pathway, the transforming growth factor β (TGFβ) signaling pathway, the epidermal growth factor receptor (EGFR)-MAPK pathway, and the phosphatidylinositol 3-kinase (PI3K) pathway.5, 16 Selected deregulated pathways in colorectal cancer and targeted therapies in clinical use or in clinical trials are summarized in Table 2.
Table 2.
Pathways Commonly Deregulated in Colorectal Cancer and Targeted Drugs in Clinical Use (bold) or in Clinical Trials
| Pathway | Specific Target | Drugs |
|---|---|---|
| EGF/MAPK | EGFR (mAb) | Cetuximab, Panitumumab |
| EGFR (TKI) | Erlotinib, Gefitinib | |
| KRAS | Tipifarnib, Lonafarnib | |
| BRAF | Sorafenib, PLX4032, XL281 | |
| MEK | Selumetinib | |
| PI3K | PI3K | BKM120, BGT226, XL147, GDC-0941 |
| mTOR | Everolimus, XL765 | |
| AKT | Perifosine | |
| WNT | Resveratrol | |
| TGFβ | TGFβ2 | AP 12009 |
| VEGF | VEGF | Bevacizumab |
| VEGFR | Vatalanib, AMG706, Pazopanib, Cediranib |
|
| HGF | HGF mAb | AMG102 |
| IGF | IGF-1 mAb | AMG479, IMC-A12 |
EGF= epidermal growth factor; MAPK= mitogen activated protein kinase; mAb= monoclonal antibody; TKI= tyrosine kinase inhibitor; TGFβ= transforming growth factor beta; PI3K= phosphatidylinositol 3-kinase; VEGF= vascular endothelial growth factor; IGF= insulin-like growth factor; HGF= hepatocyte growth factor
Key tumor suppressor genes that do not necessarily mediate their effects through signal pathway deregulation, such as TP53, and recurrent cytogenetic aberrations such as 18q loss of heterozygosity (LOH) are also well-studied in colorectal cancer and affect the malignant transformation of colon epithelial cells through specific effects on the behavior of the cells (Figure 1). The use of these molecular alterations in the management of patients with colorectal cancer will also be discussed in more detail below.
WNT Pathway
Mutations in the adenomatous polyposis coli (APC) gene occur in up to 70% of sporadic colorectal cancers and are the cause of the familial adenomatous polyposis (FAP) cancer predisposition syndrome. APC mutations can be found at the earliest stages of neoplasia and are predominantly associated with the classic tubular adenoma pathway and CIN cancers (Figure 1).6, 40, 41 The APC protein negatively regulates WNT signaling via targeting β-catenin for ubiquitin-mediated proteasomal degradation. Disruption of the APC protein results in increased WNT signaling through stabilization of nuclear β-catenin. Activating mutations in the β-catenin gene (CTNNB1) that protect the protein from APC-mediated degradation are also observed in colorectal neoplasia, although they are found more frequently in adenomas (12.5%) than invasive cancer (1.4%), suggesting that CTNNB1-mutant tumors do not frequently progress to carcinoma.42 Despite the critical and nearly universal role of WNT pathway activation in colorectal carcinogenesis, there is currently no clinical use for APC or CTNNB1 mutations for treatment selection, prognosis, or early cancer detection. There has been intense effort to develop small molecule inhibitors of this pathway, but these efforts are still confined to the pre-clinical arena. If these agents eventually reach the clinic, the assessment of APC mutations or activated β-catenin (by the detection of nuclear localization of β-catenin by immunostaining) is likely to have a role in directing the selection of patients who will respond to these agents.
TGF-β Pathway
Deregulation of TGF-β signaling, which is generally considered a tumor-suppressor pathway in the colon, occurs in the majority of colorectal cancers.43 Inactivating mutations have been observed in receptor genes (TGFBR2 and TGFBR1), post-receptor signaling pathway genes (SMAD2, SMAD4), and TGF-β superfamily members (ACVR2).17, 44, 45, 46 Functionally significant mutations in TGFBR2 are detected in as many as 30% of all colorectal cancer and are associated with the malignant transformation of late adenomas. TGFBR2 mutations are most common in MSI tumors, but also occur in approximately 15% of microsatellite stable (MSS) tumors (Figure 1).46, 47, 48 SMAD4 is located on 18q in the region commonly deleted in colorectal cancer, and is associated with adenoma formation and adenoma-carcinoma progression in mouse models, supporting a role for SMAD4 as a tumor suppressor gene.49 Furthermore, loss of SMAD4 expression as detected by immunostaining has been reported in >50% of colon cancers and is associated with lymph node metastases.50 There is still not any definite clinical role for any genetic markers in the TGF-β signaling pathway, however there is some evidence that SMAD4 expression levels may be associated with prognosis and response to 5-Fluorouricial (5-FU) and there is ongoing investigation of 18qLOH as a predictive marker, which is discussed further in the next section.51, 52
18qLOH
Loss of the long arm of chromosome 18 (18q loss of heterozygosity; LOH) is the most frequent cytogenetic alteration in colorectal cancer and is observed in up to 70% of tumors.6, 22 Two genes thought to have a role in the tumorigenic effects of this loss are deleted in colorectal carcinoma (DCC) and SMAD4. Additional mediators of the TGF-β pathway, including SMAD2 and SMAD7, are also in the 18qLOH region, suggesting that 18qLOH promotes tumorigenesis at least in part through deregulation of TGF-β signaling. It appears that deletion at 18q is associated with a worse prognosis, however efforts to definitively link 18qLOH to prognosis are limited by a lack of consistent results across studies and heterogeneous detection methods.22 Ongoing clinical trials (e.g. NCT00217737, also designated ECOG 5202) are assessing the utility of 18qLOH for treatment selection.
TP53
Mutations in the tumor-suppressor gene TP53 occur in about half of all colorectal cancers and promote the malignant transformation of adenomas (Figure 1).6 Like APC, TP53 is a key tumor suppressor that has been extensively studied in colorectal cancer but currently has no predictive or prognostic role in the clinical setting.16
Mediators of EGF signaling: EGFR/RAS/RAF/RAF/MAPK
KRAS, a member of the RAS family of proto-oncogenes, is the most frequently mutated gene in all of human cancer and arguably the most clinically important oncogene in colorectal cancer. The KRAS protein is a downstream effector of EGFR that signals through BRAF to activate the mitogen activated kinase (MAPK) pathway and promote cell growth and survival (Figure 3). Mutations in KRAS codons 12 or 13 occur in approximately 40% of colorectal cancers and lead to constitutive signaling by impairing the ability of GTPase activating proteins to hydrolyze KRAS-bound GTP.53 KRAS mutations occur after APC mutations in the adenoma-to-carcinoma progression sequence, but are still a relatively early event in tumorigenesis (Figure 1).6 Acquired KRAS mutations are maintained throughout carcinogenesis, as evidenced by the nearly perfect concordance of KRAS-mutation status in primary and metastatic colorectal cancer.54, 55 This fact is critical to the utility of KRAS mutational analysis on archived primary tumor specimens in patients with metastatic disease and usually eliminates the need for additional biopsy tissue.
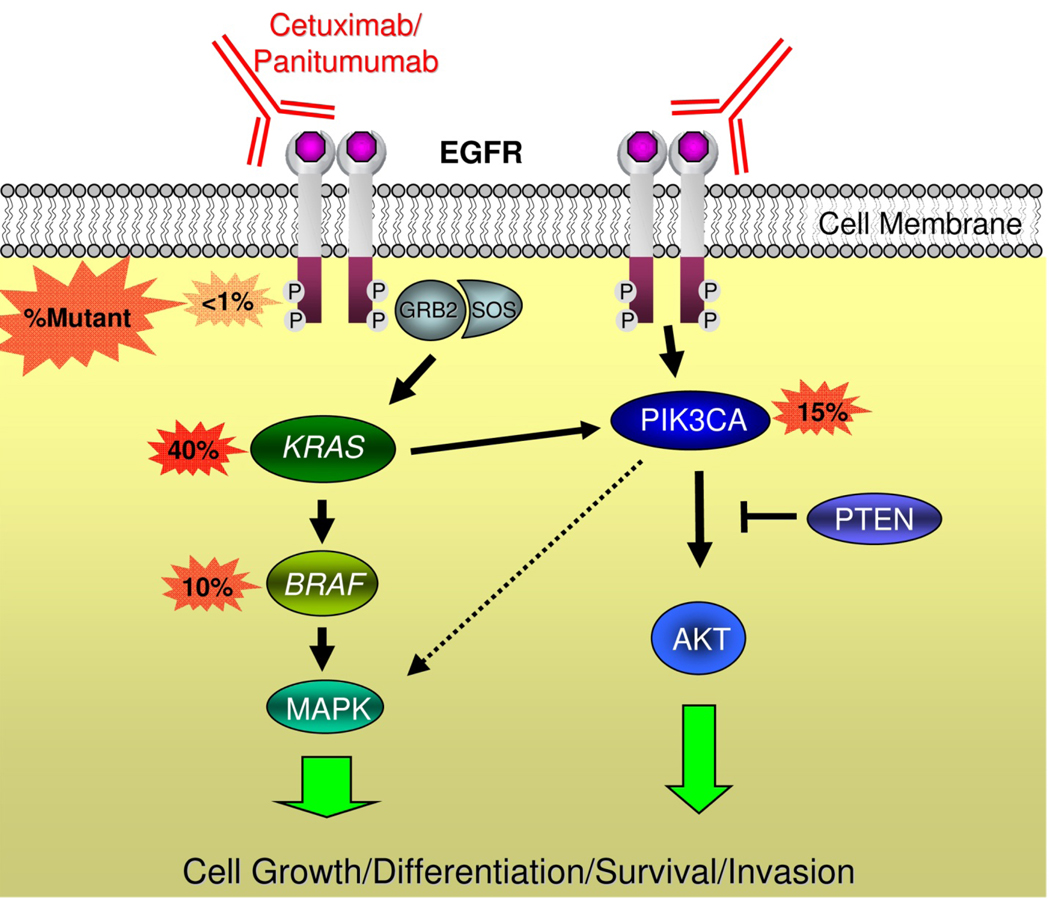
Figure 3. Mediators of EGFR signaling and anti-EGFR antibodies.
EGFR forms a homodimer after ligand activation, which results in phosphorylation/activation of the intra-cellular kinase domain and a cascade of downstream signaling including activation of the Ras/Raf/MAPK and phosphoinositol-3-kinase (PI3K) pathways that are associated with cell growth, differentiation, survival, and invasion. Monoclonal antibodies used to treat patients with metastatic colorectal cancer including cetuximab and panitumumab bind to the extracellular portion of EGFR and inhibit signaling in some patients. Activating mutations in KRAS occur in ~40% of colorectal cancers and are thought to confer resistance to these drugs by bypassing the need for upstream EGFR signals. Activating mutations in BRAF – the direct downstream effector of KRAS – occur in ~10% of colorectal cancers and also probably confer resistance to anti-EGFR monoclonal antibodies. Emerging evidence supports an additional role of oncogenic aberrations in the PI3K pathway in cetuximab and panitumumab resistance.
The BRAF gene, mutated in ~10–15% of colorectal cancers, encodes a protein kinase that is the direct downstream effector of KRAS in the Ras/Raf/MAPK signaling pathway. The majority of BRAF mutations are a single base change resulting in the substitution of glutamic acid for valine at codon 600 (V600E; sometimes referred to as “V599E”).5 KRAS and BRAF mutations are mutually exclusive, supporting the hypothesis that an activating mutation in either gene is sufficient to promote tumorigenesis via increased MAPK signaling.56 As discussed above, BRAF mutations are much more frequent in MSI tumors (~35%) compared to MSS tumors (~5%) and are very tightly linked to CIMP cancers and the serrated neoplasia pathway.56, 57 Emerging evidence supports a role for BRAF as a genetic marker for prediction, prognosis, and risk stratification.
Alterations in EGFR ligands and the EGFR gene itself are also observed in a subset of colorectal cancers. There is some data to support that upregulation of the EGFR ligands epigregulin and amphiregulin are associated with an anti-EGFR drug response.58, 59
Mediators of EGF signaling: The PI3K Pathway
Mutations in phosphatidylinositol 3-kinase (PI3K) pathway genes are observed in up to 40% of colorectal cancer and are nearly mutually exclusive of one another.60 The most frequent mutations of the PI3K pathway occur in the p110α catalytic subunit PIK3CA, which are reported in up to 32% of colorectal cancers and may promote the transition from adenoma to carcinoma (Figure 1).61 Mutations are also observed in PTEN, a tumor suppressor gene that negatively regulates PI3K signaling in as many as 30% of MSI tumors and 9% of CIN tumors.62 The PI3K pathway is modulated by EGFR signaling in part via KRAS activation, and there is a plausible role for both PIK3CA and PTEN mutations as predictive markers of anti-EGFR therapy (Figure 3).63, 64 Currently, there is not sufficient evidence from clinical studies to support the use of PI3K pathway mutations as predictive or prognostic biomarkers.
RISK STRATIFICATION AND EARLY DETECTION
One use of molecular markers in the management of colorectal cancer is in risk stratification for identifying individuals at high-risk for developing colorectal cancer and for the early detection of colon adenomas and early-stage colorectal cancers. With regard to risk stratification, the most robust molecular markers to date are germline mutations in genes that cause the hereditary colon cancer syndromes (e.g. APC mutations and Familial Adenomatous Polyposis, BMPR1A and Juvenile Polyposis, etc.) and MSI tumor status, which is an indicator of the possibility of Lynch Syndrome. The use of MSI tumor testing in the diagnosis of Lynch Syndrome will be discussed below in the context of MSI testing being a risk stratification marker because of its association with Lynch Syndrome.
Lynch Syndrome/Hereditary NonPolyposis Colon Cancer (HNPCC) syndrome
Identifying individuals with Lynch syndrome (also known as HNPCC) dramatically alters their clinical management, and can lead to effective colorectal cancer prevention programs for these individuals and their family members. However, currently the definitive molecular diagnosis of Lynch Syndrome requires expensive germline DNA mutation analysis of multiple DNA Mismatch Repair (MMR) genes (MLH1, MSH2, MSH6, and PMS2). To facilitate the most cost-effective strategies for identifying patients at high risk for Lynch syndrome who are candidates for genetic testing, the evaluation of molecular features of colorectal cancers that have occurred in these individuals or other family members can be used to predict the likelihood of identifying a germline mutation in one of the MMR genes. It is now common practice for molecular diagnostics labs to offer a step-wise series of molecular tests that are used to identify colorectal cancers that likely arose in the setting of Lynch syndrome. These tests are based on the molecular pathology of colorectal cancer (Figure 2).65 A common approach is to initially test the tumors for loss of MMR gene products (MLH1, MSH2, MSH6, PMS2) by immuohistochemistry (IHC) and for MSI by polymerase chain reaction (PCR) as the first-tier screening test (see for example http://www.mayomedicallaboratories.com/test-catalog/Clinical+and+Interpretive/17073), although there is support for the use of IHC alone as a first-line test.66 Tumors that display MSI and loss of MLH1 protein expression by IHC are then subjected to reflex testing for BRAF V600E mutation status and MLH1 promoter hypermethylation to help distinguish sporadic MSI tumors (~35% BRAF-mutant and 99% MLH1-methylated) from Lynch syndrome MSI tumors (BRAF-WT, infrequent MLH1-methylation) (Figure 2, Table 3).30, 67, 68 This strategy is most effective in excluding individuals who are unlikely to have a MMR gene mutation from undergoing germline mutation testing. It is notable that in those tumors that have MSI and loss of MSH2, MSH6, or PMS2 the likelihood of having a germline mutation is extremely high. Also of interest, it is now recognized that a strategy that relies on clinical criteria alone for the diagnosis of individuals at risk for Lynch Syndrome underdiagnose this syndrome 69. In light of the substantial effect of a missed diagnosis on an individual’s likelihood of developing cancer in the future, a strategy that employs universal testing of all colorectal cancer is being advocated by some experts in this area.70 It remains to be determined if this strategy is cost-effective and if the benefits outweigh the risks.
Table 3.
Biomarkers Used in The Diagnosis of Lynch Syndrome (HNPCC)
| Biomarker | Frequency | |
|---|---|---|
| Sporadic | Lynch Syndrome | |
| Microsatellite Instability (MSI) | 15% | >95% |
| BRAF V600E Mutations | 35% of sporadic MSI 5% of MSS 10% overall |
<1% |
| Mismatch Repair Protein Loss by IHC | 10–15%, mostly MLH1 | ~90% |
| MLH1 Promoter Hypermethylation | ~99% of sporadic MSI <1% MSS 15% overall |
<1% |
MSS= microsatellite stable; IHC= immunohistochemistry
Molecular markers and colorectal cancer early detection
Colonoscopy is the most accurate test currently for colorectal cancer screening, however, it is expensive and associated with procedure-related complications and poor patient compliance. In contrast, another commonly used colorectal cancer screening test , fecal occult blood testing (FOBT) is inexpensive and simple to perform, but has a relatively low sensitivity and specificity.71 Advances in our understanding of the molecular pathology of colorectal cancer, has led to the identification of promising early detection molecular markers for use in non-invasive colorectal cancer screening assays.72, 73 Stool-based methylated VIMENTIN (mVim) is a clinically validated marker for early colorectal cancer detection that is now commercially available in the United States (Table 1).74 The test relies on the fact that a majority of colorectal cancers (53–84%) carry an aberrantly methylated vimentin (VIM) gene. A PCR-based assay that simultaneously measured mVim and DNA integrity reported a sensitivity of 83% and a specificity of 82%, with approximately equal sensitivity in Stage I-III colorectal cancer patients .75 At this time, methods are under development to enhance the performance of stool- and plasma-based methylation assays for clinical purposes.76 The use of molecular assays, such as the fecal-methylated VIM assay, in the clinical care of patients is an area that is likely to undergo rapid advances in the near future.
GENETIC MARKERS AND PROGNOSIS
Genomic Instability and Prognosis: MSI vs. CIN
Meta-analyses across a diverse range of patients have firmly established that MSI colorectal cancer have a better prognosis and that CIN tumors have an unfavorable prognosis (Table 4).18, 25 The combined hazard ratio for MSI colorectal cancers for overall survival was estimated to be 0.65 (0.59–0.71, 95% CI) with only one of thirty-two included studies reporting an HR >1.0.25 Conversely, the overall HR associated with CIN colorectal cancer was determined to be 1.45 (1.27–1.45, 95% CI) based on 63 eligible studies and over 10,000 patients.18 Despite the clear association of MSI and CIN with prognosis, these markers have not yet been adopted into routine clinical decision making. It is most likely that MSI testing will be adopted into clinical practice before CIN testing because of the availability of a reliable assay for assessing MSI status.
Table 4.
Prognostic Biomarkers in Colorectal Cancer
| Biomarker | Mutation Frequency |
Prognosis | Evidence | Status |
|---|---|---|---|---|
| Microsatellite Instability (MSI) | 15% | Favorable | Strong | Testing available but not yet widely used |
| Chromosome Instability (CIN) | 70% | Unfavorable | Strong | No readily available test, not in clinical use |
| 18qLOH/SMAD4 Loss | 50% | Unfavorable | Moderate | No readily available test, not in clinical use |
| BRAF V600E Mutations | 10% | Probably unfavorable | Moderate | Testing available but insufficient evidence to use for prognosis |
|
KRAS Codon 12/13 Mutations |
40% | Probably unfavorable in advanced disease |
Limited | Testing widely available but insufficient evidence to use for prognosis |
| PIK3CA Mutations | 20% | Possibly unfavorable | Limited | No readily available test, not in clinical use |
18qLOH and Prognosis
Colorectal cancer patients with 18qLOH appear to have a worse prognosis compared to patients with tumors that do not carry 18qLOH. A meta-analysis of seventeen independent studies that was limited by evidence of publication bias found an overall HR of 2.00 (1.49–2.69 95% CI) for 18qLOH across all patients, and an HR of 1.69 (1.13–2.54 95% CI) in the adjuvant setting.22 Candidate genes in the 18q region, including DCC and SMAD4, have been studied individually for prognostic roles, with inconsistent results.77 The independent prognostic contribution of 18q deletion in colorectal cancer has been called into question due to the tight association between 18qLOH and CIN, and the inverse association of 18qLOH and MSI.16 This assertion is supported by a recent study that found no difference in prognosis attributable to 18qLOH in a prospectively collected cohort of 555 non-MSI tumors Stage I-IV.78 Thus, it is unclear at this time if 18qLOH represents an independent prognostic marker, or is merely a surrogate marker for CIN/MSS colorectal cancers.
Mediators of EGFR and Prognosis
Several recent studies have assessed the prognostic significance of KRAS, BRAF, and PIK3CA mutations in colorectal cancer.24, 79, 80, 81, 82, 83 Mutant KRAS was not independently associated with differences in relapse-free or overall survival in Stage II or III colorectal cancer, but mutant BRAF was prognostic for overall survival (OS) in this group of patients.24, 79 In contrast, mutant KRAS and BRAF have been reported as markers of poor prognosis in advanced colorectal cancer. In the largest study that has addressed the prognostic role of KRAS mutations in advanced colorectal cancer to date, patients with mutant KRAS cancers had a worse overall survival (HR= 1.40; 1.20–1.65 95%CI) but similar PFS compared to patients with tumors bearing wild-type KRAS.81 The potential prognostic value of KRAS mutations is of particular interest in advanced colorectal cancers because the KRAS mutational status of tumors is now being routinely collected in this setting in order to assess for eligibility for treatment with cetuximab or panitumumab. At this time, the use of KRAS mutation status for prognosis in colorectal cancer is still premature but appears to have significant potential to be adopted into clinical use in the near future.
PREDICTIVE BIOMARKERS
Although the treatment of colorectal cancer still primarily relies on the surgical resection of the primary tumor to achieve a cure, considerable progress in the medical treatment of stage III and IV colorectal cancer has occurred over the last 15 years. The adjuvant therapy of stage III colorectal cancer has become more effective as the standard regimen has advanced from 5-fluorouracil (5FU) and leucovorin to 5FU and oxaliplatin or irinotecan 84. Furthermore, the treatment of stage IV colorectal cancer patients has expanded to include targeted therapies (cetuximab, panitumumab, bevacizumab; see Table 2) in addition to 5FU, oxaliplatin, and irinotecan. With the identification of multiple effective agents for the treatment of colorectal cancer has come a need for predictive markers for selecting optimal treatment regimens for patients. This is particularly applicable to colorectal cancer because of the heterogeneity in response among colon cancers and because of the toxicity and cost of the medical treatments. The potential of genetic and epigenetic alterations to be effective predictive molecular markers has received considerable attention lately and has led to the use of some of these markers in the routine care of patients with colorectal cancer (Table 5).
Table 5.
Colorectal Cancer Biomarkers as Predictors for Drug Selection
| Biomarker | Mutation Frequency |
Drug Selection | Evidence | Status |
|---|---|---|---|---|
|
KRAS Codon 12/13 Mutations |
40% | Predicts Resistance to anti- EGFR Therapy |
Strong | Validated, In Routine Clinical Use |
|
KRAS Codon 61/117/146 Mutations |
1% | Probably Predicts Resistance to anti-EGFR Therapy |
Moderate | In Clinical Use, Not Fully Validated |
| BRAF V600E Mutations | 10% | Probably Predicts Resistance to anti-EGFR therapy, May Predict Response to BRAF-inhibitors |
Moderate | In Clinical Use, Not Fully Validated |
| PIK3CA Mutations | 20% | May Predict Resistance to anti- EGFR Therapy |
Limited | No Readily Available Test, Not in Clinical Use |
| PTEN Loss | 30% | May Predict Resistance to anti- EGFR Therapy |
Limited | No Readily Available Test, Not in Clinical Use |
| Microsatellite Instability (MSI) |
15% | May Predict adverse outcome with 5-FU and improved outcome with Irinotecan |
Moderate | Not Yet in Routine Clinical Use As a Predictive Biomarker |
| 18qLOH/SMAD4 Loss | 50% | May Predict Resistance to 5-FU | Moderate | No Readily Available Test, Not in Clinical Use |
| Topo1 Low | 50% | May Predict Resistance to Irinotecan |
Limited | No Readily Available Test, Not in Clinical Use |
5-FU= 5-Fluorouricil
The advent of cancer therapeutics that target specific molecules and pathways highlights the potential for underlying genetic and epigenetic lesions in colorectal cancer to guide personalized treatment decisions. A clear demonstration of the potential of mutant genes to direct therapy is that of mutant KRAS and treatment with cetuximab. Only ~15% of patients with metastatic colorectal cancer respond to monoclonal antibody (mAb) therapies targeting the epidermal growth factor receptor (EGFR), which prompted intense research into resistance mechanisms that could be secondary to alterations in the EGFR gene and/or mutations in downstream effectors. These studies have produced one well-validated and exceedingly robust predictive marker (mutant KRAS) and several more promising biomarkers that require further validation (mutant BRAF, PIK3CA, PTEN).85 Research efforts are also focused on identifying molecular features of colorectal cancer that predict response to adjuvant chemotherapy with cytoxic agents: 5-FU, irinotecan, and oxaliplatin.16 In this section we will discuss genetic features of colorectal cancer that have been evaluated for a role in guiding treatment selection. We have focused primarily on acquired tumor mutations as predictive markers, but it is important to note that inherited (germline) polymorphisms also influence the effects of chemotherapy on cancers and the risk for drug toxicity , particularly in the case of 5-FU and irinotecan (Reviewed in 16).
Predictors of Response to anti-EGFR mAb Therapies
EGFR-targeted monoclonal antibodies cetuximab (Erbitux®), and the fully humanized mAb panitumumab (Vectibix®), have proven to be effective in patients with metastatic colorectal cancer both as single agents and in combination with traditional chemotherapy.86, 87, 88 However, while these therapies improve both progression free survival (PFS) and overall survival (OS), they are effective in only a minority of metastatic colorectal cancer patients.85 These drugs are generally well-tolerated, but are still associated with treatment-related morbidity, including skin rash, diarrhea, and nausea, and are also expensive. To better target anti-EGFR mAb therapy to patients most likely to benefit, KRAS mutation status and additional molecular markers of cetuximab and panitumumab resistance have been extensively evaluated.5
KRAS is an accurate predictive biomarker
Results of four large phase III randomized have established unequivocally that metastatic colorectal cancer patients with KRAS mutations in codon 12 or 13 do not benefit from cetuximab or panitumumab therapy.4, 89, 90, 91 Prior to the publication of these pivotal trials, the link between KRAS mutation status and anti-EGFR mAb response was already firmly supported by several smaller studies92, 93, 94, but the data was not sufficient to warrant routine clinical testing. The recently published randomized trials have established the use of KRAS mutational analysis as a predictive marker for anti-EGFR mAb resistance in patients with metastatic colorectal cancer in most of the relevant clinical settings. These settings include the use of cetuximab or pantumimab in combination with conventional cytotoxic chemotherapy (e.g. 5-FU, FOLFOX, FOLFIRI) as first line treatment of metastatic disease90, 95, 96, and as monotherapy in relapsed/refractory patients.4, 89, 91
A second relevant question related to anti-EGFR mAb therapy is whether mutant KRAS predicts an adverse outcome in the setting of these treatments. The reported hazard ratios (HR) were almost exactly 1.0 in a total of 348 KRAS-mutant chemotherapy-resistant or refractory cancers treated with either panitumumab89 or cetuximab4 as monotherapy, confirming lack of benefit, but also suggesting no harm from anti-EGFR mAb treatment related to PFS or OS in this population. In contrast, the reported HRs were usually greater than 1.0 in studies of cetuximab or panitumumab as first line treatment in combination with FOLFOX4 (fluorouracil, leucovorin, and oxaliplatin) or FOLFIRI (fluorouracil, leucovorin, and irinotecan) chemotherapy.85 The results of the OPUS trial (Oxaliplatin and Cetuximab in First-Line Treatment of Metastatic Colorectal Cancer) in particular suggest it may be harmful to add anti-EGFR mAb treatments to 5FU, leukovorin, and oxaliplatin in patients with KRAS-mutant metastatic colorectal cancer.90
Based on the evidence from large trials, European and United States practice guidelines either recommend or require KRAS mutational analysis on colorectal cancer tumor tissue prior to the initiation of cetuximab or panitumumab treatment.1, 2, 3 The European health authority confines use of panitumumab monotherapy, and cetuximab as mono- or combination therapy, to metastatic colorectal cancer patients who are found to carry non-mutated (wild-type, WT) KRAS in the primary tumors.1 The American Society for Clinical Oncology recently published a provisional opinion stating that “All patients with metastatic colorectal carcinoma who are candidates for anti-EGFR antibody therapy should have their tumors tested for KRAS [codon 12 and 13] mutations…and [KRAS-mutant] patients should not receive anti-EGFR antibody therapy”.3 Similarly, the National Comprehensive Cancer Network (NCCN) guidelines require evidence of wild-type KRAS prior to cetuximab or panitumumab therapy in all metastatic colorectal cancer settings.2
Despite the nearly perfect negative predictive value of mutant KRAS, it is still only a minority (~30%) of KRAS codon 12/13 wild-type patients who respond to anti-EGFR mAb therapy.85 This has led to research into additional biomarkers that might predict lack of benefit in those individuals with tumors that have wild-type KRAS. There is evidence that rare KRAS mutations in codons 61 or 146 (~2% of colorectal cancer) behave similarly to codon 12/13 mutations97, but incorporating these mutations into routine clinical practice will require analysis of a larger group of patients. Other promising markers of anti-EGFR mAb resistance are BRAF V600E mutations, PIK3CA mutations, and loss of PTEN protein expression.5
BRAF: another predictor of anti-EGFR mAb response?
The biological rationale for BRAF V600E mutations as an additional biomarker of anti-EGFR mAb resistance is strong: (1) BRAF is the immediate downstream effector of KRAS in the Ras/Raf/MAPK signaling pathway (Figure 3), and (2) BRAF V600E activating mutations are 100% mutually exclusive of KRAS mutations in colorectal cancer, implying that activation of either protein is sufficient for colon tumorigenesis. Existing limited data supports BRAF V600E mutations as a negative predictor of response to anti-EGFR mAb therapy, leading to the evolving use of BRAF mutation testing in KRAS-WT patients prior to treatment as a means to further stratify patients into responders and nonresponders. A retrospective analysis showed that 0/11 tumors with mutant BRAF responded to cetuximab or panitumumab compared to 22/68 (32%) of BRAF-WT/KRAS-WT patients.98 Similar results were observed for patients treated with cetuximab plus irinotecan. None of the patients with tumors with mutant BRAF (N=13) responded compared to 24/74 (32%) patients with tumors with BRAF-WT/KRAS-WT.97 These finding were supported by work presented at the 2009 American Association of Cancer Research and American Society of Clinical Oncology annual meetings85, although not all studies have found as robust a relationship between BRAF V600E mutation status and anti-EGFR antibody response.82, 99 BRAF mutations also appear to be associated with worse prognosis independent of treatment, which can confound the assessment of its role as a predictive marker for response to EGFR directed therapies.82, 99 Despite the currently limited data, and lack of complete consensus, it is likely that BRAF mutation status has a role in anti-EGFR mAb treatment decisions and soon will be adopted into the planning for treatment with cetuximab and panitumumab.
PI3K Pathway Activation and anti-EGFR mAb Resistance
Molecular lesions in the PI3K pathway, which in colorectal cancer are primarily mutations in PIK3CA and loss of PTEN protein expression, have been proposed as additional anti-EGFR mAb resistance markers because the PI3K pathway is also stimulated by EGFR.85 However, the relationship of oncogenic alterations in PI3K signaling and cetuximab or panitumumab response is much less clear than that of KRAS and BRAF mutations. In several small studies published to date, PIK3CA mutations or PTEN loss have been associated with lack of response to cetuximab.64, 100, 101, 102 Both PIK3CA mutations and PTEN loss may coexist with KRAS or BRAF mutations, which weakens the biological rationale of the activation of this pathway as an absolute predictor of anti-EGFR mAb therapeutic response. Nonetheless, the balance of evidence points towards a probable predictive role of molecular events that activate the PI3K pathway for being negative predictive markers for EGFR monoclonal antibody based therapy. In fact, there is modest data demonstrating that when PIK3CA mutations and PTEN loss of expression are combined with KRAS and BRAF mutational analysis, up to 70% of patients unlikely to respond to cetuximab or panitumumab may be identified.85, 102 This observation has led to the idea that colon cancer may be able to be classified like breast cancers (e.g. triple negative breast cancers), and these cancers have been termed “quadruple-negative” for patients who do not have alterations in any of these four biomarkers.85, 102 However, at this time, further studies are needed to determine if mutant PIK3CA or PTEN loss should be incorporated into clinical practice.
EGFR Mutations and Amplification
The most obvious candidate biomarker for resistance to antibodies which target EGFR is the EGFR gene itself. Early studies that focused on EGFR overexpression assessed by immunohistochemistry did not show a consistent relationship with treatment response, in part because of lack of standardization of the assay, which were based on either immunostaining, fluorescent in-situ hybridization (FISH) or quantitative RT-PCR, and inter-observer variability inherent in the technique.103 EGFR gene amplification is more promising for being a predictive biomarker, but has also been fraught with technical challenges that limit the interpretation of existing data, such as dilution of tumor DNA with wild-type DNA in PCR-based assays, and lack of consistent tissue processing and scoring systems in FISH assays.5 Activating mutations in the EGFR catalytic domain are seen frequently in lung cancer and are associated with sensitivity to anti-EGFR tyrosine kinase inhibitors, but these mutations are quite rare in colorectal cancer.5 Thus, EGFR does not appear likely to be a clinically useful predictive marker for anti-EGFR monoclonal antibody therapy. Furthermore, although preliminary studies have shown that the EGFR ligands amphiregulin and epiregulin are overexpressed in colorectal cancer and may predict response to cetuximab, lack of standardization of the assays and studies that reproducibly demonstrate the same effect have prevented amphiregulin and epiregulin expression levels from being used as clinical biomarkers for directing therapy with EGFR monoclonal antibodies.58
Predictive molecular markers for response to 5-FU, irinotecan, and oxaliplatin
Currently, the tumor biomarkers that demonstrate the greatest promise for guiding adjuvant chemotherapy with conventional drugs in colorectal cancer patients include MSI and 18qLOH.
MSI
5-FU based regimens have been shown to be ineffective, or even detrimental to patients with MSI tumors.28, 104 Evidence that a functioning MMR system is required for the cytotoxic effect of fluorouracil provides a plausible biological rationale for 5-FU resistance in MSI tumors.17, 27 However, the finding of 5-FU resistance in MSI colorectal cancer is not uniform, and may vary with tumor stage.105, 106 An ongoing phase III randomized trial of patients with completely resected stage II colorectal cancer (NCT00217737) will prospectively assess the role of MSI in predicting response to adjuvant chemotherapy in localized cancers.16
MSI tumors appear to be more responsive to irinotecan-based adjuvant chemotherapy.26 Recently published results from a large randomized trial of Stage III colorectal cancer demonstrated improved outcomes (both PFS and OS) in MSI patients treated with an irinotecan-containing regimen that included 5-FU compared to 5-FU/luekovorin alone.107 In light of the prior results of the CALGB 98303 study showing no benefit of adding irinotecan to 5FU as adjuvant therapy in unselected Stage III colorectal cancer patients, the finding that MSI is a predictive biomarker for irinotecan suggests MSI could be useful for adjusting adjuvant therapy for colorectal cancer patients.108 Replication of these results in independent studies is required to validate MSI-status as an inclusion criteria for irinotecan-based adjuvant chemotherapy. Currently, neither the European Group on Tumour Markers nor the American Society of Clinical Oncology have recommendations on the use of MSI for guiding therapy in stage II or stage III colorectal cancer patients.
An important issue to consider with regards to MSI is that the majority of colorectal cancers that have MSI are sporadic colorectal cancers that have inactivated the MLH1 gene through aberrant promoter methylation. The majority of these sporadic MSI tumors also can be classified as CIMP cancers as well. It is not known whether the associations seen between 5FU and irinotecan effects in sporadic MSI tumors also apply to MSI tumors that arise in the setting of Lynch syndrome.
Loss of 18q
Loss of 18q has been associated with an adverse response to 5-FU based adjuvant chemotherapy.52, 109 There is some evidence that this effect is due to loss of the SMAD4 gene located in the 18q21 deleted region although this remains to be determined with more definitive studies.51, 52 A number of ongoing clinical trials are assessing the predictive value of 18qLOH and MSI status for the treatment of colon cancer. These include an ECOG trial of stage II colorectal cancer patients being treated with 5-FU, oxaliplatin and bevacizumab (NCT00217737), a trial in patients being treated with olaparib for metastatic disease (NCT00912743), as well as a retrospective analysis assessing MSI and 18qLOH in patients with colorectal cancer (stage II or III) treated with 5-FU or 5-FU and irinotecan (CLB-9581 or CLB-89803).
Topo1
In a large randomized trial that compared 5-FU alone to 5-FU with irinotecan and 5-FU with oxaliplatin in advanced colorectal cancer, higher expression of topoisomerase 1 (Topo1) measured by immunohistochemistry was significantly correlated with responsiveness to irinotecan.110 Conversely, cancers with low Topo1 expression (602/1269; 47%) did not appear to benefit from the addition of irinotecan (HR 0.98; 95% CI, 0.78 to 1.22). Irinotecan is a Topo1 inhibitor, thus the level of Topo1 expression has a clear biological rationale as a biomarker for predicting irinotecan response. Replication of these initial results in multiple independent studies is required before Topo1 should be considered for use as a predictive marker.
Polymorphisms and their role as molecular markers for colorectal cancer
We emphasize again that germline polymorphisms that alter pharmacokinetics and pharmacodynamics of adjuvant chemotherapy are also potential biomarkers for guiding treatment selection. For example, alterations in thymidylate synthetase and dehydropyrimidine dehydrogenase have been extensively studied in relation to 5-FU response and look promising. However, very few of these polymorphisms have been thoroughly validated and so the majority are not ready to be used clinically.111, 112 one exception to this generalization is a homozygous polymorphism that reduces the activity of UDP-glucuronosyltransferase (UGT1A1, an enzyme that detoxifies irinotecan), which is associated with a dose-related increased incidence of irinotecan toxicity.113, 114 This has led to a commercial UGT1A1 genotyping test that was approved by the Food and Drug Administration in 2005 to help guide irinotecan dosing.
CONCLUSIONS AND FUTURE DIRECTIONS
More than three decades of investigations into the molecular mechanisms of colorectal cancer carcinogenesis is finally culminating in biomarkers that are sufficiently validated for routine clinical use. KRAS-mutational analysis to guide anti-EGFR treatment stands as one of the first successes in the era of personalized medicine. MSI and BRAF-mutations already have a clear role in triaging molecular genetic testing in Lynch syndrome, and these markers are poised to take on a much greater role in prognostication and prediction of therapeutic responses for sporadic colorectal cancers. The use of assays for mutant KRAS, mutant BRAF, and MSI demonstrate how the molecular testing of colorectal cancer tissue can reduce medical costs and improve patient outcomes by targeting therapies to the appropriate patient population. Thus, it is anticipated that the use of molecular genetic markers in clinical decision making is likely to expand as more markers are identified and validated. For example, studies are in progress for assessing the efficacy of the multikinase/BRAF-inhibitor sorafinib, and specific inhibitors of PI3K signaling in the treatment of colorectal cancer.5 There is evidence that sorafinib restores sensitivity to anti-EGFR mAb therapy in BRAF-mutant cell-lines, which has prompted an ongoing Phase II National Cancer Institute sponsored clinical trial of sorafinib plus cetuximab in metastatic colorectal cancer patients (NCT00343772).98 If these initial findings are validated, the indications for mutational analysis of BRAF and KRAS would expand. Furthermore, colorectal cancer patients with tumors carrying mutant BRAF might also benefit from newer selective BRAF-inhibitors such as PLX-4032 combined with anti-EGFR mAb therapy. PIK3CA mutations or PTEN loss are likely to become clinically relevant for the treatment of colorectal cancer patients as specific PI3K pathway inhibitors (such as XL147, BGT226, GDC0941, XL765, and NVP-BEZ325) move into Phase II clinical trials.115 The expanding repertoire of drugs designed to inhibit specific oncogenes and oncogenic signaling pathways again highlights that molecular mechanisms of colorectal cancer will increasingly play a role in the clinical care of patients with colorectal cancer. The use of molecular markers for risk stratification and early detection of colorectal cancer is also showing promise and will be part of the era of molecular medicine that is rapidly emerging.
Box 1: Summary Points
Chromosome instability (CIN) and microsatellite instability (MSI) are distinct mechanisms by which colorectal cancers arise, associated with unique molecular features.
Key pathways that drive colorectal cancer are WNT signaling, TGF-β signaling , and Epidermal growth factor receptor (EGFR) signaling; Ras/Raf/MAPK and phosphatidyl inositol 3-kinase (PI3K) pathways are both stimulated by EGFR. Currently, only downstream mediators of EGFR have a clinical role as biomarkers.
KRAS mutations in codon 12/13 are a highly validated predictive marker for resistance to monoclonal antibody drugs that target EGFR; BRAF V600E mutation is likely to be a second predictive marker. Additional resistance markers including PIK3CA mutations and PTEN protein loss are being evaluated.
MSI+ cancers have a better prognosis and CIN+ cancers do worse; 18q LOH+ tumors also have a worse prognosis but are frequently with CIN. Downstream mediators of EGFR are under study for prognostication.
The role of colorectal cancer molecular biomarkers in clinical decision making is likely to expand as more targeted drugs become available.
ACKNOWLEDGEMENTS
This work was supported by a Burroughs Welcome Fund Clinical Scientist in Translational Research Award, NCI RO1CA115513, and P30 CA015704 (WMG). We thank Jonathan Tait for help with reviewing the manuscript.
Footnotes
License for Publication: The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive license (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Gut editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our license (http://group.bmj.com/products/journals/instructions-for-authors/licence-forms).
Competing Interest: None to declare.
REFERENCES
- 1.European Medicines Agency. Committee for Medicinal Products for Human Use May 2008 Plenary Meeting monthly report. 2008 http://www.emea.europa.eu/pdfs/human/press/pr/27923508en.pdf.
- 2.National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology: Colon Cancer V.2.2010. 2010 doi: 10.6004/jnccn.2009.0056. http://www.nccn.org/professionals/physician_gls/PDF/colon.pdf. [DOI] [PubMed]
- 3.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 4.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 5.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 7.Dukes C. The classification of cancer of the rectum. J Pathol Bacteriol. 1932;35:323. [Google Scholar]
- 8.Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacher JW, Flanagan LA, Smalley RL, Nassif NA, Burgart LJ, Halberg RB, et al. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis Markers. 2004;20:237–250. doi: 10.1155/2004/136734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein NS. Serrated pathway and APC (conventional)-type colorectal polyps: molecular-morphologic correlations, genetic pathways, and implications for classification. Am J Clin Pathol. 2006;125:146–153. [PubMed] [Google Scholar]
- 11.Jass JR. Hyperplastic polyps and colorectal cancer: is there a link? Clin Gastroenterol Hepatol. 2004;2:1–8. doi: 10.1016/s1542-3565(03)00284-2. [DOI] [PubMed] [Google Scholar]
- 12.Baker K, Zhang Y, Jin C, Jass JR. Proximal versus distal hyperplastic polyps of the colorectum: different lesions or a biological spectrum? J Clin Pathol. 2004;57:1089–1093. doi: 10.1136/jcp.2004.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol. 2009;4:343–364. doi: 10.1146/annurev.pathol.4.110807.092317. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.Little MP, Vineis P, Li G. A stochastic carcinogenesis model incorporating multiple types of genomic instability fitted to colon cancer data. J Theor Biol. 2008;254:229–238. doi: 10.1016/j.jtbi.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 17.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–950. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 19.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 20.Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 21.Grady WM. Genomic instability and colon cancer. Cancer Metastasis Rev. 2004;23:11–27. doi: 10.1023/a:1025861527711. [DOI] [PubMed] [Google Scholar]
- 22.Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41:2060–2070. doi: 10.1016/j.ejca.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 23.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 24.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 25.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 26.Fallik D, Borrini F, Boige V, Viguier J, Jacob S, Miquel C, et al. Microsatellite instability is a predictive factor of the tumor response to irinotecan in patients with advanced colorectal cancer. Cancer Res. 2003;63:5738–5744. [PubMed] [Google Scholar]
- 27.Jo WS, Carethers JM. Chemotherapeutic implications in microsatellite unstable colorectal cancer. Cancer Biomark. 2006;2:51–60. doi: 10.3233/cbm-2006-21-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sargent D, Marsoni S, Thibodeau SN, Labianca R, Hamilton SR, Torri V, Monges G, Ribic C, Grothey A, Gallinger S. Confirmation of deficient mismatch repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): a pooled molecular reanalysis of randomized chemotherapy trials. Journal of Clinical Oncology. 2008;26 Abstr. 4008. [Google Scholar]
- 29.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 30.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Cunningham JM, Winters JL, Guenther JC, French AJ, Boardman LA, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003;63:5209–5212. [PubMed] [Google Scholar]
- 32.Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 33.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 34.Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup? Int J Clin Oncol. 2008;13:498–503. doi: 10.1007/s10147-008-0854-3. [DOI] [PubMed] [Google Scholar]
- 38.Matsuzaki K, Deng G, Tanaka H, Kakar S, Miura S, Kim YS. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–8569. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 40.Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–865. doi: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 41.Miyaki M, Iijima T, Kimura J, Yasuno M, Mori T, Hayashi Y, et al. Frequent mutation of beta-catenin and APC genes in primary colorectal tumors from patients with hereditary nonpolyposis colorectal cancer. Cancer Res. 1999;59:4506–4509. [PubMed] [Google Scholar]
- 42.Samowitz WS, Powers MD, Spirio LN, Nollet F, van Roy F, Slattery ML. Beta-catenin mutations are more frequent in small colorectal adenomas than in larger adenomas and invasive carcinomas. Cancer Res. 1999;59:1442–1444. [PubMed] [Google Scholar]
- 43.Chittenden TW, Howe EA, Culhane AC, Sultana R, Taylor JM, Holmes C, et al. Functional classification analysis of somatically mutated genes in human breast and colorectal cancers. Genomics. 2008;91:508–511. doi: 10.1016/j.ygeno.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deacu E, Mori Y, Sato F, Yin J, Olaru A, Sterian A, et al. Activin type II receptor restoration in ACVR2-deficient colon cancer cells induces transforming growth factor-beta response pathway genes. Cancer Res. 2004;64:7690–7696. doi: 10.1158/0008-5472.CAN-04-2082. [DOI] [PubMed] [Google Scholar]
- 45.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 46.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 47.Grady WM, Rajput A, Myeroff L, Liu DF, Kwon K, Willis J, et al. Mutation of the type II transforming growth factor-beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- 48.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 49.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T, Watanabe T, Kazama Y, Tanaka J, Kanazawa T, Kazama S, et al. Loss of Smad4 protein expression and 18qLOH as molecular markers indicating lymph node metastasis in colorectal cancer--a study matched for tumor depth and pathology. J Surg Oncol. 2008;97:69–73. doi: 10.1002/jso.20896. [DOI] [PubMed] [Google Scholar]
- 51.Alhopuro P, Alazzouzi H, Sammalkorpi H, Davalos V, Salovaara R, Hemminki A, et al. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin Cancer Res. 2005;11:6311–6316. doi: 10.1158/1078-0432.CCR-05-0244. [DOI] [PubMed] [Google Scholar]
- 52.Boulay JL, Mild G, Lowy A, Reuter J, Lagrange M, Terracciano L, et al. SMAD4 is a predictive marker for 5-fluorouracil-based chemotherapy in patients with colorectal cancer. Br J Cancer. 2002;87:630–634. doi: 10.1038/sj.bjc.6600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 54.Artale S, Sartore-Bianchi A, Veronese SM, Gambi V, Sarnataro CS, Gambacorta M, et al. Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. J Clin Oncol. 2008;26:4217–4219. doi: 10.1200/JCO.2008.18.7286. [DOI] [PubMed] [Google Scholar]
- 55.Zauber P, Sabbath-Solitare M, Marotta SP, Bishop DT. Molecular changes in the Ki-ras and APC genes in primary colorectal carcinoma and synchronous metastases compared with the findings in accompanying adenomas. Mol Pathol. 2003;56:137–140. doi: 10.1136/mp.56.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 57.Lubomierski N, Plotz G, Wormek M, Engels K, Kriener S, Trojan J, et al. BRAF mutations in colorectal carcinoma suggest two entities of microsatellite-unstable tumors. Cancer. 2005;104:952–961. doi: 10.1002/cncr.21266. [DOI] [PubMed] [Google Scholar]
- 58.Jacobs B, De Roock W, Piessevaux H, Van Oirbeek R, Biesmans B, De Schutter J, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009;27:5068–5074. doi: 10.1200/JCO.2008.21.3744. [DOI] [PubMed] [Google Scholar]
- 59.Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene. 29:3033–3043. doi: 10.1038/onc.2010.89. [DOI] [PubMed] [Google Scholar]
- 60.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 61.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 62.Danielsen SA, Lind GE, Bjornslett M, Meling GI, Rognum TO, Heim S, et al. Novel mutations of the suppressor gene PTEN in colorectal carcinomas stratified by microsatellite instability- and TP53 mutation- status. Hum Mutat. 2008;29:E252–E262. doi: 10.1002/humu.20860. [DOI] [PubMed] [Google Scholar]
- 63.Razis E, Briasoulis E, Vrettou E, Skarlos DV, Papamichael D, Kostopoulos I, et al. Potential value of PTEN in predicting cetuximab response in colorectal cancer: an exploratory study. BMC Cancer. 2008;8:234. doi: 10.1186/1471-2407-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 65.Baudhuin LM, Burgart LJ, Leontovich O, Thibodeau SN. Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer. 2005;4:255–265. doi: 10.1007/s10689-004-1447-6. [DOI] [PubMed] [Google Scholar]
- 66.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bettstetter M, Dechant S, Ruemmele P, Grabowski M, Keller G, Holinski-Feder E, et al. Distinction of hereditary nonpolyposis colorectal cancer and sporadic microsatellite-unstable colorectal cancer through quantification of MLH1 methylation by real-time PCR. Clin Cancer Res. 2007;13:3221–3228. doi: 10.1158/1078-0432.CCR-06-3064. [DOI] [PubMed] [Google Scholar]
- 68.Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn. 2008;10:301–307. doi: 10.2353/jmoldx.2008.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 70.South CD, Yearsley M, Martin E, Arnold M, Frankel W, Hampel H. Immunohistochemistry staining for the mismatch repair proteins in the clinical care of patients with colorectal cancer. Genet Med. 2009;11:812–817. doi: 10.1097/GIM.0b013e3181b99b75. [DOI] [PubMed] [Google Scholar]
- 71.Smith RA, Cokkinides V, Brooks D, Saslow D, Brawley OW. Cancer screening in the United States, 2010: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer J Clin. 60:99–119. doi: 10.3322/caac.20063. [DOI] [PubMed] [Google Scholar]
- 72.Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 138:2127–2139. doi: 10.1053/j.gastro.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 73.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192–206. doi: 10.1053/j.gastro.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 74.Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, 3rd, Sontag S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 75.Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 76.Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carethers JM, Hawn MT, Greenson JK, Hitchcock CL, Boland CR. Prognostic significance of allelic lost at chromosome 18q21 for stage II colorectal cancer. Gastroenterology. 1998;114:1188–1195. doi: 10.1016/s0016-5085(98)70424-x. [DOI] [PubMed] [Google Scholar]
- 78.Ogino S, Nosho K, Irahara N, Shima K, Baba Y, Kirkner GJ, et al. Prognostic significance and molecular associations of 18q loss of heterozygosity: a cohort study of microsatellite stable colorectal cancers. J Clin Oncol. 2009;27:4591–4598. doi: 10.1200/JCO.2009.22.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuchs C, Ogino S, Meyerhardt JA. KRAS mutation, cancer recurrence, and patient survival in stage III colon cancer: findings from CALGB 89803. J Clin Oncol. 2009;27 A-4037. [Google Scholar]
- 80.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 82.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 83.Zlobec I, Kovac M, Erzberger P, Molinari F, Bihl MP, Rufle A, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. doi: 10.1002/ijc.25265. [DOI] [PubMed] [Google Scholar]
- 84.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 85.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 86.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 87.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 88.Saltz LB, Meropol NJ, Loehrer PJ, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 89.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 90.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 91.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 92.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 93.Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 95.Douillard J, Siena S, Cassidy J. Randomized phase 3 study of panitumumab with FOLFOX4 compared to FOLFOX4 alone as 1st-line treatment (tx) for metastatic colorectal cancer (mCRC): The PRIME trial. Eur J Cancer. 2009;7:S6. doi: 10.1093/annonc/mdu141. (suppl; abstr 10LBA) [DOI] [PubMed] [Google Scholar]
- 96.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 97.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 99.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 100.Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prenen H, De Schutter J, Jacobs B, De Roock W, Biesmans B, Claes B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15:3184–3188. doi: 10.1158/1078-0432.CCR-08-2961. [DOI] [PubMed] [Google Scholar]
- 102.Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, et al. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One. 2009;4:e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shia J, Klimstra DS, Li AR, Qin J, Saltz L, Teruya-Feldstein J, et al. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol. 2005;18:1350–1356. doi: 10.1038/modpathol.3800417. [DOI] [PubMed] [Google Scholar]
- 104.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 106.Liang JT, Huang KC, Lai HS, Lee PH, Cheng YM, Hsu HC, et al. High-frequency microsatellite instability predicts better chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV sporadic colorectal cancer after palliative bowel resection. Int J Cancer. 2002;101:519–525. doi: 10.1002/ijc.10643. [DOI] [PubMed] [Google Scholar]
- 107.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saltz LB, Niedzwiecki D, Hollis D, Goldberg RM, Hantel A, Thomas JP, et al. Irinotecan fluorouracil plus leucovorin is not superior to fluorouracil plus leucovorin alone as adjuvant treatment for stage III colon cancer: results of CALGB 89803. J Clin Oncol. 2007;25:3456–3461. doi: 10.1200/JCO.2007.11.2144. [DOI] [PubMed] [Google Scholar]
- 109.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Braun MS, Richman SD, Quirke P, Daly C, Adlard JW, Elliott F, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. doi: 10.1200/JCO.2007.15.5580. [DOI] [PubMed] [Google Scholar]
- 111.Ezzeldin HH, Diasio RB. Predicting fluorouracil toxicity: can we finally do it? J Clin Oncol. 2008;26:2080–2082. doi: 10.1200/JCO.2007.15.5481. [DOI] [PubMed] [Google Scholar]
- 112.Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, Dippon J, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 113.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 114.Palomaki GE, Bradley LA, Douglas MP, Kolor K, Dotson WD. Can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? An evidence-based review. Genet Med. 2009;11:21–34. doi: 10.1097/GIM.0b013e31818efd77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]