Abstract
Nicotinamidases are salvage enzymes that convert nicotinamide to nicotinic acid. These enzymes are essential for the recycling of nicotinamide into NAD+ in most prokaryotes, most single cell and multicellular eukaryotes, but not in mammals. The significance of these enzymes for nicotinamide salvage and for NAD+ homeostasis has increased interest in nicotinamidases as possible antibiotic targets. Nicotinamidases are also regulators of intracellular nicotinamide concentrations, thereby regulating signaling of downstream NAD+ consuming enzymes, such as the NAD+-dependent deacetylases (sirtuins). Here, we report several high resolution crystal structures of the nicotinamidase from Streptococcus pneumoniae (SpNic) in unliganded and ligand-bound forms. The structure of the C136S mutant in complex with nicotinamide provides details about substrate binding while a trapped nicotinoyl-thioester complexed with SpNic reveals the structure of the proposed thioester reaction intermediate. Examination of the active site of SpNic reveals several important features including a metal ion that coordinates the substrate and the catalytically relevant water molecule, and an oxyanion hole which both orients the substrate and offsets the negative charge that builds up during catalysis. Structures of this enzyme with bound nicotinaldehyde inhibitors elucidate the mechanism of inhibition and provide further details about the catalytic mechanism. In addition, we provide a biochemical analysis of the identity and role of the metal ion that orients the ligand in the active site and activates the water molecule responsible for hydrolysis of the substrate. These data provide structural evidence for several proposed reaction intermediates and allow for a more complete understanding of the catalytic mechanism of this enzyme.
Nicotinamide adenine dinucleotide (NAD+) and its phosphorylated and reduced forms, NADP+, NADH, and NADPH, are central to cellular metabolism and energy production. Maintenance of NAD+ concentrations is important for cell and organism viability, and the strategies of cells to replete NAD+ from nicotinamide (NAM), nicotinic acid (NA), nicotinamide riboside and via de novo routes are quite elaborate (1). The complexity of NAD+ biosynthesis in most organisms is likely linked to the importance of the dinucleotides in central metabolism, and targeting NAD+ biosynthesis as an antibiotic approach has recently been the subject of increased investigation (1-3). In addition to biosynthetic production, salvage pathways operate to offset depletion of NAD+ stocks by NAD+ consuming enzymes and non-enzymatic hydrolysis, which can occur even at physiological temperatures and pH (4, 5).
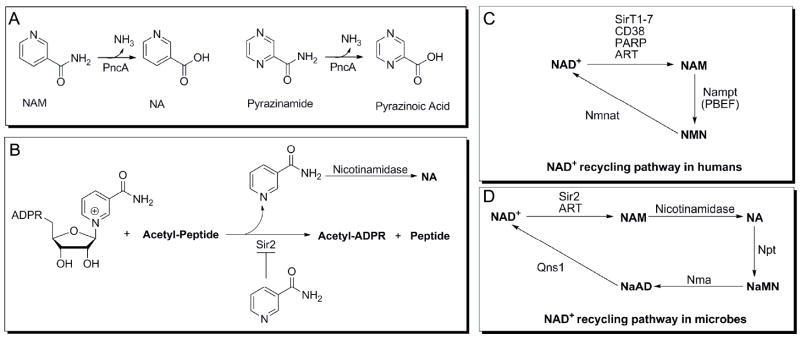
An important difference between human NAD+ homeostasis and that of most prokaryotes, unicellular eukaryotes and multicellular eukaryotes involves the mechanism of NAD+ production and/or salvage. In most organisms nicotinamide is recycled back to NAD+ by first being converted to nicotinic acid by the enzyme nicotinamidase, the genes of which are also known as pyrazinamidase/nicotinamidase (PncA) (Figure 1A). Mammalian genomes do not encode nicotinamidases but instead convert NAM directly into nicotinamide mononucleotide (NMN) using nicotinamide phosphoribosyltransferase (Nampt). NMN is then adenylated by nicotinomide mononucleotide adenyltransferase to form NAD+ (6, 7). Mammals also retain the capacity to utilize nicotinic acid to make NAD+, doing so using the Preiss Handler pathway. This pathway is biochemically similar to the way most organisms recycle nicotinic acid (Figure 1B) (7).
Figure 1.
Nicotinamidase chemistry and function. (A) PncA hydrolyzes nicotinamide to give nicotinic acid (top reaction) and is also able to catalyze the conversion of the drug pyrazinamide to pyrazinoic acid (bottom reaction). (B) Sirtuin chemistry. Sirtuins deacetylate an acetylated peptide in an NAD+-dependent manner, releasing nicotinamide in the process. Heightened levels of nicotinamide inhibit sirtuin chemistry. (C) and (D) NAD+ recycling. A schematic of the NAD+ recycling pathways in humans (C) and microbes (D).
The importance of the nicotinamidase activity to organisms that are pathogenic to humans, combined with its absence in human NAD+ biosynthetic pathways has increased interest in this enzyme as a possible drug target. Recent work on the spirochaete Borrelia burgdorferi indicates that host nicotinic acid levels are too low to support pathogen survival and that a plasmid-encoded nicotinamidase is essential for viability (8). Similarly, a nicotinamidase deficient mutant from Brucella abortus failed to replicate in cells but was rescued by the addition of nicotinic acid (9). In addition, an increased nicotinamidase activity has been observed in Plasmodium falciparum infected erythrocytes (10). A need for nicotinamidase activity for viability is consistent with the idea that some organisms lack a de novo NAD+ biosynthetic pathway and therefore rely upon recycling and salvage of host NAM for growth (1, 2, 11, 12).
Nicotinamidase activity was first reported by Williamson and Hughes in extracts from Lactobacillus arabinosus (13) and this activity was later observed in many other microorganisms (14-19). Nicotinamidases have also been confirmed in plants (20, 21), Caenorhabditis elegans (22), and Drosophila melanogaster (23). In addition to proposed roles in NAD+ homeostasis, nicotinamidases are reported to regulate the NAD+ dependent deacetylase activity of sirtuins (22, 24, 25). This is accomplished through modulation of in vivo concentrations of nicotinamide, a potent inhibitor of sirtuins (Figure 1C) (26-28). Increased nicotinamidase expression increases Sir2-dependent gene silencing (24, 29, 30) and increases replicative lifespan in yeast (25), D. melanogaster (23, 31) and C. elegans (22, 32).
In addition to its known biological functions, nicotinamidase activity is required for activation of the front-line tuberculosis prodrug, pyrazinamide. Nicotinamidase encoded by Mycobacterium tuberculosis hydrolyzes pyrazinamide to form the active form of the drug, pyrazinoic acid (Figure 1A). Despite the effectiveness of pyrazinamide, drug resistant strains of M. tuberculosis have emerged that are unable to metabolize this drug. Resistance arises from point mutations on the nicotinamidase enzyme (33) and these mutations have been mapped to catalytic residues as well as to residues predominantly structural in nature (34, 35).
With considerable interest in nicotinamidases for their biological effects, their potential as antibiotic targets, and a long history of investigation, it is somewhat surprising that the structural basis for the catalytic function of these enzymes has remained largely undetermined. Until recently, only unliganded crystal structures of nicotinamidases, from Saccharomyces cerevisiae (36) and Pyrococcus horikoshii (37), had been reported. While the active site within both structures was identified through similarity to known structures and by the presence of a coordinated metal, few conclusions could be made about ligand binding or the reaction mechanism. Recently a structure of the Acenitobacter baumanii nicotinamidase with nicotinic acid bound at the active site was reported in which the pyridine nitrogen of the ligand was determined to be coordinated to the central metal ion (38). The results suggest that nicotinamidases utilize metal coordination of the pyridine nitrogen to orient the substrate and possibly also to activate it for catalysis.
In this manuscript we report crystal structures, both unliganded and of several trapped intermediates, of the nicotinamidase from the human pathogen, Streptococcus pneumoniae (SpNic). In addition, we provide an analysis of the role and identity of the active site metal ion. These structures provide insight into key active site interactions and provide snapshots of several presumed reaction intermediates. The structure of the C136S mutant with nicotinamide bound provides details about substrate binding while the nicotinyl thioester observed in the structure of nicotinic acid treated crystals provides evidence for a thioester intermediate. The structures of the SpNic-aldehyde complexes reveal a covalent adduct with tetrahedral geometry, analogous to another putative reaction intermediate. These structures, along with the biochemical evidence presented, allow us to propose a catalytic mechanism for the nicotinamidase catalyzed reaction and provide a likely rationale for the inhibition by aldehyde analogues of nicotinamide.
MATERIALS AND METHODS
Cloning and Mutagenesis
The PncA gene for S. pneumoniae TIGR4 was amplified by PCR from genomic DNA (ATCC 6314D) and engineered to contain the NdeI and BamHI restriction endonuclease recognition sites. The PCR product was inserted into the pSTBlue vector (Novagen) using the AccepTor vector kit (Novagen). The gene was then digested with NdeI and BamHI restriction endonucleases and ligated into similarly digested pET28a; the presence of the gene was verified by sequencing. The C136S mutant plasmid was made at the Cornell Protein Characterization and Production Facility by using site-directed mutagenesis of the native gene. Briefly, site-directed mutagenesis was performed on SpNic by a standard PCR protocol using PfuTurbo DNA polymerase (Invitrogen) and DpnI (New England Biolabs) to digest the methylated parental DNA prior to transformation. The presence of the mutated residue was verified by sequencing.
Protein Expression and Purification
Rosetta 2(DE3) pLysS (Novagen) E. coli cells were transformed by the plasmid DNA. Cells were grown in LB medium at 37 °C with shaking (180 rpm) to an OD600 of 0.6. SpNic expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside and cells were incubated at 37 °C with shaking for an additional 10 h. Cells were harvested by centrifugation, resuspended in lysis buffer (300 mM NaCl, 50 mM Na2PO4 and 5 mM imidazole, pH 7.6) and lysed by sonication. The lysate was then centrifuged at 30 000 g for 60 min at 4 °C. The cleared lysate was loaded onto Ni-NTA resin (Qiagen) that had been pre-equilibrated with the lysis buffer. After loading, the lysate was washed with lysis buffer and then with wash buffer (300 mM NaCl, 50 mM Na2PO4, 10% glycerol and 25 mM imidazole, Ph 7.6). Finally, the protein was eluted with 300 mM NaCl, 50 mM Na2PO4 and 250 mM imidazole, pH 7.6 and then buffered exchanged using an Econo-Pac 10DG column (BioRad) into 30 mM NaCl and 10 mM Tris, pH 7.6. The protein was concentrated using a centrifugal filter with a 10 kDa molecular weight cutoff (Amicon) to a final concentration of 25 mg/mL as measured by the method of Bradford (39). Protein purity was verified to be greater than 95% by SDS-PAGE.
Synthesis of 5-Methoxynicotinaldehyde
5-Methoxynicotinaldehyde was synthesized in two steps from 3,5-dibromopyridine as previously reported (40).
Protein Crystallization, Data Collection and Structure Determination
Crystallization experiments were conducted using the hanging drop vapor diffusion method at 18 °C by combining equal volumes of protein and reservoir solution. Initial crystallization conditions were determined by sparse matrix screening (Hampton Research, Emerald Biostructures). After optimization, diffraction quality crystals were grown from 18 – 22 % polyethylene glycol 3350, 0.2 – 0.3 M NaCl and 0.2 M sodium malonate, pH 6.3 at 18 °C. To obtain the SpNic-small molecule complexes, the crystals were soaked for 12 hours in the crystallization solution containing 2 mM ZnCl2, and 2 mM of the ligand (nicotinaldehyde, 5-methoxynicotinaldehyde, NA or NAM). Prior to flash-freezing in liquid nitrogen, crystals were soaked in a solution containing the crystallization buffer, 2 mM ZnCl2, 2 mM ligand and 18% glycerol. Data sets were collected at the Advanced Photon Source (APS) beamline 24-ID-C using an ADSC Quantum 315 detector at a wavelength of 0.9795 Å. The data collection statistics are provided in Table 1.
Table 1.
Summary of Data Collection Statistics
| SpNic-Zn | SpNic-Zn-nicotin-aldehyde | SpNic-Zn-5-methoxy-nictotinaldehyde | SpNic-Zn-nicotinic acid | SpNic-Cys136Ser-Zn-NAM | |
|---|---|---|---|---|---|
| resolution (Å) | 1.95 | 1.63 | 1.9 | 1.84 | 1.60 |
| wavelength (Å) | 0.9795 | 0.9795 | 0.9795 | 0.9795 | 0.9795 |
| Space group | P21 | P21 | P212121 | P21 | P21 |
| a (Å) | 60.8 | 60.8 | 61.2 | 61.2 | 61.2 |
| b (Å) | 120.9 | 120.0 | 114.5 | 121.2 | 120.8 |
| c (Å) | 63.2 | 62.8 | 120.9 | 63.2 | 62.8 |
| β (°) | 114.3 | 114.9 | 90 | 94.3 | 114.7 |
| unique reflections | 54280 | 96559 | 67241 | 71957 | 106320 |
| average I/σ(I) | 11.3 (2.2) | 14.7 (1.9) | 14.3 (4.7) | 16.5 (4.5) | 18.9 (3.8) |
| redundancy | 3.4 (2.1) | 4.7 (2.5) | 5.0 (4.9) | 5.4 (4.9) | 4.0 (3.4) |
| completeness(%) | 88.2 (73.7) | 95.1 (79.4) | 99.0 (99.9) | 99.5 (98.8) | 97.7 (86.2) |
| Rsyma (%) | 13.2 (29.2) | 9.3 (17.7) | 9.8 (35.3) | 9.9 (28.9) | 5.1 (18.9) |
Numbers in parentheses correspond to the highest resolution shell
Rsym = ΣΣi|Ii − ⟨I⟩|/ Σ⟨I⟩, where ⟨I⟩ is the mean intensity of the N reflections with intensities Ii and common indices h, k, l
The structure of SpNic was solved by molecular replacement using the program MolRep (41) with an all-alanine monomer of P. horikoshii pyrazinamidase (PDB code 1IM5) as the search model. The models were refined by iterative cycles of restrained refinement using Refmac5 (42) and CNS (43) and by manual model building using the interactive graphics program Coot (44). Water molecules were added using Coot only after the refinement of the protein structure had converged. Ligands were directly built into the corresponding difference electron density and the model was then subject to an additional round of refinement. Refinement statistics for the model are listed in Table 2.
Table 2.
Summary of Data Refinement Statistics
| SpNic-Zn | SpNic-Zn-nicotin-aldehyde | SpNic-Zn-5-methoxy-nicotinaldehyde | SpNic-Zn-NA | SpNic-Cys136Ser-Zn-NAM | |
|---|---|---|---|---|---|
| resolution (Å) | 1.95 | 1.63 | 1.9 | 1.84 | 1.60 |
| total no. of atoms | 5851 | 6043 | 5980 | 5996 | 6028 |
| no. of ligand atoms | 4 | 60 | 68 | 72 | 40 |
| no. of water atoms | 534 | 510 | 548 | 567 | 840 |
| reflections in working set | 51163 | 77657 | 62379 | 66829 | 95621 |
| reflections in test set | 2719 | 4097 | 3328 | 3542 | 5312 |
| R factora | 21.8 | 20.4 | 20.8 | 20.9 | 16.4 |
| Rfree b | 26.0 | 23.3 | 23.8 | 23.6 | 20.0 |
| rmsd bonds (Å) | 0.006 | 0.006 | 0.007 | 0.006 | 0.006 |
| rmsd angles (°) | 0.92 | 0.90 | 0.93 | 0.95 | 0.92 |
| mean B factor (Å2) | 25.0 | 29.7 | 26.2 | 19.8 | 15.8 |
| Ramachandran | |||||
| most favored (%) | 92.3 | 92.4 | 92.7 | 91.3 | 91.9 |
| additionally allowed (%) | 7.7 | 7.6 | 7.3 | 8.7 | 8.1 |
| generously allowed (%) | 0 | 0 | 0 | 0 | 0 |
| disallowed (%) | 0 | 0 | 0 | 0 | 0 |
R factor = Σhkl∥Fobs| − k|Fcalc∥/Σhkl|Fobs|, where Fobs and Fcalc are observed and calculated structure factors respectively.
For Rfree, the sum is extended over a subset of reflections excluded from all stages of refinement
Estimation of Coordinate Error
While the variances and covariances of parameters used in structural refinement can be obtained through the inversion of the full least-squares matrix, this calculation is computationally very challenging and difficult to realize for protein structures, given the number of parameters needed for refinement. Alternatively, approximations can be used to compute an estimate of coordinate error in a model. To determine the approximate coordinate error for the Zn2+ and water atoms bound in the SpNic active site, the method of Cruickshank (45), called the diffraction-component precision index (DPI), was used.
Total Elemental Analysis of Protein Samples
Elemental analysis of protein samples was performed by the Cornell Nutrient Analysis Laboratory (46) following EPA protocol 3051-6010. This method involves microwave assisted nitric acid digestion of samples followed by analysis by inductively coupled plasma – atomic emission spectrometry. Prior to treatment of samples, the 6-His tag was cleaved from the protein using TEV protease. An aliquot was set aside to use as an untreated sample and the remaining was incubated with 2 mM EDTA and 2 mM 1,10-phenanthroline for 24 hours at 4 °C. Samples were buffer exchanged on a Centricon 10 kDa cutoff centrifugal device (Millipore) into 30 mM Tris, 10 mM NaCl. Three subsequent centrifugation runs were carried out using a 20:1 ratio of new buffer to concentrated sample on each run. Metal salts were added to a final concentration of 2 mM to 5 mg/mL solutions of protein and incubated for 16 h. These samples were buffer exchanged to remove unbound metal ions as described above and sent for analysis at a concentration of 2 mg/mL.
Metal Dependence Measurements
SpNic protein used for metal dependent kinetics analysis had the 6-His tag removed, treated with chelators and buffer exchanged as detailed above. After addition of Zn(OCH3)2, Fe(II)SO4, MnCl2, CoCl2, NiSO4, or Fe(III)Cl3 (in concentrations ranging from 0 to 2 mM for each metal salt), the solutions were incubated for 6 h and buffer exchanged using spin columns to remove excess metals. This step is necessary as the metal ions can interfere with the coupling enzyme. The rate of reaction was monitored on a 96-well plate using a glutamate dehydrogenase coupled assay (40). Control reactions using ammonium sulfate with each enzyme solution without added NAM were run to ensure that the coupling enzyme was not limiting the reaction.
RESULTS
Structure of Unliganded SpNic
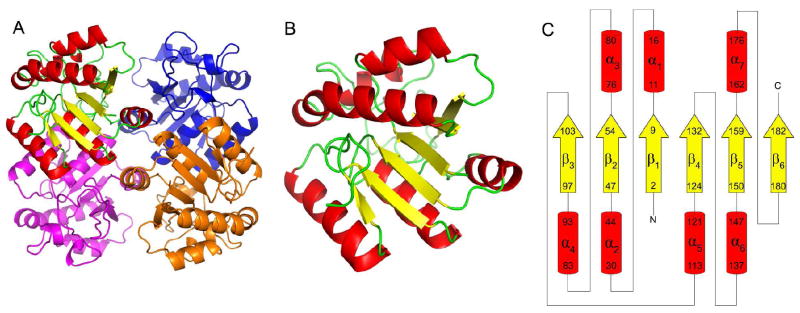
S. pneumoniae nicotinamidase is a 191 amino acid protein with a molecular weight of 21.4 KDa. The structure of SpNic is a homotetramer (Figure 2A), consistent with molecular weight estimates determined by size exclusion chromatography (data not shown). One complete tetramer was observed in the asymmetric unit of the crystal. SpNic is an α/β protein with a six-stranded β-sheet flanked by three helices on one side and four helices on the opposite side (Figures 2B and 2C). The interface between protomers is dominated by polar contacts of α5 on one side and by π-stacking and polar interactions of α6 and α7. The four active sites of the SpNic tetramer are located in solvent-accessible pockets in each protomer that are formed primarily by three loop regions containing residues 55 through 75 (between β1 and α3), residues 104 through 112 (between β3 and α5) and residues 133 through 136 (between β4 and α6). While one of the open sides of the active site is bordered by two residues (Thr176 and His172) of α7 from a neighboring protomer, all interactions necessary for substrate binding and catalysis, including those that coordinate the metal ion, are provided by a single SpNic protomer.
Figure 2.
Structure of SpNic. (A) SpNic tetramer. (B) SpNic monomer and topology diagram of SpNic. The yellow arrows represent β-strands and red cylinders represent α-helices.
Organization of Metal Binding Site
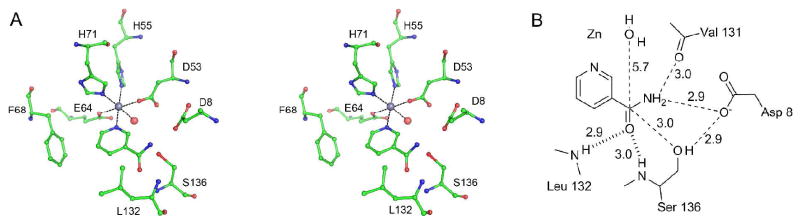
The metal center is coordinated by two histidines (His55 and His71), an aspartate (Asp53) and a glutamate (Glu64). Two additional coordination interactions are provided by a single water molecule and by the pyridyl nitrogen of the ligand (Figure 3), or by two water molecules in the absence of a ligand, in each case furnishing full octahedral coordination to the metal ion. The magnitude of the electron density peak at the metal ion binding site was low when crystals were grown without exogenous metal added. Crystallization solution supplemented with Zn2+ ions, however, led to a clear, strong peak. Supplementation of the well solution with other ions, including Mn2+, Fe2+, Fe3+, Co2+, Ni2+ or Ca2+ did not lead to improvement over crystals grown in the absence of exogenous metals.
Figure 3.
SpNic active site. A) Stereoview of the active site of SpNic shown with NAM bound as observed in the Cys136Ser–NAM structure. The grey and red spheres represent zinc and water respectively. Phe-14 (not shown) is situated in front of NAM in the plane of the paper and makes an additional π-stacking interaction with the NAM ring. B) Schematic of the active site of the Cys136Ser SpNic mutant in complex with NAM. The heavy dashed lines represent interactions provided by the residues that make up the oxyanion hole. The distances shown are heavy atom to heavy atom distances in angstroms.
Metal Ion Dependence of SpNic
To determine the metal dependence of SpNic, the enzyme was first treated with EDTA and phenanthroline, buffer exchanged to remove chelators and then treated with various metal ion solutions. These solutions were further buffer exchanged to remove unbound metals and then tested for activity. Addition of Fe2+, Mn2+, Co2+, and Zn2+ restored various levels of enzyme activity, with Zn2+ showing the highest level (Table 4). Note that the treatment of the enzyme to remove and reintroduce metals may have resulted in some small degree of protein loss, accounting for the only 90% recovery of activity of the zinc treated enzyme. Addition of Fe3+ or Ni2+ resulted in no detectable activity. Moreover, analysis of metal content of untreated native enzyme showed the presence of Zn2+ ions only (Table 3). Similarly, soaks of SpNic by metals after treatment with chelators showed that, while both Fe2+ and Mn2+ can bind, Zn2+ is observed at the highest concentration under equivalent conditions. Note that for Fe2+ and Zn2+ the observed concentration of metal was higher than that of enzyme. This indicates that, in these cases, some nonspecific binding of the metal to the enzyme is occurring.
Table 4.
Recovery of SpNic activity by addition of metal ions
| Sample | Relative Rate1 |
|---|---|
| Untreated | 1.02 |
| Metals Removed | 0.0 ± 0.01 |
| Zn2+ | 0.90 ± 0.04 |
| Mn2+ | 0.27 ± 0.02 |
| Fe2+ | 0.13 ± 0.03 |
| Co2+ | 0.13 ± 0.02 |
| Fe3+ or Ni2+ | 0.0 ± 0.02 |
The data are represented as mean of triplicate test ± standard deviation
This value is for the activity of the enzyme ‘as purified’ and may not necessarily reflect the maximal rate of turnover for this enzyme.
Table 3.
Analysis of Metal Content
| Sample | S1 (μM) | [SpNic]1 (μM) | Mn (μM) | Fe (μM) | Zn (μM) | Co, Cu, Ca, Mg, K (μM) |
|---|---|---|---|---|---|---|
| Untreated | 203 | 34 | < det | < det | 5 | < det |
| No metal added | 119 | 20 | < det | < det | < det | < det |
| Zn2+ | 144 | 24 | < det | < det | 142 | < det |
| Mn2+ | 144 | 24 | 18 | < det | 17 | < det |
| Fe2+ | 122 | 20 | < det | 51 | < det | < det |
< det = below detection limit
S refers to the sulfur content in the enzyme sample. The [SpNic] was calculated using the measured sulfur content and the known ratio of the total mass of sulfur to the total mass of the enzyme as determined by analysis of the sequence.
Active Site Water Molecule and Metal-Water Relationship
In addition to four bonds to the enzyme side chains and an interaction with the pyridyl nitrogen of the ligand, a water molecule is coordinated to the metal in the SpNic active site (Figure 3). This water is located about 5 - 6 Å from the carbonyl carbon of the ligand in the SpNic structures. Water molecules coordinated to metals can be activated for catalysis and the degree of activation of the water has implications for the mechanism of the enzymatic reaction. The length of the metal-water bond can indicate the type of species present in the structure. The distances between the metal ion and the water in the SpNic structures vary from 2.2 Å to 2.4 Å. The method of Cruickshank (45) was used to estimate the coordinate error present in the model and from this information and the individual B-factors, approximate coordinate errors for the water and metal ion were calculated (see Materials and Methods). The estimated coordinate error for both the metal ion and the water varied between 0.12 Å and 0.19 Å.
Structure of C136S SpNic with Bound Nicotinamide
To assess the roles of active site residues, the structures of several SpNic complexes were determined. The structure of an inactive form of SpNic, the C136S mutant, reveals several important features of the active site (Figures 3 and 4A). The orientation of the ligand in the active site is facilitated by several π-electron interactions of the pyridyl ring with Phe14, Phe68 and Tyr106 and through coordination of the pyridyl nitrogen to the observed metal ion. Both Phe68 and Tyr106 (not shown in Figure 3A, but is positioned behind nicotinamide in this view) make herringbone interactions, while the aromatic ring of Phe14 (not shown in Figure 3A, but is positioned in front of nicotinamide in this view) stack with the pyridine ring of nicotinamide. Additional contacts are made to the amide nitrogen of nicotinamide by the sidechains of Asp8 and Ser136 as well as by the backbone carbonyl of Val131. A putative oxyanion hole, which forms H-bonds with the backbone amines of Leu132 and Ser136 in this structure, is also observed (Figure 3A). In addition, the carbonyl carbon of nicotinamide is situated less than 3 Å from the suspected catalytic residue (136) while an aspartate residue (Asp8) is approximately equidistant from the amide nitrogen and the oxygen of Ser136 (Figure 3B).
Figure 4.
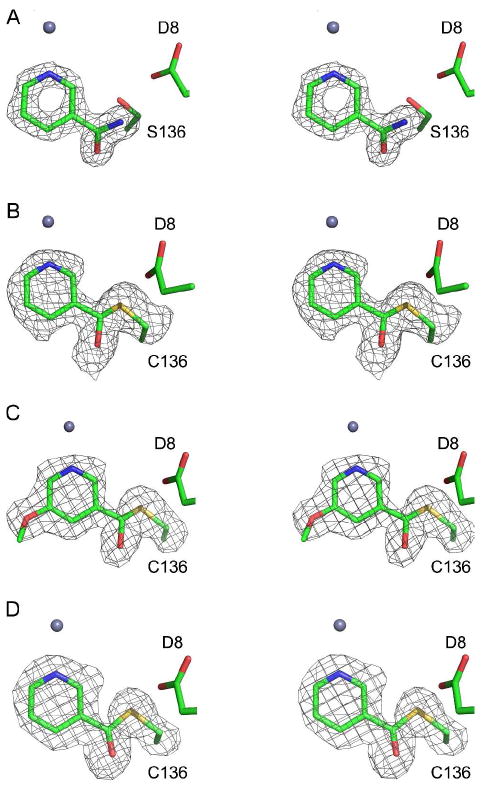
Electron densities of bound ligands to SpNic. Each section shows a stereo diagram of the various ligand-bound complexes; the zinc ion (grey sphere) and Asp8 are shown for reference. All electron density shown is from 1FO – FC maps and density shown, contoured at 2.5σ, was calculated prior to adding the ligand to the model. For the covalent complex maps, the Cys136 side chain was also omitted from the model in order to illustrate the linkage (A) Cys136Ser SpNic mutant with NAM bound. (B) SpNic complex with nicotinaldehyde. (C) SpNic complex with 5-methoxynicotinaldehyde. (D) SpNic thioester complex formed in the presence of nicotinic acid.
Structures of SpNic-Aldehyde Complexes
To further probe the active site of SpNic, we solved the crystal structures of SpNic in complex with the aldehyde inhibitors nicotinaldehyde and 5-methoxynicotinaldehyde. While the organization of the active site remains essentially unchanged, the observed geometry about the carbonyl carbon varies in the differently liganded structures of SpNic. In both of the nicotinaldehyde complexes, tetrahedral geometry is observed at what was the carbonyl carbon center (Figures 4B and C and Figure 5). The difference density observed in these structures allows for the unambiguous placement of the proposed thiohemiacetal in the active site (Figures 4B and 4C).
Figure 5.
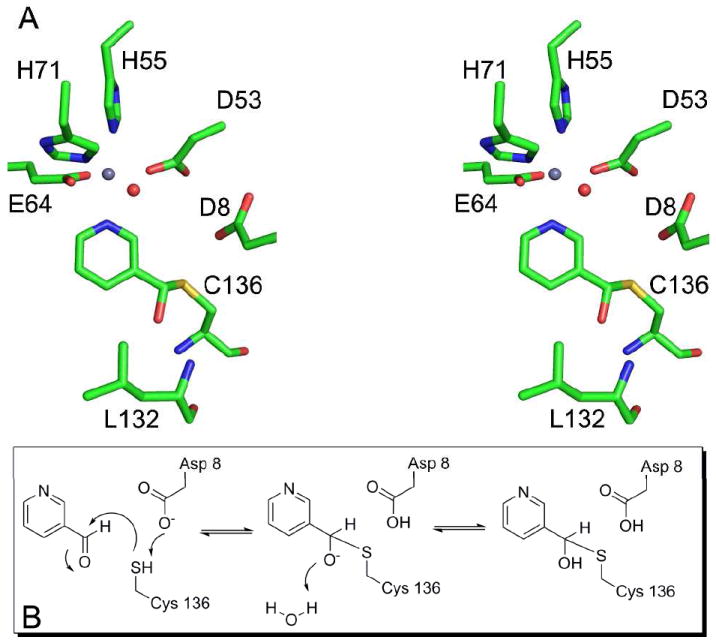
Mechanism of SpNic inhibition by nicotinaldehyde. A) Stereo diagram of the structure of the SpNic-nicotinaldehyde complex showing the trapped tetrahedral adduct. In the structure carbon atoms are colored green, oxygen atoms red, nitrogen atoms blue, sulfur is yellow and the zinc atom is grey. The red sphere is the water molecule that is coordinated to the zinc ion. B) The mechanism of inhibition whereby a tetrahedral adduct is formed by reaction with Cys136 to the carbonyl of the inhibitor resulting in a stalled complex due to the inability of the aldehyde hydrogen to act as a leaving group.
Structure of SpNic Treated with Nicotinic Acid
As was observed for the aldehyde soaks of SpNic, treatment of this enzyme with nicotinic acid had little effect on the overall organization of the active site. The observed ligand density, however, was distinct from the other liganded structures. The electron density observed from data collected on soaks of SpNic treated with nicotinic acid corresponded to a covalent adduct with sp2 hybridization at the carbonyl carbon. A covalent nicotinyl thioester linkage between the ligand and Cys 136 of SpNic was a good fit to the observed density (Figure 4D).
DISCUSSION
Structural Similarity to Other Proteins
The crystal structure of SpNic was compared to other proteins contained within the Protein Data Bank using the DALI server (47). The overall fold of SpNic is shared by several classes of enzymes, most of which belong to the cysteine hydrolase family. These enzymes predominantly catalyze the hydrolysis of amides, esters or ethers and share a conserved catalytic cysteine residue. Structurally the most closely related enzymes are the N-carbamoylsarcosine amidases (PDB codes 3EEF and 1NBA), and the isochorismatases (1NF9, 2FQ1 and 2A67). N-carbamoylsarcosine amidohydrolases (CSHases) hydrolyze N-carbomoylsarcosine to sarcosine, carbon dioxide and ammonia. While the fold of this enzyme is quite similar to that of SpNic, the shape and nature of substrates accommodated are different. In addition, CSHase catalyzes the removal of an amide group and does not employ a metal ion in catalysis or substrate binding (48). The isochorismatases catalyze the conversion of isochorismate to 2,3-dihydroxybenzoate and pyruvate via the hydrolysis of a vinyl ether bond. Despite the structural similarities between these enzymes and PncA, isochorismatases lack the conserved cysteine residue and catalyze unrelated chemistry (49).
Key Active Site Residues
There are several features of the SpNic active site that are significant for substrate binding and catalysis. Structrurally, the pyridine ring of the ligand is oriented in the active site through coordination to the metal ion and through π-interactions made with the aromatic side chains Phe14, Phe68 and Tyr106; additional polar contacts are made with the amide moiety through both side chain and backbone atoms. The SpNic-aldehyde complex structures indicate that Cys136 is the catalytic residue responsible for nucleophilic attack of the carbonyl carbon. Asp8, nearly equidistant from Cys136 and the −NH2 of the amide, is poised to both deprotonate the catalytic cysteine and to transfer a proton to the amine leaving group (Figure 3). Studies of PncA mutants, predominantly carried out in Mycobacterium tuberculosis, have shown that a complete loss of activity occurs upon mutation of either the active site cysteine or aspartate (34, 35, 50). One interesting case, the Borrelia bergdorferri PncA, is annotated as having an N-terminally truncated PncA, which lacks the aspartate residue equivalent to Asp8. Studies on the activity of this enzyme showed that an additional 48 base pairs 5′ of the designated start codon (16 amino acid residues) were needed to reconstitute PncA activity (8). When aligned with SpNic, this leading sequence contained an aspartate residue equivalent to Asp8 in SpNic.
Another active site mutation known to eliminate PncA activity is Lys96 in M. tuberculosis (equivalent to Lys103 in SpNic) (34, 35). It has been speculated that, along with the cysteine and aspartate residues, the lysine completes a catalytic triad in the PncA active site (51). There has been no evidence put forth, however, to indicate a direct role for the lysine residue in catalysis. Our structures indicate that the role of Lys103 is likely ancillary. This residue makes salt bridges with Asp8 and Asp53 (one of the residues that coordinates the metal ion) as well as interacting with Cys136 in the SpNic active site. Its likely role, therefore, is to orient these residues before and during catalysis and to help to offset the negative charges that develop along the reaction pathway. The proximity of Lys103 to Asp8 is also likely to influence the pKa of the carboxylate group, facilitating the proton transfer step.
A further notable feature of the SpNic active site is the presence of a putative oxyanion hole. In this pocket, the backbone amides of Leu132 and Cys136 (Ser136 in the C136S mutant) make favorable contacts to the carbonyl oxygen of the ligand (Figure 3). This site is structurally configured by a cis-peptide bond between Val131 and Leu132 that is conserved amongst nicotinamidases as well as in two other proteins with a similar fold, N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. (52) and YcaC from Escherichia coli (53). This oxyanion hole presumably acts by both contributing to the proper orientation of the ligand in the active site, donating hydrogen bonds to the carbonyl oxygen, and by stabilizing negative charge that builds up along the reaction coordinate.
Identity and Role of the Metal Ion
Previous structural and biochemical studies on the PncA from M. tuberculosis (35) and P. horikoshii (51) have shown that this enzyme binds a metal ion. For the former, either Fe2+ or Mn2+ was sufficient to reconstitute activity and a 1:1 stoichiometry of Fe2+:Mn2+ was found bound in the native enzyme (35). Kinetic analysis of the P. horikoshii PncA, however, showed a 19-fold increase in enzyme activity in the presence of zinc (51). In the case of SpNic, elemental analysis indicates that both Zn2+ and Fe2+ can bind to the enzyme, although zinc was the only metal isolated from the native protein. Clearly, Zn2+ can occupy the active site of SpNic as both the ligand and metal electron densities in our structures were very clear when crystals were grown in the presence of ZnCl2. The absence of electron density for the metal in the structure solved from crystals grown without added zinc also indicates that metal may freely associate/dissociate from the active site under different conditions. In addition, our analysis of the metal dependence of SpNic kinetics shows that, while several divalent metals can participate in catalysis, the most catalytically active is Zn2+ (Table 4) These data suggest that this enzyme is most suited to utilize zinc in the metal site, but can also accommodate other divalent ions.
The number and nature of the amino acid residues coordinated to the metal differ between the P. horikoshii PncA, S. cerevisiae PncA and SpNic. In both P. horikoshii and S. cerevisiae, the metal ion is coordinated by two histidine residues and an aspartate (51, 54). An additional glutamate residue is present in the SpNic enzyme (Figure 3). Mutational studies of the M. tuberculosis PncA suggest that this enzyme also coordinates the metal through four sidechain interactions, however, these residues are putatively identified as three histidines and one aspartate (35). These findings indicate that the identity of the bound metal may depend upon the organism and is dictated by the nature of the coordinating residues. One possible explanation for this phenomenon is a divergent evolved specificity for particular metal(s) in distinct organisms due to differing environmental conditions. In this case, the ability for SpNic to utilize different metals for catalysis could have provided a competitive advantage and thus provided evolutionary pressure towards promiscuity.
Catalytic Water Molecule
In addition to four bonds to the enzyme and an interaction with the pyridyl nitrogen of the ligand, a water molecule is coordinated to the metal in the SpNic active site (Figure 3). In order to complete the hydrolysis of NAM, an −OH donor is needed, which presumably originates from a water molecule. Since the metal bound water molecule is the only water less than 10 Å from the catalytic cysteine, it is reasonable to assume that this molecule may act as the −OH donor. While the distance from this water to the carbonyl carbon of the ligand is approximately 5.5 Å, all of our SpNic-ligand complexes show the carbonyl of the ligand oriented in such a way that this water molecule has a clear line of attack to the carbonyl carbon (Figure 4).
Water molecules coordinated to metals can be activated for catalysis. The degree of activation of the water has implications for the mechanism of the enzymatic reaction. In some cases the water is deprotonated upon coordination to the metal thus being activated as a hydroxide ion. The nature and number of coordinated ligands to a metal affects the overall ligand-metal bond strengths, with higher coordination leading to weaker bonds (55). In the case of water coordinated to a zinc ion, bond lengths of less than 2 Å generally indicate a metal-hydroxide moiety, while bonds greater than 2 Å in length indicate zinc-bound water (55). The metal-water bond length in our structures ranges from 2.3 to 2.5 Å with an estimated coordinate error at each atom of 0.1 to 0.2 Å. In addition, it has been observed that hydrogen bonds to the metal-bound water/hydroxide moiety orient it for attack (55). In the SpNic structure both Glu64 and Asp53 are ideally positioned to accept hydrogen bonds from the water molecule coordinated to zinc (Figure 3A). Considering the octahedral coordination, the metal-water bond length and the presence of two hydrogen bond acceptors, it is likely that, while the water may be activated for nucleophilic attack, it is coordinated as a water molecule and not as a hydroxide ion.
Upon completion of the hydrolysis of nicotinamide and release of the product, the active site must be repopulated with a water molecule to complete the catalytic cycle. This water molecule may reenter the active site from two different directions. Firstly, despite the relatively hydrophobic nature of the entrance to the active site, water molecules can gain access in the same manner as the substrate. This is evident in the unliganded structure of SpNic, wherein several water molecules are seen occupying the site where the ligand is known to bind. The second alternative is a pathway through the protein at a loop region that ‘caps’ the metal coordination site. This loop, which is solvent exposed on one face, contains the glutamate residue (E64) that is coordinated to the metal ion and the active site water. Two other water molecules, both with access to the bulk solvent, are also within hydrogen-bond distance of the carboxylate group of this residue. A slight movement of Glu64, therefore, may allow for the replacement of the catalytic water molecule to complete the catalytic cycle.
SpNic-Inhibitor Complexes and Mechanism of Inhibition
Nicotinaldehydes are known competitive inhibitors of nicotinamidase enzymes (19, 40). To examine the mechanism of inhibition, we solved the structure of SpNic in complex with both nicotinaldehyde and 5-methoxynicotinaldehyde (Figures 4B and 4C). These structures reveal a covalent adduct, formed between the carbonyl carbon and Cys136, that has tetrahedral geometry at the (formerly) carbonyl carbon. This observation is consistent with the hypothesis that Cys136 acts as the nucleophile during catalysis. With the exception of the amide nitrogen, the inhibitors are structurally identical to the substrate and therefore share the same enzyme-substrate interactions in the active site. The methoxy group at the 5- position of 5-methoxynicotinaldehyde extends into a relatively open region of the active site, causing a slight shift in Phe68, but otherwise appears to have little effect on the orientation of the molecule or the active site residues.
The observation of a covalently linked tetrahedral structure suggests that the enzyme reacts with the inhibitor but is unable to proceed past a relatively stable intermediate. In the absence of a good leaving group, the intermediate formed by reaction of the inhibitor with SpNic persists in the active site for a long enough time period to form the product observed in the structures. The presence of the oxyanion hole, which could stabilize the negative charge of a tetrahedral intermediate, suggests that the observed SpNic-thiohemiacetal adducts are formed as outlined in Figure 5B. This mechanism involves deprotonation of Cys136 by the nearby Asp8 followed by attack at the carbonyl carbon of the inhibitor. The charge buildup is stabilized by the oxyanion hole. In the absence of a leaving group, the intermediate either proceeds in the reverse direction to reform the inhibitor or gains a proton from the solvent, leading to the structure observed.
Structure of Trapped Thioester
The nicotinamidase reaction has been proposed to proceed through a thioester intermediate formed after attack of the substrate by the catalytic cysteine and concomitant liberation of ammonia (38, 51). While biochemical and structural evidence supports this hypothesis, there has been no direct evidence reported for this type of intermediate. The structure of SpNic from crystals soaked with nicotinic acid, however, suggests that such a reaction intermediate is likely formed along the nicotinamidase reaction pathway. This structure shows a covalently linked intermediate with planar geometry at the carbonyl carbon center. The difference density for the ligand allows for unambiguous placement of a nicotinyl thioester linked to Cys136 of SpNic (Figure 4D). This structure not only provides evidence for a thioester intermediate, but also indicates that the final steps in the reaction are fully reversible. This observation is supported by 18O exchange experiments wherein facile exchange of 18O was observed in the presence of both unlabelled and 18O-nicotinic acid (40). These data, together with the C136S SpNic-NAM and SpNic-inhibitor complexes, suggest a reaction mechanism whereby the putative thioester intermediate is accessible in both the forward and reverse directions.
Implications for Catalysis
The structures presented herein elucidate several important features of substrate binding and catalysis, and provide evidence for two putative intermediates in the nicotinamidase reaction pathway. Our analysis suggests that SpNic catalyzes a ping-pong reaction that proceeds through two half reactions separated by the reactive thioester intermediate. The proposed mechanism for the SpNic catalyzed reaction is given in Figure 6C. Nicotinamide is first oriented in the SpNic active site through π-stacking with several aromatic side chains and by coordination to the active site metal ion, which may also serve to activate the ligand for nucleophilic attack (Figures 6A and 3A). Additional interactions occur between the carbonyl oxygen of the substrate and the backbone amine groups that constitute the oxyanion hole. Catalysis is initiated by deprotonation of the catalytic cysteine, Cys136, by the nearby Asp8, followed by attack of the carbonyl carbon by the nucleophilic thiol of Cys136. This leads to the formation of the putative tetrahedral intermediate. The negative charge that builds up on the carbonyl oxygen of this species is stabilized by the oxyanion hole. This proposed intermediate is analogous in structure to that observed between SpNic and nicotinaldehyde (Figures 4B, 4C and 5A). The tetrahedral intermediate then collapses to form a thioester, as the amino group of the ligand is protonated by Asp8 and leaves as ammonia. This aspartate residue is observed in the C136S SpNic-nicotinamide structure to be nearly equidistant from the nucleophile and the leaving group, aligned ideally for proton transfer between these two groups.
Figure 6.
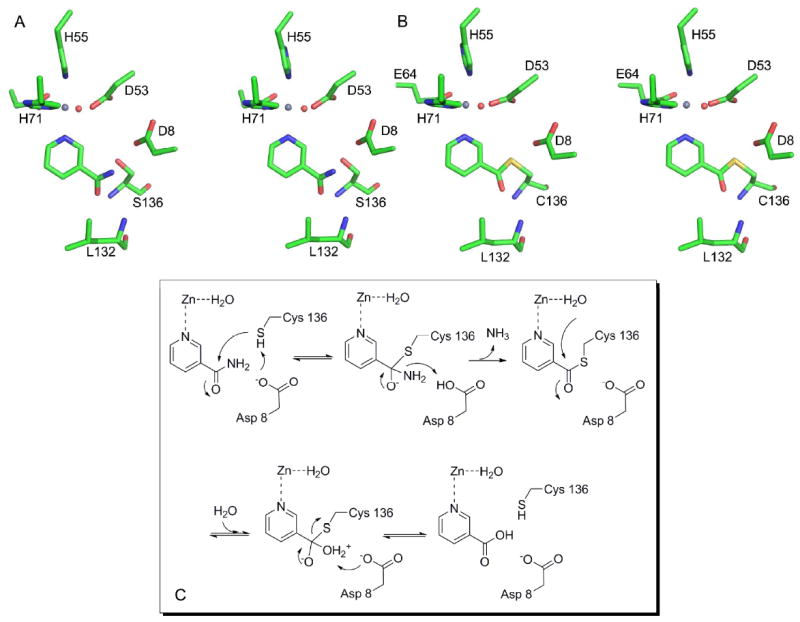
Mechanism of SpNic Catalyzed Hydrolysis of Nicotinamide. A) Stereo diagram showing the Cys136Ser mutant in complex with NAM. B) Stereo diagram showing the trapped thioester intermediate formed by soaking native SpNic with NA. For both A) and B) carbon atoms are colored green, oxygen atoms red, nitrogen atoms blue, sulfur is yellow and the zinc atom is grey. The red sphere is the water molecule that is coordinated to the zinc ion. C) The proposed mechanism of the SpNic catalyzed reaction. See text for details of the steps.
The thioester intermediate, presumably equivalent to what is observed for the nicotinic acid treated SpNic, can then undergo nucleophilic attack by a water molecule to lead to a second tetrahedral intermediate. While our structures cannot unequivocally determine the source of the water that ultimately hydrolyzes the nicotinamide, the most likely source of this nucleophile is the water coordinated to the zinc ion. This water is ideally positioned for attack at the carbonyl carbon and is properly oriented by Asp53 and Glu64 to carry out this chemistry. The orientation of the carbonyl group of the intermediate makes it unlikely that a water molecule from the bulk solvent could affect this hydrolysis, as has been proposed in an alternative mechanism (38). In addition, coordination of a water molecule to the metal ion activates it for attack. Despite the structural evidence of a fully protonated water molecule coordinated to the zinc ion, it cannot be ruled out, however, that deprotonation of the water molecule occurs prior to attack, yielding a hydroxide moiety as the nucleophile.
The final stages of the reaction, leading from the second tetrahedral intermediate to product, are analogous to the first steps but occur in the reverse order. Deprotonation of the bound water by Asp8 prevents reversal of the reaction as collapse of the intermediate expels the thiol group of Cys136. The product, nicotinic acid, dissociates, likely driven by unfavorable interactions between Cys136 and Asp8 of the enzyme and the acid group of the product. The catalytic cycle is completed by the return of the proton from Asp8 to Cys136.
In summary, we have presented several crystal structures of the nicotinamidase enzyme from the human pathogen, S. pneumoniae, and provided a biochemical analysis of the role and identity of the active site metal. The structures allow us to define many of the key active site interactions and provide details about the mechanism by which aldehyde analogs of nicotinamide inhibit this enzyme. These structures also provide a glimpse of the putative reaction intermediates and provide much needed insights into the catalytic mechanism of this important enzyme.
Acknowledgments
We would like to thank the NE-CAT staff at beamlines 24-ID-C and 24-ID-E of the Advanced Photon Source for assistance with data collection. JBF acknowledges the Tri-Institutional Training Program in Chemical Biology for financial support. Anthony Sauve and Steven Ealick would like to acknowledge support from the Milstein Program in Chemical Biology for a pilot grant to support this project.
ABBREVIATIONS
- NAD
nicotinamide adenine dinucleotide
- NAM
nicotinamide
- NA
nicotinic acid
- NMN
nicotinamide mononucleotide
- PncA
nicotinamidase
- SpNic
Streptococcus pneumoniae nicotinamidase
- GDH
glutamate dehydrogenase
Footnotes
This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The coordinates of the SpNic structures have been deposited in the Protein Data Bank under accession numbers 3O90, 3O91, 3O92, 3O93, and 3O94 for unliganded, nicotinaldehyde complex, 5-methoxynicotinaldehyde complex, nicotinoyl-thioester complex, and C136S-nicotinamide complex, respectively.
References
- 1.Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp The r. 2008;324:883–893. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- 2.Gerdes SY, Scholle MD, D’Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM, Campbell JW, Anantha S, Shatalin KY, Chowdhury SA, Fonstein MY, Osterman AL. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velu SE, Mou L, Luan CH, Yang ZW, DeLucas LJ, Brouillette CG, Brouillette WJ. Antibacterial nicotinamide adenine dinucleotide synthetase inhibitors: amide- and ether-linked tethered dimers with alpha-amino acid end groups. J Med Chem. 2007;50:2612–2621. doi: 10.1021/jm061349l. [DOI] [PubMed] [Google Scholar]
- 4.Oppenheimer NJ. Nad Hydrolysis - Chemical and Enzymatic Mechanisms. Molecular and Cellular Biochemistry. 1994;138:245–251. doi: 10.1007/BF00928468. [DOI] [PubMed] [Google Scholar]
- 5.Handlon AL, Xu C, Mullersteffner HM, Schuber F, Oppenheimer NJ. 2’-Ribose Substituent Effects on the Chemical and Enzymatic-Hydrolysis of Nad(+) J Am Chem Soc. 1994;116:12087–12088. [Google Scholar]
- 6.Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andirs F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Biochem. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, Kurokawa D, Watanabe K, Makino S, Shirahata T, Watarai M. Brucella abortus nicotinamidase (PncA) contributes to its intracellular replication and infectivity in mice. FEMS Microbiol Lett. 2004;234:289–295. doi: 10.1016/j.femsle.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 10.Zerez CR, Roth EF, Schulman S, Tanaka KR. Increased nicotinamide adenine dinucleotide content and synthesis in Plasmodium faciparum-infected human erythrocytes. Blood. 1990;75:1705–1710. [PubMed] [Google Scholar]
- 11.Li Y, Bao W. Why do some yeast species require niacin for growth? Different modes of NAD synthesis. FEMS Yeast Res. 2007;7:657–664. doi: 10.1111/j.1567-1364.2007.00231.x. [DOI] [PubMed] [Google Scholar]
- 12.Kurnasov OV, Polanuyer BM, Anata S, Sloutsky R, Tam A, Gerdes SY, Osterman AL. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J Bacteriol. 2003;184:6906–6917. doi: 10.1128/JB.184.24.6906-6917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes DE, Williamson DH. The synthesis of cozymase from nicotinic acid and its derivatives by Lactobacillus arabinosus. Biochem J. 1952:330–338. doi: 10.1042/bj0510330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson WJ, Gadd REA. Inhibition of nicotinamide deamidase from Micrococcus lysodeikticus by analogues of nicotinamide. Int J Biochem. 1974;5:633–641. [Google Scholar]
- 15.Foster JE, Kinney DM, Moat AG. Pyridine nucleotide cycle of Salmonella typhimurium: isolation and characterization of pncA, pncB and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J Bacteriol. 1979;137:1165–1175. doi: 10.1128/jb.137.3.1165-1175.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi JG, Handler P. J Biol Chem. 1961;237:929–935. [PubMed] [Google Scholar]
- 17.Pardee AB, Benz EJ, Peter DAS, Krieger JN, Meuth M, Trieshmann HW. Hyperproduction and purification of nicotinamide deamidase, a microconstituitive enzyme of Escherichia coli. J Biol Chem. 1971;246:6792–6796. [PubMed] [Google Scholar]
- 18.Tanigawa Y, Shimoyama M, Dohi K, Ueda I. Purification and properties of nicotinamide deamidase from Flavobacterium peregrinum. J Biol Chem. 1972;247:8036–8042. [PubMed] [Google Scholar]
- 19.Yan C, Sloan DL. Purification and characterization of nicotinamide deamidase from yeast. J Biol Chem. 1987;262:9082–9087. [PubMed] [Google Scholar]
- 20.Joshi JG, Handler P. Biosynthesis of trigonelline. J Biol Chem. 1960;235:2981–2986. [PubMed] [Google Scholar]
- 21.Wang G, Pichersky E. Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 2007;49:1020–1029. doi: 10.1111/j.1365-313X.2006.03013.x. [DOI] [PubMed] [Google Scholar]
- 22.van der Horst A, Schavemaker JM, Pellis-van Berkel W, Burgering BM. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128:346–349. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Balan V, Miller GS, Kaplun L, Balan K, Chong ZZ, Li F, Kaplun A, VanBerkum MF, Arking R, Freeman DC, Maiese K, Tzivion G. Life span extension and neuronal cell protection by Drosophila nicotinamidase. J Biol Chem. 2008;283:27810–27819. doi: 10.1074/jbc.M804681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallo CM, Smith DL, Smith JS. Nicotinamide clearence by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 27.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 28.Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- 29.Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 30.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The Biochemistry of Sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 31.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 33.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;6:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 34.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA Mutations in Pyrazinamide-Resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Deng JY, Bi LJ, Zhou YF, Zhang ZP, Zhang CG, Zhang Y, Zhang XE. Characterization of Mycobacterium tuberculosis nicotinamidase/pyrazinamidase. FEBS J. 2008;275:753–762. doi: 10.1111/j.1742-4658.2007.06241.x. [DOI] [PubMed] [Google Scholar]
- 36.Hu G, Taylor AB, McAlister-Henn L, Hart PJ. Crystal structure of the yeast nicotinamidase Pnc1p. Arch Biochem Biophys. 2007;461:66–75. doi: 10.1016/j.abb.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du X, Wang W, Kim R, Yakota H, Nguyen H, Kim SH. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry. 2001;40:14166–14172. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 38.Fyfe PK, Rao VA, Zemla A, Cameron S, Hunter WN. Specificity and mechanism of Acinetobacter baumanii nicotinamidase: implications for activation of the front-line tuberculosis drug pyrazinamide. Angewandte Chemie (International ed) 2009;48:9176–9179. doi: 10.1002/anie.200903407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradford M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 40.French JB, Cen Y, Vrablik TL, Hanna-Rose W, Sauve AA. Characterization of nicotinamidases: Class-wide inhibition by nicotinaldehydes and mechanism of inhibition and catalysis. submitted. 2010 doi: 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. 2000;D 56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project-Number 4. The CCP-4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D 50 [Google Scholar]
- 43.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D 54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Cruickshank DWJ. Remarks about protein structure precision. Acta Cryst. 1999;D55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 46.Cornell Nutrient Analysis Laboratory. Ithaca: 2008. http://cnal.cals.cornell.edu. [Google Scholar]
- 47.Holm L, Sander C. Protein structure comparison by alignment of distance matrixes. J Mol Biol. 1993;233:123–128. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 48.Zajc A, Romao MJ, Turk B, Huber R. Crystallographic and fluorescence studies of ligand binding to N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. J Mol Biol. 1996;263:269–283. doi: 10.1006/jmbi.1996.0574. [DOI] [PubMed] [Google Scholar]
- 49.Parsons JF, Calabrese K, Eisenstein E, Ladner JE. Structure and Mechanism of Pseudomonas aeruginosa PhzD, an Isochorismatase from the Phenazine Biosynthetic Pathway. Biochemistry. 2003;42:5684–5693. doi: 10.1021/bi027385d. [DOI] [PubMed] [Google Scholar]
- 50.Lemaitre N, Callebautt I, Frenois F, Jarlier V, Sougakoff W. Study of the structure-activity relationships for the pyrazinamidase (PncA) from Mycobacterium tuberculosis. Biochem J. 2001;353:453–458. doi: 10.1042/0264-6021:3530453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du X, Wang W, Kim R, Yakota H, Nguyen H, Kim S-H. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochem. 2001;40:14166–14172. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 52.Zajc A, Romao MJ, Turk B, Huber R. Crystallographic and fluorescence studies of ligand binding to N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. J Mol Biol. 1996;263:269–283. doi: 10.1006/jmbi.1996.0574. [DOI] [PubMed] [Google Scholar]
- 53.Colovos C, Cascio D, Yeates TO. The 1.8 Å crystal structure of the ycaC gene product from Escherichia coli reveals an octameric hydrolase of unknown specificity. Structure. 1998;6:1329–1337. doi: 10.1016/s0969-2126(98)00132-4. [DOI] [PubMed] [Google Scholar]
- 54.Hu G, Taylor AB, Mcalister-Henn L, Hart PJ. Crystal structure of the yeast nicotinamidase Pnc1p. Arch Biochem Biophys. 2007;461:66–75. doi: 10.1016/j.abb.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christianson DW, Cox JD. Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu Rev Biochem. 1999;68:33–57. doi: 10.1146/annurev.biochem.68.1.33. [DOI] [PubMed] [Google Scholar]