Abstract
Transient Receptor Potential (TRP) channels are rapidly gaining attention as important receptors and transducers of diverse sensory and environmental cues. Recent progress in the field has provided new insights into the structure and function of the ankyrin repeat motifs present in the N-terminal cytosolic domain of many TRP channels. The topics addressed in this review include the structural features of canonical ankyrin repeats, new clues into the functions these repeats perform in cells, and how this information can be applied to develop further experiments on TRP channels and other proteins containing ankyrin repeats.
TRP proteins form a large family of ion channels particularly important in animals from worms to man, with more than 30 members in mammals.1 The founding family member, the Drosophila trp gene - named for the “transient receptor potential” phenotype its mutation causes in light perception by photoreceptors - was first cloned in 1989.2 The functions assigned to TRP channels in physiology are constantly expanding. They are key transducers of sensory signals, and are generally important in sensing information about the environment in both neuronal and non-neuronal cells (see ref. 3 for comprehensive reviews). Some TRP channels are found in excitable cells of the sensory nervous system, while others are expressed in various other tissues and organs. TRP channels have been implicated in many physiological processes, including calcium and magnesium homeostasis, neuronal growth, temperature sensation and pain perception, to name a few. For example, TRPV1 senses painfully hot temperatures, low extracellular pH and the capsaicin of “hot” chili peppers, while TRPA1 senses many painful chemical stimuli, including the pungent compounds in wasabi and cinnamon. TRPV1 and TRPA1 are expressed in partially overlapping sets of nociceptor neurons that transmit pain signals to the brain.
At a molecular level, TRP channels assemble as tetramers through a channel domain that spans the membrane six times and has sequence similarity to voltage-gated ion channels. TRP channels also have large N- and C-terminal cytosolic domains that flank the transmembrane channel domain. These cytosolic domains participate in channel gating and regulation and sense information about the cellular state - ion and metabolite levels, for example. The TRP channel family is divided into seven subfamilies based on sequence similarity1: TRPA (ankyrin), TRPC (canonical), TRPM (melastatin), TRPML (mucolipin), TRPP (polycystin), TRPV (vanilloid), and TRPN (NOMPC). Several of these subfamilies - TRPA, TRPC, TRPN and TRPV - have ankyrin repeat sequence motifs in their N-terminal cytosolic domains. Recent work from my lab and others has taken a structural biology approach to elucidate the roles of these ankyrin repeats in TRP channel function.4–9 The emerging structural and functional features of these TRP channel ankyrin repeats are the focus of this short review.
Ankyrin repeats and their structure
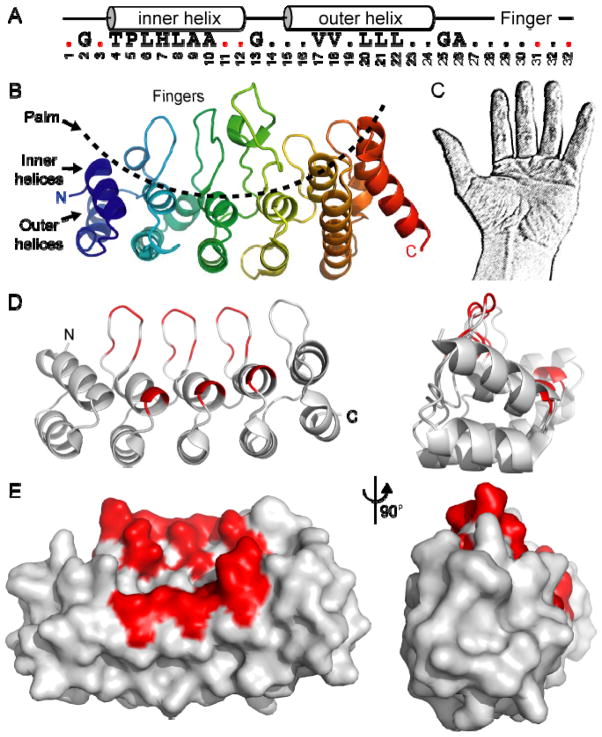
Ankyrin repeats were first identified as repeating sequence motifs in two transcription factors - yeast Swi6/Cdc10 and Drosophila Notch10 - and acquired their moniker when it was discovered that the cytoskeletal ankyrin protein contains 24 such repeats.11 Ankyrin repeat motifs have now been found in thousands of proteins from every branch of the tree of life, although they are most common in eukaryotes. In fact, ankyrin repeat-containing proteins are the ninth most common InterPro family in the human genome, with nearly 300 genes containing ankyrin repeats.12 Ankyrin repeats are found in many types of proteins besides TRP channels, including several large families of transcription factors (e.g. NFκB and Notch) and regulatory proteins (e.g. cyclin-dependent kinase inhibitors) for example. The ankyrin repeat motif is a short sequence, typically 33 amino acid residues, which forms an anti-parallel helix-turn-helix structure followed by a β-hairpin loop (Fig. 1A). At least three repeats, often more, are found in tandem and stack together to yield a hand-shaped domain with a concave palm surface formed of the inner helices and fingers, and a convex surface analogous to the back of the hand (Fig. 1). The structural features of ankyrin repeats and their consensus sequences have been extensively reviewed.13,14
Fig. 1.
Structure of ankyrin repeats. (A) Consensus ankyrin repeat sequence motif, with the resulting secondary structure above, and the repeat position number underneath. Consensus residues form the hydrophobic core as multiple repeats stack to form a domain, as well as the twists and turns through residues such as glycine and proline, leaving the protein surface highly variable. Red dots represent variable residues in DARPin libraries.37 (B) Ribbon diagram of the six ankyrin repeats of TRPV1 with an N- to C-terminal rainbow coloring. The inner helices and fingers form the concave palm surface, while the outer helices form a convex surface. (C) The shape of an ankyrin repeat domain is often likened to a hand, and analogously, the palm and fingers are the most commonly used interaction surfaces. (D–E) Structure of a five-repeat DARPin,37 with the variable residues in red, represented as a ribbon (D) or surface (E).
Ankyrin repeat domains fit broadly into two categories. Many proteins have a small set of approximately four to seven ankyrin repeats forming a structural domain. Within the TRP channel family, that includes the TRPC proteins which likely have four or five repeats, and the TRPV channels which have six repeats (although a few invertebrate TRPV channels may only have five). Some proteins have highly regular repeats with sequences close to the consensus motif; an example is Gankyrin, an oncogenic protein associated with the proteasome, with seven regular repeats (Fig. 2A). In contrast, others have a number of deviations from consensus, such as insertions, deletions or kinks in the domain. MYPT1, a phosphatase regulatory subunit, is one example.15 MYPT1 has eight ankyrin repeats with a sharp kink in the middle of the domain that allows it to wrap around the C-terminus of the PP1δ phosphatase. The ankyrin repeats of TRPV ion channels provide another example of irregular repeats, with several small insertions leading to elongated fingers and outer helices, and altered packing causing a pronounced twist between the fourth and fifth repeat (Fig. 2C; see refs. 6,8,9 for detailed descriptions). A second category of domains have a very large number of ankyrin repeats. The cytoskeletal component ankyrin, with 24 repeats, certainly fits in this category. The TRPA1 and TRPN-subfamily channels also fit in this second category, with 17–29 repeats.16,17 Most proteins with a large number of repeats tend to have sequence motifs very close to consensus. This leads to a very regular domain structure (Fig. 2D), the implications of which are discussed below.
Fig. 2.
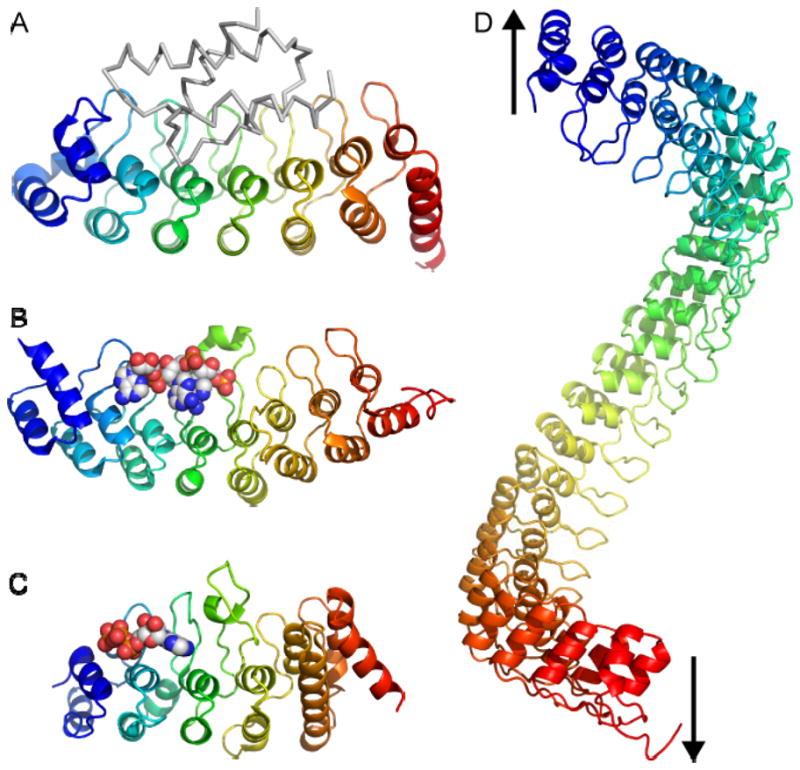
Function of ankyrin repeats: beyond protein-protein interaction. (A) Structure of Gankyrin bound to the S6 ATPase of the 26S proteasome (grey).48 (B) Structure of the ankyrin repeats of RNase L bound to an oligoadenylate trimer (spheres).20 (C) Structure of the ankyrin repeats of TRPV1 bound to ATP (spheres).7 (D) Model of a 24-repeat domain, such as that of ankyrin, with the arrows indicating the direction of the possible spring (based on ref. 49). All ankyrin repeat domains have an N- to C-terminal rainbow coloring.
Ligand binding properties of ankyrin repeats
To date, no enzymatic activity has been linked to ankyrin repeats, and they are generally labeled as protein-protein interaction motifs. Several structures are available for ankyrin repeats of various cellular proteins interacting with their protein targets (Table 1; also see earlier review14). Some of these interactions are dependent on covalent modifications, like the interaction of the ankyrin repeat domain of the methylated lysine 9 of histone H3 with the histone methyltransferases G9A and G9A-like protein (GLP).18 The structures and functions of ankyrin repeat-interacting proteins are very diverse, and have no shared properties. The only commonality is the binding site on the ankyrin repeat, which generally includes the surface formed by the concave surface of the palm and fingers (Fig. 2).
Table 1.
Structures of ankyrin repeats
| Proteins (number of repeats) | Interacting partner in the structure(s) | Species and PDB ID Codes | |
|---|---|---|---|
|
Ankyrin repeat domains from natural proteins
| |||
| Ankyrin-R (12) | Human (1N11) | ||
| Bcl-3 (7) | Human (1K1A 1K1B) | ||
| Myotrophin (4) | Rat (1MYO 2MYO) | ||
| PAPβ (4) | Mouse (1DCQ) | ||
| Q5ZSV0 (9) | L. pneumophila (2AJA) | ||
| SWI6 (5) | S. cerevisiae (1SW6) | ||
| TRPV2 (6) | Human (2F37), rat (2ETA 2ETB 2ETC) | ||
| UPLC1 GAP domain (4) | Human (2B0O) | ||
| P15INK4B (4) | Mouse (1D9S) | ||
|
| |||
|
Ankyrin repeat domains from natural proteins in complex with
protein target
| |||
| 53BP2 (4) | P53 | Human (1YCS) | |
| GABPβ1 subunit (5) | GABPα subunit | Mouse (1AWC) | |
| IκBα (6) and IκBβ (6) | NFκB | Human IκBα (1IKN 1NFI), mouse IκBβ (1K3Z 1OY3) | |
| MYPT1 (8) | Phosphatase PP1δ | Human (1S70) | |
| Ribonuclease L (8) | Oligoadenylate trimer | Human (1WDY) | |
| TRPV1 (6) | ATP | Rat (2NYJ 2PNN) | |
|
| |||
|
Ankyrin repeat domains from natural proteins with and without
protein target
| |||
| Without | With | ||
| Gankyrin or Nas6p (7) | S6 ATPase of the 26S proteasome | Human (1QYM 1TR4 1UOH) | Mouse (2DVW 2DWZ) |
| S. cerevisiae Nas6p (1IXV 1WG0) | S. cerevisiae Nas6p (2DZN 2DZO) | ||
| Notch/Notch-1/Lin-2 (4–8) | CSL/Lag-1 and MAML/Lag-3 | Human (2F8Y 1YYH 2HE0) | Human (2F8X) |
| Drosophila (1OT8), Mouse (1YMP), | C. elegans (2FO1) | ||
| P16INK4A (4) | Cyclin-dependent kinase (CDK) 6 | Human (1A5E 2A5E 1DC2) | Human (1BI7) |
| P18INK4C (5) | CDK6 | Human (1BU9 1IHB; mutants 1MX2 1MX4 1MX6) | Human (1G3N) |
| P19INK4D (5) | CDK6 | Human (1BD8), mouse (1AP7) | Human (1BI8), mouse (1BLX) |
| G9A-like protein (7) | Histone H3 tail methylated at K9 | Human (3B7B) | Human (3B95) |
|
| |||
|
Artificial ankyrin repeat domains
| |||
| 3ANK (3) | 1N0Q | ||
| 4ANK (4) | 1N0R | ||
| E3_19 (5) | 2BKG | ||
| DARPin against HER2 (4) | 2JAB | ||
| NI3C (5) | 2QYJ | ||
| SANK E3_5 (5) | 1NK0 | ||
|
| |||
|
Artificial ankyrin repeat domains bound to their protein
targets
| |||
| DARPin (5) | AcrB | 2J8S | |
| DARPin (5) | Caspase-2 | 2P2C | |
| DARPin (5) | Maltose binding protein | 1SVX | |
| DARPin (5) | APH (3′)-IIIA | 2BKK | |
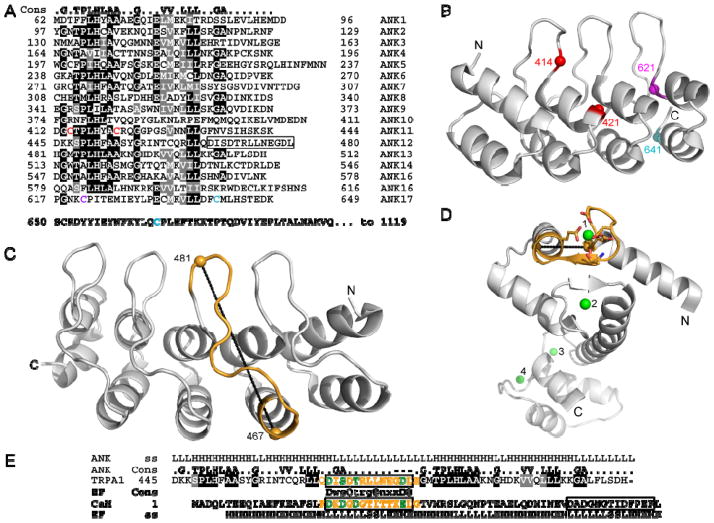
Less well known is that non-proteinaceous ligands have also been identified for ankyrin repeats. The ankyrin repeats of RNase L regulate its activity through the interaction with oligoadenylates (Fig. 2B).19,20 More recently, a bound ATP molecule was observed when the structure of the TRPV1 ankyrin repeats was determined by x-ray crystallography (Fig. 2C).7 The palm surface of the TRPV1 repeats forms an atypical nucleotide binding pocket. Biochemical studies demonstrated that ATP and calmodulin bind competitively to the same site. ATP prevents desensitization to repeated applications of capsaicin, while calmodulin is required for desensitization. These findings contribute to a model for the calcium-dependent regulation of TRPV1 channel sensitivity through ligand binding to its ankyrin repeats.7 A third example is TRPA1, which is activated by many pungent compounds like isothiocyanates in wasabi and mustard, or cinnamaldehyde.21 These compounds have very different structures, but two groups noted that they all are electrophiles and determined that the compounds covalently modify intracellular cysteines in the ankyrin repeat domain and adjacent linker region of TRPA1.22,23 Two sets of cysteines were identified as important in the response to these agents - cysteines 414 and 421 on the predicted palm surface of repeat 11, and cysteines 621, 641 and 665 on the most C-terminal predicted repeat and adjacent linker (numbering follows the human sequence; Fig. 3A–B).22,23 These results implicate the TRPA1 ankyrin repeats in the channel’s sensing and gating functions, although how the covalent attachments lead to channel activation is still unclear. TRPA1 is also directly gated by calcium, and residues in the finger loop of predicted ankyrin repeat 12 have been implicated in this mechanism (Fig. 3).24 The sequence motif important for calcium gating of TRPA1 has similarity to an EF-hand motif, a common calcium-binding motif.24 However, structural comparisons of an EF-hand motif of calmodulin and the ankyrin repeat finger suggest that these two structures are mutually exclusive (Fig. 3C–E). However, finger loop 12 and adjacent finger loops contain several negatively-charged residues, suggesting that the TRPA1 ankyrin repeat could form a non-EF-hand calcium-binding site. In summary, the TRPA1, TRPV1 and RNase L data broaden the definition of ankyrin repeat function to that of ligand-interaction motif. Therefore, when seeking to identify potential ligands, one should keep in mind that ankyrin repeat binding partners are clearly not restricted to proteins.
Fig. 3.
The ankyrin repeats of TRPA1. (A) Sequence of the ankyrin repeats of human TRPA1 (accession number O75762) aligned against the ankyrin repeat consensus (top). Similar and identical residues are shaded grey and black, respectively. The EF-hand-like sequence motif is boxed. Colored cysteines are important in activation of TRPA1 by electrophiles as identified by the Patapoutian (red)22 or Julius group (cyan)23 or both (magenta). (B) The structure of four canonical ankyrin repeats is used to illustrate the position of important TRPA1 cysteines either in internal repeat 11 (red) or terminal repeat 17 (magenta and cyan). The cysteines are colored as in (B). (C) The structure of five canonical ankyrin repeats is used to illustrate the position of the EF-hand-like sequence motif in repeat 12 of TRPA1. Positions 467 and 481, which are adjacent to highly conserved ankyrin repeat residues, and therefore predicted to closely match this canonical structure, are 25.6 Å apart (dotted line). Note that the finger loop of TRPA1 repeat 12 contains three additional residues compared to the canonical finger loop illustrated here. (D) The structure of calcium-bound calmodulin (PDB ID Code 3CLN) with four calcium ions in green and the first EF-hand motif colored gold. The black dotted line represents a 9.1 Å distance between residues 20 and 34 (10.7 Å separate these residues in apo-calmodulin; PDB ID Code 1DMO). (E) The sequences of TRPA1 and calmodulin are aligned at their EF-hand sequence motifs (box), along with the corresponding EF-hand and ankyrin repeat consensus sequences and secondary structures (ss; L = loop, H = helix, S = sheet). In the EF-hand consensus, capital letters indicate conserved negatively charged residues; small letters, residues that fit the consensus; x, any residue; and Φ, hydrophobic residues. Segments in yellow are illustrated in parts C and D, and residues in green provide coordinating groups to the calcium in an EF-hand structure.
As mentioned above, no common features can readily be detected from such broad diversity of ankyrin repeats ligands. Still, a few generalizations can be drawn from the ankyrin repeat-ligand interactions that have been characterized to date. First, as noted above, all interactions for which structures are available involve the palm surface of the ankyrin repeats. Some structures with larger ligands also include additional adjacent interfaces, such as the loops connecting inner and outer helices. But the palm and fingers remain a significant element of the binding site in all cases for which structures are available. Second, in all cases where structures have been determined both in the presence and absence of ligand, the ankyrin repeats essentially do not change conformation. Some ankyrin repeat domains, notably the NFκB regulator IκBα, are partially disordered until they bind their target ligand.25 But in general, information about the ligand-binding state of the ankyrin repeats is transmitted to the rest of the functional macromolecular complex through some other mechanism than a conformational change within the ankyrin repeats. Finally, there is no example yet of oligomerizing ankyrin repeat domains. Earlier reports on the TRPV5 and TRPV6 ion channels provided tantalizing evidence suggesting that their ankyrin repeats did in fact tetramerize.26 However, biochemical and structural studies with the isolated ankyrin repeat domains of TRPV5 and TRPV6 have failed to show any evidence of oligomerization.9
Ankyrin repeats as springs?
Domains with a large tandem array of ankyrin repeats are intriguing for at least two reasons: (i) the shape of a domain with many ankyrin repeats resembles a coil (Fig. 2D) and (ii) several TRP channels with a long ankyrin repeats domains - TRPA1 and the TRPN subfamily - have been implicated in mechanosensation (see ref. 27 for a recent review). In Drosophila, the TRPN-subfamily “no mechanoreceptor potential C” (NOMPC) channel is essential for the fly’s mechanosensory physiology.16 Furthermore, C. elegans TRPN1 and Drosophila NOMPC are both found in mechanosensory organs,16 and TRPN1 is a critical component in hearing and mechanosensation in zebrafish.28 TRPN proteins all have 29 ankyrin repeat motifs, and the predicted elongated coil shape of their ankyrin repeat domain led to the hypothesis that it acts as a spring that directly senses mechanical stimuli.29 But mice and humans do not have any TRPN-subfamily channel, and the identity of the mammalian mechanosensitive channel involved in hearing remains unknown. The TRPA1 channel, which has 17 ankyrin repeats, is expressed in mammalian inner ear hair cells, making it a tantalizing candidate for the mechanosensitive channel responsible for hearing.30 However, the TRPA1 knock-out mice are not hearing-impaired,31,32 although the knock-out strategies, which eliminated the pore segment, leave open the remote possibility that other portions of the protein (including the ankyrin repeats) are the essential parts for TRPA1 function in hearing. Although interest in TRPA1 as the potential mechanotransducer channel in hearing is waning, TRPA1 is clearly implicated in mechanosensation in mice and C. elegans.31,33
Molecular dynamics calculations5 and experimental evidence from atomic force microscopy4 have in fact demonstrated the elastic properties of ankyrin repeats in response to pulling on both ends of the domain. As expected for a mechanical spring, both studies have found that: (i) ankyrin repeat domains pulled from both ends stretch reversibly with forces on the order of 100 pN; (ii) the inferred spring constants of ~2–20 pN/nm are consistent with spring constants of biological mechanoreceptors; and (iii) increasing the number of repeats decreases the measured or calculated stiffness.4,5 These studies therefore support the hypothesis that long ankyrin repeat domains function as springs when pulled from both ends. In summary, although there is still no physiological evidence that the ankyrin repeats are directly involved in sensing mechanical stimuli, the hypothesis is certainly worth pursuing experimentally.
An additional question that arises from this hypothesis is whether these long ankyrin repeat domains have ligands, and whether ligand binding would impact the mechanical properties of the domain. For such a domain to sense mechanical stress as a spring requires both ends to be tethered. This could be done through a covalent link at one or both ends of the ankyrin repeat domain - for instance, with the transmembrane domain of the TRPA1 or TRPN channels C-terminal to the ankyrin repeat domain. A tether could also be formed through a non-covalent interaction with the ankyrin repeat domain. Moreover, these tethers have to provide a way to sense mechanical stimuli. For example, if a non-covalent interaction tethers the N-terminal end of a TRPN channel to the cytoskeleton, movement of the plasma membrane relative to the cytoskeleton attachment could cause stretch of the ankyrin repeats and be relayed to the transmembrane domain to cause channel opening. Alternatively, a non-covalent tether of the ankyrin repeat domain to the membrane could also allow the channel to sense changes in membrane tension between the ankyrin repeat anchor and the transmembrane domain. In addition, ligands interacting through the palm surface could influence or modulate the mechanical properties of the ankyrin repeat domain. Thus far, few ligands have been identified for TRPA1 and TRPN channels and their ankyrin repeats, aside from the covalent cysteine-modifying agents and calcium ions that gate TRPA1. But experimental tests of the mechanical spring hypothesis will need to include the identification of ankyrin repeat ligands as potential tethers and modulators of mechanical properties.
Experimenting with ankyrin repeats
It is clear that much remains to be discovered about the functions of ankyrin repeats in sensing and signaling by TRP channels. The abundant knowledge base about ankyrin repeat structure can and should be leveraged when designing experiments to identify new ligands and functions for ankyrin repeat domains in TRP channel biology. A few examples of how this information can be applied are described below.
The folding of ankyrin repeats is highly cooperative,34 requiring the presence of at least two or three consecutive repeats to generate a stable fold.35 For example, two missense mutations in the ankyrin repeats of the C. elegans osm-9 gene, a TRPV channel, result in a phenotype very similar to the null phenotype.36 Both mutations substitute the conserved glycine that precedes an inner helix with an acidic residue (position 2 in Fig. 1A), and likely disrupt the sharp turn linking an ankyrin repeat finger to the following inner helix (Fig. 1B). Therefore, in generating recombinant protein constructs containing ankyrin repeats, one should generally avoid breaking up a domain, or mutating ankyrin repeat consensus residues, because this would probably result in an unstable protein. However, identifying the N- and C-terminal boundaries of ankyrin repeat domains can be challenging. The terminal capping repeats are less well conserved than internal repeats since they have to pack against only one, rather than two, adjacent repeat. As a result, hydrophobic residues important for packing of adjacent repeats - particularly at positions 6, 8, 10, 17, 20, 21 and 22 (Fig. 1A) - are often substituted with solvent-interacting polar residues in capping repeats.14,37 These substitutions often prevent terminal repeats from being identified by sequence motif search algorithms. Supplementing sequence motif searches with secondary structure predictions and detection of sequence conservation using protein sequence alignments can be helpful in identifying terminal repeats. Using secondary structure predictions algorithms like NPS@,38 additional pairs of predicted helices adjacent to recognized repeats can be identified, which may correspond to capping repeats. Also, regions with low probability of secondary structure often mark domain boundaries. Similarly, domain boundaries can sometimes be identified as locations where the sequence conservation changes drastically. To this end, several multiple sequence alignments can be generated with different groupings of sequences using algorithms like ClustalW.39 Comparing alignments of orthologs from different species to alignments of homologs - subfamilies, families or superfamilies - can be particularly useful in identifying breaks in sequence similarity. Such an approach was used to determine that the TRPV proteins have six ankyrin repeats,6,40 although only three or four are recognized by motif search algorithms like Prosite41 and SMART.42
Properly folded ankyrin repeats form a very stable scaffold. Libraries of designed ankyrin repeat proteins (DARPins) have been generated using consensus residues in most positions, but varying residues on the palm surface (colored red in Fig. 1A).37 The resulting DARPins, containing four, five or six ankyrin repeats, are very stable, with melting temperatures ranging from 66 °C to 85 °C.37 Such libraries can then be screened to identify DARPins that bind and/or inhibit targets of interest. DARPins have been isolated that act as potent inhibitors of kinases, proteases and drug export systems, and the structures of several of these complexes have been determined (Table 1; see ref. 43 for a recent review). DARPins could become favorable alternatives to antibodies in some situations, and are often easier and less expensive to produce in large quantities. The successful use of DARPins highlights the fact that ankyrin repeats can, in principle, evolve - in nature or in the lab - to form stable protein domains that interact with just about any possible target.
The work on DARPins also serves to illustrate methods that can be used to ascertain that an isolated ankyrin repeat domain construct is stable and properly folded - short of determining its three-dimensional structure. Circular dichroism (CD) spectroscopy requires small amounts of purified protein samples to assess the secondary structure content of a protein as well as its thermal stability. Reviews listed in ref. 44 provide an excellent primer on CD methods and protocols. Many published spectra are available for both natural and designed ankyrin repeats (see ref. 37 and references therein). These spectra can serve as benchmarks because ankyrin repeat domains are expected to have similar secondary structure content, and therefore similar CD spectra. Size exclusion chromatography and other hydrodynamic measurements such as light scattering and analytical ultracentrifugation can also be used to determine the aggregation state of these recombinant proteins. A similar battery of tests was performed on recombinant ankyrin-B in the atomic force microscopy studies of its spring-like behavior.4 These techniques are therefore valuable tools to validate recombinant ankyrin repeat domains constructs generated from natural proteins. Most of these techniques do require purified proteins. Fortunately, many (although not all) ankyrin repeat domains can readily be purified from an E. coli-based overexpression system. Ref. 45 provides information on expression and purification of protein protocols based on the integrated experiences of structural genomics groups, while ref. 46 is an excellent primer on protein characterization and structural biology. Especially once validated, ankyrin repeat domain constructs are powerful research tools and could be used, for example, as baits in two-hybrid screens and pull-downs to identify new ligands. The validation process is likely to reduce the risk of finding false positive hits because of misfolding or aggregation problems, for example. Again, the ligand search should not be limited to proteins. New techniques involving chemical screens and metabolomics will likely be an important component in future research on TRP channels and ankyrin repeats. Many of these techniques (recently reviewed in ref. 47) have so far been applied to proteins with enzymatic activity, but a challenge for the future is to apply them to other biochemical activities such as regulatory binding interactions, as is the case for ankyrin repeats.
Another strategy in investigating the role of ankyrin repeats in TRP channels and other proteins is to disrupt the interactions of ankyrin repeats with their targets by mutagenesis, and determine the effect of such mutations on protein function. However, for many ankyrin repeat domains, the ligands and/or their binding sites remain unknown. In those cases, the generalizations about ankyrin repeats described above can be helpful in choosing which residues to mutate. The ankyrin repeat consensus motif is derived from the residues conserved to maintain the stable ankyrin repeat fold; that is, they are conserved for form (Fig. 1). In contrast, surface residues are particularly variable, underscoring the malleability of different ankyrin repeat surfaces to evolve interactions with distinct targets. Residues at positions 1, 3, 11, 12, 31 and 33 within each ankyrin repeat (Fig. 1A, D and E), which largely shape the palm surface, are good mutagenesis targets: variations at these positions, which were used to generate DARPin libraries, have little effect on protein stability,43 and since they form the palm surface, these residues are likely to be involved in interactions with the ligand(s). Furthermore, mutating residues conserved in orthologs across species but predicted to be exposed at the surface could be particularly effective: if they are not conserved to maintain the protein fold, then they are likely performing an important function, such as interacting with a ligand. For example, the TRPV1 ATP-binding site is formed by palm surface residues highly conserved across species.8 Another example is the patch of aromatic residues on ankyrin repeat fingers 2 and 3 that is particularly striking on TRPV2 but also conserved in other TRPV ion channels.6 In sum, the general strategy should be to use the current knowledge about ankyrin repeat structure and stability to select residues that are unlikely to affect protein folding and its intrinsic stability, but rather affect the ligand-binding function of the ankyrin repeats.
Conclusions
Recent progress in TRP channel research is shinning the spotlight on the structure and function of their ankyrin repeats. The ankyrin repeats of TRP channels offer a broad sampling of the diverse features of ankyrin repeat-containing proteins: a variety of ligand types; some long and regular domains and other shorter domains; and insertions, deletions and other irregularities in some repeats. Just like the ankyrin repeat domains of proteins like Notch, Gankyrin or IκB share more similarity within protein families than across, the ankyrin repeats of TRPA, TRPC, TRPN and TRPV each have their own characteristic features. Although much remains to be learned, a wealth of information is available on ankyrin repeat structure that can and should harnessed to design better experiments. It is hoped that the information and sources provided here will prove helpful to biologists in their studies of ankyrin repeats in TRP channels and beyond.
Acknowledgments
I thank Paul Garrity, Marcos Sotomayor and members of my lab, particularly Christopher Phelps, for discussions. This work was supported by a Klingenstein Fellowship Award and NIH 1R01GM081340 to RG.
References
- 1.Montell C. Sci STKE. 2005;2005:re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- 2.Montell C, Rubin GM. Neuron. 1989;2:1313–1323. doi: 10.1016/0896-6273(89)90069-x. [DOI] [PubMed] [Google Scholar]
- 3.Clapham DE. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]; Voets T, Talavera K, Owsianik G, Nilius B. Nat Chem Biol. 2005;1:85–92. doi: 10.1038/nchembio0705-85. [DOI] [PubMed] [Google Scholar]; Venkatachalam K, Montell C. Annual Review of Biochemistry. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gaudet R. Structural Insights into the Function of TRP Channels. In: Liedtke W, Heller S, editors. TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades. CRC Press; Boca Raton, FL: 2006. [Google Scholar]
- 4.Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 5.Sotomayor M, Corey DP, Schulten K. Structure (Camb) 2005;13:669–682. doi: 10.1016/j.str.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Jin X, Touhey J, Gaudet R. J Biol Chem. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]; McCleverty CJ, Koesema E, Patapoutian A, Lesley SA, Kreusch A. Protein Sci. 2006;15:2201–2206. doi: 10.1110/ps.062357206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 8.Phelps CB, Procko E, Lishko PV, Wang RR, Gaudet R. Channels. 2007;1:148–151. doi: 10.4161/chan.4716. [DOI] [PubMed] [Google Scholar]
- 9.Phelps CB, Huang RJ, Wang RR, Gaudet R. Biochemistry. 2008 doi: 10.1021/bi702109w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breeden L, Nasmyth K. Nature. 1987;329:651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- 11.Lux SE, John KM, Bennett V. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Sedgwick SG, Smerdon SJ. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]; Li J, Mahajan A, Tsai MD. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 14.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 16.Walker RG, Willingham AT, Zuker CS. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 17.Story GM, Peier AM, Reeve AJ, Eid SR, Mosbacher J, Hricik TR, Earley TJ, Hergarden AC, Andersson DA, Hwang SW, McIntyre P, Jegla T, Bevan S, Patapoutian A. Cell. 2003;112:819–829. doi: 10.1016/s0092-8674(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 18.Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1384. advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong B, Silverman RH. J Biol Chem. 1997;272:22236–22242. doi: 10.1074/jbc.272.35.22236. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT. Embo J. 2004;23:3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandell M, Story GM, Hwang SW, Viswanath V, Eid SR, Petrus MJ, Earley TJ, Patapoutian A. Neuron. 2004;41:849–857. doi: 10.1016/s0896-6273(04)00150-3. [DOI] [PubMed] [Google Scholar]; Jordt SE, Bautista DM, Chuang HH, McKemy DD, Zygmunt PM, Hogestatt ED, Meng ID, Julius D. Nature. 2004;427:260–265. doi: 10.1038/nature02282. [DOI] [PubMed] [Google Scholar]
- 22.Macpherson LJ, Dubin AE, Evans MJ, Marr F, Schultz PG, Cravatt BF, Patapoutian A. Nature. 2007;445:541–545. doi: 10.1038/nature05544. [DOI] [PubMed] [Google Scholar]
- 23.Hinman A, Chuang H-h, Bautista DM, Julius D. Proceedings of the National Academy of Sciences. 2006;103:19564–19568. doi: 10.1073/pnas.0609598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doerner JF, Gisselmann G, Hatt H, Wetzel CH. J Biol Chem. 2007;282:13180–13189. doi: 10.1074/jbc.M607849200. [DOI] [PubMed] [Google Scholar]; Zurborg S, Yurgionas B, Jira JA, Caspani O, Heppenstall PA. Nat Neurosci. 2007;10:277–279. doi: 10.1038/nn1843. [DOI] [PubMed] [Google Scholar]
- 25.Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. J Mol Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Q, Gyftogianni E, van de Graaf SF, Hoefs S, Weidema FA, Bindels RJ, Hoenderop JG. J Biol Chem. 2004;279:54304–54311. doi: 10.1074/jbc.M406222200. [DOI] [PubMed] [Google Scholar]; Erler I, Hirnet D, Wissenbach U, Flockerzi V, Niemeyer BA. J Biol Chem. 2004;279:34456–34463. doi: 10.1074/jbc.M404778200. [DOI] [PubMed] [Google Scholar]
- 27.Christensen AP, Corey DP. Nat Rev Neurosci. 2007;8:510–521. doi: 10.1038/nrn2149. [DOI] [PubMed] [Google Scholar]
- 28.Sidi S, Friedrich RW, Nicolson T. Science. 2003;301:96–99. doi: 10.1126/science.1084370. [DOI] [PubMed] [Google Scholar]
- 29.Howard J, Bechstedt S. Curr Biol. 2004;14:R224–226. doi: 10.1016/j.cub.2004.02.050. [DOI] [PubMed] [Google Scholar]; Corey DP, Sotomayor M. Nature. 2004;428:901–903. doi: 10.1038/428901a. [DOI] [PubMed] [Google Scholar]
- 30.Corey DP, Garcia-Anoveros J, Holt JR, Kwan KY, Lin SY, Vollrath MA, Amalfitano A, Cheung EL, Derfler BH, Duggan A, Geleoc GS, Gray PA, Hoffman MP, Rehm HL, Tamasauskas D, Zhang DS. Nature. 2004;432:723–730. doi: 10.1038/nature03066. [DOI] [PubMed] [Google Scholar]
- 31.Bautista DM, Jordt SE, Nikai T, Tsuruda PR, Read AJ, Poblete J, Yamoah EN, Basbaum AI, Julius D. Cell. 2006;124:1269–1282. doi: 10.1016/j.cell.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Kwan KY, Allchorne AJ, Vollrath MA, Christensen AP, Zhang DS, Woolf CJ, Corey DP. Neuron. 2006;50:277–289. doi: 10.1016/j.neuron.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 33.Kindt KS, Viswanath V, Macpherson L, Quast K, Hu H, Patapoutian A, Schafer WR. Nat Neurosci. 2007;10:568–577. doi: 10.1038/nn1886. [DOI] [PubMed] [Google Scholar]
- 34.Barrick D, Ferreiro DU, Komives EA. Curr Opin Struct Biol. 2008;18:27–34. doi: 10.1016/j.sbi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosavi LK, Minor DL, Jr, Peng ZY. Proc Natl Acad Sci U S A. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colbert HA, Smith TL, Bargmann CI. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. Journal of Molecular Biology. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 38.Combet C, Blanchet C, Geourjon C, Deleage G. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 39.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wissenbach U, Niemeyer BA, Fixemer T, Schneidewind A, Trost C, Cavalie A, Reus K, Meese E, Bonkhoff H, Flockerzi V. J Biol Chem. 2001;276:19461–19468. doi: 10.1074/jbc.M009895200. [DOI] [PubMed] [Google Scholar]
- 41.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stumpp MT, Amstutz P. Curr Opin Drug Discov Devel. 2007;10:153–159. [PubMed] [Google Scholar]
- 44.Kelly SM, Jess TJ, Price NC. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]; Martin SR, Schilstra MJ, John, Correia J, WilliamDetrich H., III . Methods in Cell Biology. Academic Press; 2008. Circular Dichroism and Its Application to the Study of Biomolecules. [DOI] [PubMed] [Google Scholar]
- 45.Graslund S, Nordlund P, Weigelt J, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Nat Methods. 2008;5:135–146. [Google Scholar]
- 46.Minor DL., Jr Neuron. 2007;54:511–533. doi: 10.1016/j.neuron.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saghatelian A, Cravatt BF. Nat Chem Biol. 2005;1:130–142. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura Y, Nakano K, Umehara T, Kimura M, Hayashizaki Y, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structure. 2007;15:179–189. doi: 10.1016/j.str.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Michaely P, Tomchick DR, Machius M, Anderson RG. Embo J. 2002;21:6387–6396. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]