Abstract
Coreceptor switching from CCR5 to CXCR4 is less common in subtype C HIV-1 infection than in subtype B for reasons that are unclear. We have examined sequential virus samples from a subtype C-infected child who had evidence of coreceptor switching. Ninety-three sequences revealed three distinct coexistent virus lineages and only some members of one lineage evolved to use CXCR4. These lineages also had diverse alternative coreceptor patterns including the ability to use FPRL1, CCR3, CCR8, APJ, CMKLR1, RDC-1, CXCR6, CCR1, GPCR1, GPR15 and CCR6. Coreceptor switching was associated with extensive and rapid sequence divergence in the V1/V2 region in addition to V3 changes. Furthermore, interlineage recombination within the C2 region resulted in low predictability of a V3 sequence-based phenotype algorithm, and highlighted the importance of V1/V2 as well as V3 sequences in coreceptor usage. These results suggest that the evolution to coreceptor switching in subtype C infection requires more mutations than other subtypes, and this contributes to the reduced incidence of R5X4 viruses.
Keywords: Coreceptor switching, HIV-1 subtype C, alternative coreceptors, V1/V2 region
INTRODUCTION
HIV-1 gains entry into host cells by binding to CD4 and a coreceptor, predominantly CCR5 and less frequently CXCR4. Accelerated CD4 decline and disease progression is associated with the emergence of viruses able to use CXCR4 in greater than 50% of subtype B infected individuals 1–3. However, use of CXCR4 is much rarer in subtype C infection 4–6. Coreceptor switching has been extensively studied in subtype B infection, but only a few reports 6–10 deal specifically with coreceptor switching in subtype C infection. Even fewer studies have collected longitudinal samples from subtype C infected subjects with documented coreceptor switching 11–13. These studies point mostly to V3 characteristics such as increased positive charge, changes at amino acid 11 and 25 and increased loop length associated with CXCR4 usage, similar as in subtype B. Despite all these similarities subtype specific conformational differences within the stem and loop of V3 suggest other mechanisms and possibly the result of infrequent development of CXCR4 in subtype C14. The V3 region is an important determinant of coreceptor use, but other regions such as V1/V2 and V4-V5 have also been implicated 15–17. Previous work from this laboratory has identified potential barriers to the acquisition of CXCR4 use in subtype B infection including extensive mutation in the V3 region of envelope, compensatory mutations elsewhere in envelope to maintain viral fitness, and loss of entry efficiency via CCR5 as CXCR4 use is gained 17, 18.
In this study, we have examined HIV-1 envelope evolution in serial samples from a subtype C-infected patient with prior evidence of CCR5 to CXCR4 coreceptor switching to determine if these same barriers were similar or even higher in subtype C HIV-1. The ability to correlate env sequence with phenotypic characteristics in entry assays also allowed us to assess the value of the phenotype predictor based on V3 sequences (C-PSSM) 19 and investigate viral entry using alternative coreceptors such as FPRL1, CCR3, CCR8 and others 20.
We show that extensive env mutation, particularly in the V1/V2 region, must take place for subtype C viruses to use CXCR4, and that more compensatory mutations outside of the V3 region may be necessary to maintain viral fitness. The co-existence of CCR5-and CXCR4-using viruses late in infection further suggests that there was no loss of fitness for CCR5-mediated entry. We interpret these results to mean that barriers to CXCR4 use are higher for subtype C than subtype B viruses, but the nature of the barriers (mutational distance, maintenance of fitness) is similar.
MATERIALS AND METHODS
Viral isolation and coreceptor usage
The patient (TM18) was part of a cohort of perinatally infected children who had survived for >4 years and was classified as a slow progressor 21. This patient received no anti-retroviral therapy, as this was unavailable to most children at the time, developed advanced AIDS and has subsequently died. Three whole blood samples were obtained (TM18 A-C) at one-year intervals during 1999–2002. Ethical clearance was obtained from the University of Witwatersrand Committee for Research on Human Subjects. Levels of virus in plasma were measured using the Versant HIV-1 RNA 3.0 assay (bDNA from Bayer Nucleic Acid Diagnostics) and CD4 counts were determined using a FACS count (Becton Dickinson, San Jose, CA). The initial isolate used CCR5 for viral entry and subsequently gained the ability to use CXCR4 as previously described 6, 7, 21.
V1-V4 molecular clones
Viral RNA was extracted from plasma using a MagNaPure LC Isolation station and the Total NucleicAcid isolation kit (Roche AppliedScience, Penzberg, Germany). The V1-V4 region was amplified using primers ED5 (5′-ATG GGA TCA AAG CCT AAA GCC ATG TG-3′) and ES8 (5′-CAC TTC TCC AAT TGT CCC TCA-3′). PCR products were purified using the High Pure PCR Product Purification kit (Roche Diagnostics GmbH, Mannheim, Germany) and cloned into the pGEMTeasy vector (Promega, USA).
Full-length env clones
The gp160 envelope (env) gene was amplified from plasma as previously described using primer pair envA and envM 22. The 3KB PCR fragments were cloned into an expression vector (pcDNA3.1, Invitrogen) and co-expressed with the NL4.3 Env-negative, luciferase-positive reporter plasmid 23 into 293T cells. Trans-complementation resulted in the production of pseudovirions capable of a single round infection cycle. The primary coreceptor use of the full-length env clones was determined by infecting NP2.CD4 cell lines expressing either CCR5 or CXCR4 24. The ability of these pseudovirions to use alternative coreceptors for entry in vitro was also tested using NP2.CD4 cell lines expressing CCR3, CCR8, FPRL-1, APJ, CMKLR1, RDC-1, CXCR6, CCR1, GPR1 or CCR6 [described in 20]. Pseudovirion infectivity using a specific coreceptor was evaluated by luciferase activity. Relative light units (RLU) >5000 (>3-fold above background levels) was scored as positive entry via the specific coreceptor expressed.
Sequencing analysis
The V1-V4 clones and full-length env clones were sequenced and submitted to Genbank database. Sequence quality was assessed by comparing all selected clones with sequences of different subtypes, as well as sequences from subtype C with documented coreceptor usage obtained from the HIV database at Los Alamos National Laboratory (http://www.hiv.lanl.gov). Sample clustering within a phylogenetic tree indicated the env clones were representative of HIV-1 subtype C isolates with various coreceptor usage and donor age, and were more closely linked to each other than to any other patient isolate, excluding dual infection or sample contamination (data not shown).
Sequences were aligned with ClustalX and manually edited using BioEdit (version 7). All sequences were checked for intra-patient recombination by inferring separate phylogenetic trees for the V1/V2, C2, V3, C3 and V4 region. If sequences clustered differently in these trees they were removed and further analysed to identify possible parental sequences and determine recombination break points using Simplot (version3.2). The recombinants were further visualised with the Highlighter tool (http://www.hiv.lanl.gov).
Phylogenetic analysis, genetic distances, divergence and positive selection were determined using MEGA (version 3.1; Molecular Evolutionary Genetics Analysis) with complete deletion of gaps or missing data. To estimate genetic diversity within a time point, pairwise nucleic acid distance between all sequences from a specific time point were calculated. The genetic divergence within each time point was determined as the pairwise genetic distance from a founder sequence. This founder sequence was approximated from the time point A sequences.
Positive selection was assessed by the non-synonymous or synonymous distance (dn and ds, respectively) with a ratio of dn/ds (ω) >1 indicating selection. Potential N-glycosylation sites were determined using N-GLYCOSITE 25. The V3 region of full-length env clones (with known phenotype) was used to validate the C-PSSM (subtype C position specific scoring matrix) 19 and applied to predicted the phenotype of the V1-V4 molecular clones.
RESULTS
We analyzed sequential subtype C HIV-1 isolates from a previously described pediatric patient [21, 99ZATM18]. The patient was infected by mother-to-child transmission, and showed phenotypic evidence of coreceptor switching from R5 (TM18A; age 5.0 years) to R5/R5X4 (TM18B; age 6.6 years and TM18C; age 7.4 years) over a period of 2.4 years (Table 1). The coreceptor switch to CXCR4 was associated with a dramatically reduced CD4 count and modest reduction in viral load. Genotypic and phenotypic characteristics of plasma virus were evaluated using both V1-V4 env clones and full-length env clones from each sampling time point (Table 1).
Table 1.
Clinical information and summary of clones generated from each time point with associated phenotype.
| Samples | Age (years) | CD4 count | CD4 % | CD4:CD8 ratio | Viral Load | Biological Phenotype$ | Clones V1-V4 env# | Full length env* |
|---|---|---|---|---|---|---|---|---|
| TM18A | 5.0 | 1 239 | 20 | 0.31 | 699 740 | R5 | predicted CCR5, n=19 | R5, n=10 |
| TM18B | 6.6 | 202 | 11 | 0.13 | 500 000 | R5/R5X4 | predicted CCR5, n=9 predicted CXCR4, n=13 |
R5, n=3 R5X4, n=3 |
| TM18C | 7.4 | 7 | 1 | 0.01 | 177 797 | R5/R5X4 | predicted CCR5, n=6 predicted CXCR4, n=21 |
R5, n=6 R5X4, n=3 |
Phenotype prediction of V1-V4 clones
Since coreceptor usage for these V1-V4 env clones was not known, a phenotype predictor for HIV-1 subtype C samples (C-PSSM) was used [19, summarized in Table 1]. The C-PSSM results predicted that all 19 time point A clones were R5-like, confirming the initial report of predominant CCR5 use at this time point. It also predicted a mixture of CCR5 and CXCR4-using V3 loops for time point B and C.
Functional analysis of full-length env clones
In order to examine biological phenotypes, full-length envelope (env) genes were amplified from plasma samples of TM18 time points A, B and C. The coreceptor use of 25 functional env clones representing 10 clones from time point A, 6 from B and 9 from time point C is shown in Table 1. The results confirm CCR5 use at time point A with a mixture of viruses able to use CCR5-only or CCR5 and CXCR4 at time point B and C.
Three coexistent lineages observed
A phylogenetic tree of the V1-V4 region was constructed representing sequences from the V1-V4 env clones (with predicted phenotype) and functional env clones (with known coreceptor usage) (Figure 1). The tree topology indicated an atemporal structure (sequences from different time points intermixed in the tree) with three main clusters observed subsequently referred to as lineages (Figure 1, lineages indicated and only non-recombinant clones shown). Most time point A clones within lineage 2 and 3 had recombination events and are not shown in the tree (Figure 1, discussed later).
Figure 1.
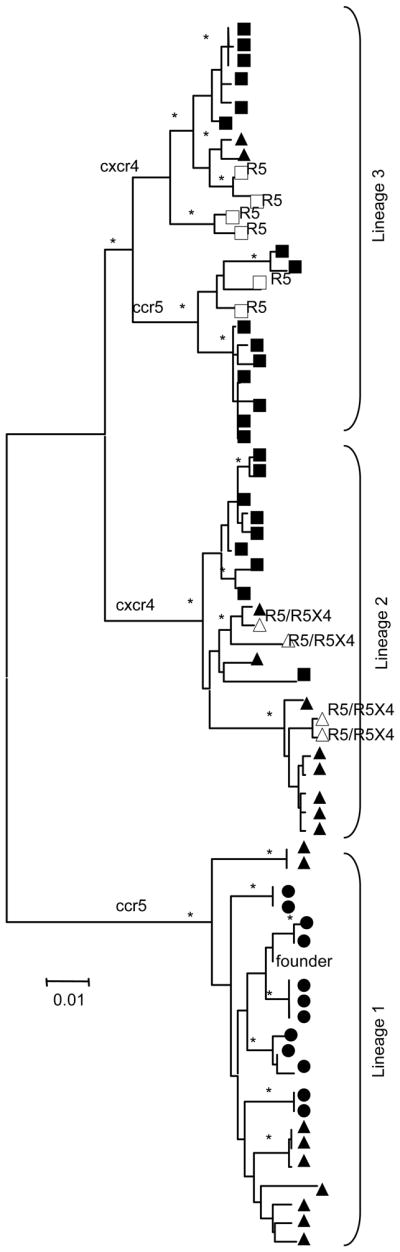
Phylogenetic tree of the V1-V4 region clones depicting an atemporal topology with three lineages. The neighbor joining tree was drawn using Kimura-2-parameter and bootstraps values >85% indicated (*) on branches with only non-recombinant clones represented. Circle indicated time point A, triangles time point B and squares time point C samples. Open symbols indicate full-length env clones with biologically determined coreceptor usage indicated next to symbol. C-PSSM predicted coreceptor usage is indicated in small letters on major branches.
V3 diversity during coreceptor switching
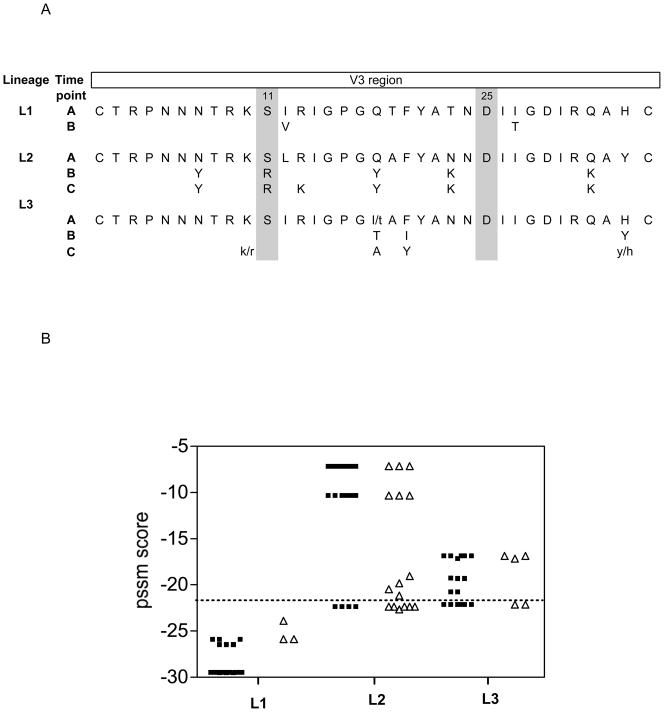
To investigate the genotypic changes associated with the observed phenotype changes we first focused on the V3 region, since previous studies have established this region as a major determinant of coreceptor usage. A phylogenetic tree of the V3 region showed similar grouping of viruses into the three lineages (data not shown), with viral sequences from time point A, B and C present in all three lineages, further suggesting an atemporal tree as seen in Figure 1. Consensus V3 amino acid sequences were compiled for each time point within a lineage, highlighting the V3 mutational pathway associated with that lineage (Figure 2A). The distribution of C-PSSM scores within a lineage was also plotted (Figure 2B), differentiating between clones with known and predicted coreceptor usage.
Figure 2.
Characterization of the V3 region for lineage 1, 2 and 3. (A) Consensus V3 sequence for each time point within a lineage, highlighting the amino acid changes that occurred over time compared to the time point A within that lineage. (B) Distribution of C-PSSM scores for the three lineages representing clones with known and predicted coreceptor usage. Open triangles represent clones with confirmed coreceptor usage determined biologically and black circles clones with predicted coreceptor usage.
Lineage 1 represent clones from time point A and B (Figure 2A) that were biologically determined, as well as predicted by C-PSSM, to use only CCR5. The V3 was very similar to the consensus C sequence and characterised by a GPGQ crown motif, V3 amino acid charge of +3 and S/D at position 11/25, typically seen in CCR5-using isolates. No time point C clones were observed in this lineage.
Lineage 2 is more complex, with R5 and R5X4 clones from different time points, establishing that this lineage consists of variants that underwent a coreceptor switch over time. V3 amino acid changes associated with the coreceptor switch included changes within the crown GPGQ to GPGY, increased V3 charge (to +7), as well as increased amino acid substitution to tyrosine (Figure 2A). The C-PSSM also reflected a distribution of CCR5 and CXCR4-like sequences and there was a good correlation correctly predicted sequences with known coreceptor usage (Figure 2B).
Lineage 3 is another CCR5-using cluster of variants that appeared to expand predominantly between time point B and C. Entry assays indicate this is a CCR5-using lineage with V3 sequence suggesting a distinct group compared to lineage 1, already present in time point A. Characteristics such as a different crown motif and diverse mutational pathway further suggesting a distinct population (Figure 2A). The C-PSSM predicted most of the clones as CXCR4-using including some full-length env clones with known R5 phenotype (Figure 2B, open triangles above perforated line). The V3 region of lineage 3 had characteristics associated with predicted CXCR4 usage (see Figure 2A), but the entry phenotype implies that other env regions contributed to the preferential use of CCR5.
Alternative coreceptor usage in HIV-1 subtype C
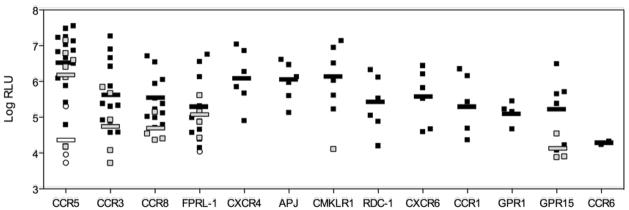
We next determined if representative env clones from each lineage could use alternative coreceptors. This was determined through single round infection of an NP2.CD4 cell line expressing one of the following alternative coreceptors: CCR3, CCR8, FPRL-1, APJ, CMKLR1, RDC-1, CXCR6, CCR1, GPR1 or CCR6. All env clones could use CCR5 and many could infect via CCR3, CCR8 or FPRL1 (Figure 3). Env clones from lineage 1 exclusively used CCR5, whereas lineage 3 could use in addition to CCR5 also CCR3, CCR8 and FPRL1. Lineage 2 consisted of two groups, one with early time point A clones able to use CCR5 as well as CCR3 and CCR8, and the second group able to use CXCR4 as well as many other alternative coreceptors including APJ, CMKLR1, RDC-1, CXCR6, CCR1 and GPR1. Therefore, the three lineages defined by sequence similarity were also distinguished by their ability to use alternative coreceptors.
Figure 3.
Entry mediated by full-length env clones representing the three different lineages described in Figure 1 using CCR5, CXCR4 and alternative coreceptors. Open circles indicate lineage 1 clones; black squares represent lineage 2 and gray squares represent lineage 3. The mean log RLU value for viral entry via a specific coreceptor is also shown for each lineage. Pseudoviruses were characterized as being able to use a coreceptor if RLU were >5000.
Recombination
Since three genetically distinct lineages were present during the course of infection, the possibility of recombination between these lineages was addressed. Recombination was tested within this data set by drawing multiple trees of the various regions within the env gene (including V1/V2, C2, V3, C3 and V1-V4) (data not shown), indicating 28 of the 93 sequences had a recombination event within the V1/V2 or C2 region.
For insight into the role of recombination in coreceptor usage, we further investigated these recombinant env clones with biologically determined phenotypes (Table 2). In vitro recombination during PCR is possible and can usually be excluded with single genome analysis (SGA), an approach that was beyond the scope of this study. Nonetheless, PCR-based recombination is considered unlikely in the recombinants analyzed because they were not identical to the parental sequence and, in some cases, the parental sequence was isolated from a different time point. If recombination occurred within the V1/V2 region, tropism still correlated with the V3 region. However, ten envelope clones from lineage 2 had C2 regions similar to lineage 3 R5 viruses. Features within the C2 region were therefore sufficient to confer CCR5 and limited alternative coreceptor use despite the rest of the env sequence being associated with CXCR4 use. This data highlight the limitation of prediction models based on only V3 sequences (as was also observed in Figure 2B within Lineage 3), particularly when other regions within env are contributing to coreceptor usage.
Table 2.
Interlineage recombination events within full-length env clones from lineage 1 and 2. Phenotype was biologically determined and number of env clones associated with similar recombination shown. The region where recombination occurred is highlighted with the lineage number that the region grouped with in a phylogenetic tree.
| Lineage | Phenotype | V1V2 | C2 | V3 | C3 | |
|---|---|---|---|---|---|---|
| 1 | CCR5 | n=3 | 3 | 1 | 1 | 1 |
| 2 | CCR5 | n=10 | 2 | 3 | 2 | 2 |
| 2 | CXCR4 | n=1 | 3 | 2 | 2 | 2 |
Viral evolution during disease progression within HIV-1 subtype C
The remaining 65 non-recombinant sequences (lineage 1, n=21; lineage 2, n=21; and lineage 3, n=23, Figure 1) were further analysed to determine the contribution each lineage had to the viral diversity and divergence during disease progression. The nucleotide genetic diversity within each lineage ranged from 2–3%, with diversity between lineage 1 and 2 13%, between lineage 1 and 3 12%, and between lineage 2 and 3 7%.
We addressed whether this diversity was equally distributed across the envelope region and under positive selection. The amino acid divergence from the founder sequence was measured and positive selection (indicated by dN/dS>1) was determined for each lineage. Lineage env sequences 1 (represented by only time point A and B) showed limited diversification (3%) for the 1.6 years studied, with no positive selection. By contrast, the remaining two lineages had diverged (within the 2.4 years of study) 24% and 22% respectively within the V1-V4 region, and the V1/V2 region more than 30% divergent from the founder sequence. Positive selection was seen in the V1/V2, C2 and C3 region. In addition, the V3 region of lineage 2 was very different from the founder R5-like sequence, and also showed evidence of positive selection. The major divergence observed in lineage 2 and 3, and the separate clustering within phylogenetic trees, suggests independent or variable evolution within these lineages and possibly different selective pressures.
DISCUSSION
Coreceptor switching from CCR5 to CXCR4 is less frequently observed in subtype C HIV-1 infection 5–7. To gain insight into this process, we investigated coreceptor utilization during disease progression within an HIV-1 subtype C individual. The results show multiple pathways of viral evolution each resulting in different patterns of coreceptor usage. Three distinct but coexisting lineages were observed within this patient. Each lineage consisted of viruses from different time points and had distinct phenotypic characteristics; lineage 1 was stable R5, lineage 2 diverged into a population able to use CXCR4; and lineage 3 showed equivalent divergence but remained R5.
The emergence of CXCR4 use required major divergence from the earliest consensus sequence as well as positive selection within the V1/V2 and V3 regions. An increase in divergence has been described for coreceptor switching in subtype B infection 26, but the extent of divergence was much less than observed here. Although the current results are based on one patient, the dramatic sequence divergence provides some plausible possibilities why so few subtype C viruses switch to use CXCR4. Despite the divergence lineage 2 and 3 had undergone in the V1/V2 region, only lineage 2 gained the ability to use CXCR4. We have previously reported V1/V2 compensatory changes and increased divergence in the V4-V5 region (in addition to V3 changes) associated with coreceptor switching in subtype B 17, 18. To determine whether these observations were unique to the current samples, we compared the amino acid divergence of subtype B and C sequences with known coreceptor usage available in the Los Alamos database to the consensus B and C sequences (see Supplemental Digital Content 1, http://links.lww.com/QAI/A96). This revealed that subtype B and C CXCR4-using V3 sequences have diverged further from the consensus sequence compared to CCR5 sequences (p<0.0001). In subtype B CXCR4-using V4-V5 sequences (p=0.02) and subtype C V1/V2 sequences (p=0.0002) had also moved significantly in mutational space, further highlighting subtype-specific differences in coreceptor usage 20. This extreme divergence within the V1/V2 region could be partially attributed to neutralization escape variants since it has been reported that early neutralizing antibodies target this region in HIV-1 subtype C 27. Previous reports linked V1/V2 sequence changes to coreceptor preference, but these results were mostly based on subtype B isolates and were not uniformly observed 28–32. We therefore suggest that the extensive V1/V2 changes needed (in addition to V3 changes) in HIV-1 subtype C isolates might be one barrier preventing CXCR4 switching. The finding that late R5 viruses isolated from patient TM18 had extensive V1/V2 divergence but not the V3 changes associated with CXCR4 use further suggests that the V1/V2 changes need to occur prior to V3 mutations, a more extreme case of the compensatory changes required to counteract the fitness losses associated with V3 mutations observed in prior studies 17.
Lineage 3 reflect what happens more commonly in subtype C infection, with most patients progressing to AIDS maintaining R5 viruses 33. It is possible that the multiple variants observed in this patient contributed to rapid CD4 T cell depletion resulting in loss of target cells, which tipped the fitness advantage back to late R5 viruses with altered cell tropism 26, 34. Late R5 viruses have shown altered biological properties compared to early R5 isolates, including enhanced viral fitness, altered mode of using CCR5 (less CCR5/CD4 expression dependant) and sensitivity to inhibitors 34, 35. Despite the substantial divergence from consensus within the V1/V2 region and noticeable changes in the V3 region, lineage 3 env clones maintained CCR5 usage. Inhibition studies with TAK779 suggest that these viruses and the early CCR5 viruses from lineage 2 have similar sensitivities to the CCR5 inhibitor (data not shown). Further analysis would be needed to determine if the V3 changes observed within lineage 2 would confer CXCR4 usage to lineage 3. Previously, we have shown that in addition to compensating for the loss of fitness, V1/V2 mutations often increased entry efficiency on CCR5 targets 17. These earlier results propose that lineage 3 may have evolved to improve entry via CCR5 and that this process is reflected by the divergence from the year 5 consensus sequence.
The coexisting lineages also had distinctive alternative coreceptor profiles. Viruses able to use CCR5 could in general also utilize CCR3, CCR8 and FPRL1. By contrast, env clones that gained the ability to use CXCR4 also expanded coreceptor use to include APJ, CMKLR1, RDC-1, CXCR6, CCR1, GPR1 and CCR6, comparable to recent findings 20, 36. In addition, we also observed that some of these clones could use D6 efficiently for entry (data not shown). Nonetheless, CCR5 is still the preferred coreceptor used in pediatric HIV-1 infection 37, 38. It is interesting to note that lineage 1 exclusively used CCR5 and the ability to utilize alternative coreceptors evolved with disease progression. In addition, the expansion to use alternative coreceptors did not necessarily correlate with the ability to use CXCR4, exemplified by lineage 3. These observations imply that improved CCR5 use is associated with use of some alternative coreceptors, while acquisition of CXCR4 use correlates with further expansion of alternative coreceptor use. Prior studies of HIV-1 subtype C clones indicate that regions outside V3 including V1/V2 and additional N-glycosylation sites, may be responsible for alternative coreceptor usage 39–41. In this study lineage 2 and 3 shared longer V1/V2 loops, increased glycosylation sites and major divergence from the consensus with positive selection in V1/V2 and C2 region, which possibly resulted in their ability to use alternative coreceptors in vitro.
Recombination occurs frequently during HIV-1 replication with the C2 region shown to be a recombination hotspot 42, 43. In this study most recombination occurred within the V1-C2 region of env. Interestingly, we observed in some viruses that features within the C2 region were enough to confer CCR5 use despite the remainder of env being associated with CXCR4 use. Recombination between viruses of different tropism has previously been reported 44–48 and, combined with these results, supports the importance of the V1/V2 and C2 region in coreceptor usage. However, these recombinant viruses highlight the problem of predicting virus phenotype based on only the V3 region sequence.
There is renewed interest in coreceptor determination with the approval of a CCR5 antagonist, particularly for easy-to-use methods such as phenotype predictors. The usefulness of these predictors in clinical settings is still debatable 49–51. Here the C-PSSM had good predicative value for the early R5 and R5X4 viruses, but incorrectly predicted some of the late R5 viruses as CXCR4-using (from lineage 3)7. We showed the applicability of C-PSSM as a supportive tool in sequence analysis, but the current results indicate that caution is needed, particularly in predicting late stage X4 viruses.
Our results provide important insights into the lower incidence of coreceptor switching in subtype C infection. More sequence divergence is needed to acquire CXCR4 use than in subtype B infection, and extensive mutation of the V1/V2 region is required in subtype C but not in subtype B. These conclusions are limited by the number of patient samples examined, but the current results suggest that it will be important and illuminating to extend these studies to longitudinal samples from additional patients.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 AI052778 and U19 AI076981 from the National Institute of Allergy and Infectious Diseases (NIAID). Initial viral isolation studies done at the NICD were funded through the South African AIDS Vaccine Initiative (SAAVI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health. This is manuscript number 20547 from The Scripps Research Institute.
Disclosure of funding: NIH grant RO1AI52778 and U19AI076981.
Sources of support: NIH and the La Jolla Foundation for Microbicides Research.
Footnotes
Meetings where data was presented: Keystone Symposium on HIV Biology and Pathogenesis, Santa Fe, New Mexico Jan 12 – Jan 17, 2010.
Nucleotide sequence accession numbers. These will be supplied on acceptance of the manuscript.
References
- 1.Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 2.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1--infected individuals. J Exp Med. 1997;185(4):621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scarlatti G, Tresoldi E, Bjorndal A, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3(11):1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 4.Abebe A, Demissie D, Goudsmit J, et al. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. Aids. 1999 Jul 30;13(11):1305–1311. doi: 10.1097/00002030-199907300-00006. [DOI] [PubMed] [Google Scholar]
- 5.Ping LH, Nelson JA, Hoffman IF, et al. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73(8):6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cilliers T, Nhlapo J, Coetzer M, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol. 2003 Apr;77(7):4449–4456. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coetzer M, Cilliers T, Ping LH, Swanstrom R, Morris L. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology. 2006 Dec 5–20;356(1–2):95–105. doi: 10.1016/j.virol.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 8.Batra M, Tien PC, Shafer RW, Contag CH, Katzenstein DA. HIV type 1 envelope subtype C sequences from recent seroconverters in Zimbabwe. AIDS Res Hum Retroviruses. 2000 Jul 1;16(10):973–979. doi: 10.1089/08892220050058399. [DOI] [PubMed] [Google Scholar]
- 9.Singh A, Page T, Moore PL, et al. Functional and genetic analysis of coreceptor usage by dualtropic HIV-1 subtype C isolates. Virology. 2009 Oct 10;393(1):56–67. doi: 10.1016/j.virol.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michler K, Connell BJ, Venter WD, Stevens WS, Capovilla A, Papathanasopoulos MA. Genotypic characterization and comparison of full-length envelope glycoproteins from South African HIV type 1 subtype C primary isolates that utilize CCR5 and/or CXCR4. AIDS Res Hum Retroviruses. 2008 May;24(5):743–751. doi: 10.1089/aid.2007.0304. [DOI] [PubMed] [Google Scholar]
- 11.Coetzer M, Cilliers T, Papathanasopoulos M, et al. Longitudinal Analysis of HIV Type 1 Subtype C Envelope Sequences from South Africa. AIDS Res Hum Retroviruses. 2007 Feb;23(2):316–321. doi: 10.1089/aid.2006.0207. [DOI] [PubMed] [Google Scholar]
- 12.Kassaye S, Johnston E, McColgan B, Kantor R, Zijenah L, Katzenstein D. Envelope coreceptor tropism, drug resistance, and viral evolution among subtype C HIV-1-infected individuals receiving nonsuppressive antiretroviral therapy. J Acquir Immune Defic Syndr. 2009 Jan 1;50(1):9–18. doi: 10.1097/QAI.0b013e31818ffdff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White EJ, McColgan B, Kassaye S, Zijenah L, Katzenstein D. Unusual five amino acid insert within subtype C HIV-1 envelope contributes to dual-tropism (X4R5) Aids. Apr 24;24(7):1063–1064. doi: 10.1097/QAD.0b013e328331f717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel MB, Hoffman NG, Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J Virol. 2008 Jan;82(2):903–916. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrillo A, Ratner L. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J Virol. 1996 Feb;70(2):1301–1309. doi: 10.1128/jvi.70.2.1301-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koito A, Harrowe G, Levy JA, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994 Apr;68(4):2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pastore C, Nedellec R, Ramos A, Pontow S, Ratner L, Mosier DE. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J Virol. 2006 Jan;80(2):750–758. doi: 10.1128/JVI.80.2.750-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coetzer M, Nedellec R, Salkowitz J, et al. Evolution of CCR5 use before and during coreceptor switching. J Virol. 2008 Dec;82(23):11758–11766. doi: 10.1128/JVI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen MA, Coetzer M, van ’t Wout AB, Morris L, Mullins JI. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope v3 sequences. J Virol. 2006 May;80(10):4698–4704. doi: 10.1128/JVI.80.10.4698-4704.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nedellec R, Coetzer M, Shimizu N, et al. Virus Entry via the Alternative Coreceptors CCR3 and FPRL1 Differs by Human Immunodeficiency Virus Type 1 Subtype. J Virol. 2009 Jun 24; [Google Scholar]
- 21.Choge I, Cilliers T, Walker P, et al. Genotypic and phenotypic characterization of viral isolates from HIV-1 subtype C-infected children with slow and rapid disease progression. AIDS Res Hum Retroviruses. 2006 May;22(5):458–465. doi: 10.1089/aid.2006.22.458. [DOI] [PubMed] [Google Scholar]
- 22.Gao F, Morrison SG, Robertson DL, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996 Mar;70(3):1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995 Feb 1;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 24.Soda Y, Shimizu N, Jinno A, et al. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem Biophys Res Commun. 1999 May 10;258(2):313–321. doi: 10.1006/bbrc.1999.0633. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Gaschen B, Blay W, et al. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004 Dec;14(12):1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 26.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999 Dec;73(12):10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore PL, Ranchobe N, Lambson BE, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009 Sep;5(9):e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozek K, Thielen A, Sierra S, Kaiser R, Lengauer T. V3 loop sequence space analysis suggests different evolutionary patterns of CCR5- and CXCR4-tropic HIV. PLoS One. 2009;4(10):e7387. doi: 10.1371/journal.pone.0007387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman NG, Seillier-Moiseiwitsch F, Ahn J, Walker JM, Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J Virol. 2002 Apr;76(8):3852–3864. doi: 10.1128/JVI.76.8.3852-3864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollakis G, Kang S, Kliphuis A, Chalaby MI, Goudsmit J, Paxton WA. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J Biol Chem. 2001 Apr 20;276(16):13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]
- 31.Dong XN, Chen X, Chen Y, et al. Short communication: HIV type 1 phenotype, tropism, and sequence patterns: association and preference. AIDS Res Hum Retroviruses. 2005 Mar;21(3):234–238. doi: 10.1089/aid.2005.21.234. [DOI] [PubMed] [Google Scholar]
- 32.Masciotra S, Owen SM, Rudolph D, et al. Temporal relationship between V1V2 variation, macrophage replication, and coreceptor adaptation during HIV-1 disease progression. Aids. 2002 Sep 27;16(14):1887–1898. doi: 10.1097/00002030-200209270-00005. [DOI] [PubMed] [Google Scholar]
- 33.Bjorndal A, Sonnerborg A, Tscherning C, Albert J, Fenyo EM. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retroviruses. 1999 May 1;15(7):647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 34.Gray L, Sterjovski J, Churchill M, et al. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology. 2005 Jul 5;337(2):384–398. doi: 10.1016/j.virol.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 35.Repits J, Oberg M, Esbjornsson J, et al. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J Gen Virol. 2005 Oct;86(Pt 10):2859–2869. doi: 10.1099/vir.0.81111-0. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu N, Tanaka A, Oue A, et al. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. Aids. 2009 Apr 27;27(7):761–769. doi: 10.1097/QAD.0b013e328326cc0d. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan WM, Dorr P, Perros M, et al. Lack of alternative coreceptor use by pediatric HIV-1 R5 isolates for infection of primary cord or adult peripheral blood mononuclear cells. Arch Virol. 2008;153(2):363–366. doi: 10.1007/s00705-007-1099-6. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YJ, Moore JP. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J Virol. 1999 Apr;73(4):3443–3448. doi: 10.1128/jvi.73.4.3443-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dash PK, Siddappa NB, Mangaiarkarasi A, et al. Exceptional molecular and coreceptor-requirement properties of molecular clones isolated from an Human Immunodeficiency Virus Type-1 subtype C infection. Retrovirology. 2008;5:25. doi: 10.1186/1742-4690-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cilliers T, Willey S, Sullivan WM, et al. Use of alternate coreceptors on primary cells by two HIV-1 isolates. Virology. 2005 Aug 15;339(1):136–144. doi: 10.1016/j.virol.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 41.Gorry PR, Dunfee RL, Mefford ME, et al. Changes in the V3 region of gp120 contribute to unusually broad coreceptor usage of an HIV-1 isolate from a CCR5 Delta32 heterozygote. Virology. 2007 May 25;362(1):163–178. doi: 10.1016/j.virol.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jetzt AE, Yu H, Klarmann GJ, Ron Y, Preston BD, Dougherty JP. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol. 2000 Feb;74(3):1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galetto R, Moumen A, Giacomoni V, Veron M, Charneau P, Negroni M. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J Biol Chem. 2004 Aug 27;279(35):36625–36632. doi: 10.1074/jbc.M405476200. [DOI] [PubMed] [Google Scholar]
- 44.Pollakis G, Abebe A, Kliphuis A, et al. Phenotypic and genotypic comparisons of CCR5- and CXCR4-tropic human immunodeficiency virus type 1 biological clones isolated from subtype C-infected individuals. J Virol. 2004 Mar;78(6):2841–2852. doi: 10.1128/JVI.78.6.2841-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J Virol. 2006 Mar;80(5):2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kemal KS, Foley B, Burger H, et al. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12972–12977. doi: 10.1073/pnas.2134064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mild M, Esbjornsson J, Fenyo EM, Medstrand P. Frequent intrapatient recombination between human immunodeficiency virus type 1 R5 and X4 envelopes: implications for coreceptor switch. J Virol. 2007 Apr;81(7):3369–3376. doi: 10.1128/JVI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salemi M, Burkhardt BR, Gray RR, Ghaffari G, Sleasman JW, Goodenow MM. Phylodynamics of HIV-1 in lymphoid and non-lymphoid tissues reveals a central role for the thymus in emergence of CXCR4-using quasispecies. PLoS ONE. 2007;2(9):e950. doi: 10.1371/journal.pone.0000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007 Dec;25(12):1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 50.Low AJ, Dong W, Chan D, et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. Aids. 2007 Sep 12;21(14):F17–24. doi: 10.1097/QAD.0b013e3282ef81ea. [DOI] [PubMed] [Google Scholar]
- 51.Poveda E, Briz V, Roulet V, et al. Correlation between a phenotypic assay and three bioinformatic tools for determining HIV co-receptor use. Aids. 2007 Jul 11;21(11):1487–1490. doi: 10.1097/QAD.0b013e32826fb741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.