Abstract
Cell-penetrating peptides (CPPs) are short amino acid sequences that promote their own translocation across cell plasma membrane. When linked with cargo such as polypeptides, nucleic acid, or liposomes, CPPs can facilitate the transport of these entities across the cell membrane. Therefore, CPPs are receiving increased interest in drug delivery and gene therapy. The majority of CPPs identified so far are polycationic peptides which interact with heparin sulfate chains of plasma membrane for internalization. Here, we report the identification and characterization of a conformationally constrained 13 amino acid peptide (CVQWSLLRGYQPC, designated as S41) which is clearly distinct from classical polycationic peptides. Immunofluorescence assay was employed to test the cellular uptake of S41 in mouse neuroblastoma cell line Neuro2A (N2A) and rat cerebellar granule neurons (CGNs). Internalization of S41 was further examined in N2A cells by means of mutational analysis, flow cytometry and confocal microscopy. Our results demonstrate that S41 can enter cells through lipid rafts dependent endocytosis.
Keywords: CPPs, S41, Phage-display, Lipid-rafts, Endocytosis
Cell-penetrating peptides (CPPs) are short peptide sequences with the ability to translocate across cell plasma membrane [1]. One of the first described CPPs was found within the human immunodeficiency virus (HIV-1) transactivator of transcription (TAT). TAT possesses the property of penetrating into cultured mammalian cells [2,3]. The minimum region of TAT required for its internalization was determined to be a positively charged fragment between amino acids 47–57 [4]. Subsequently, a variety of polycationic CPPs have been identified, such as AntP (the third helix of the homeotic protein of Drosophila melanogaster Antennapedia) [5], vp22 (derived from the herpes virus structural protein VP22) [6], and nano-arginine (R9) [7,8]. CPPs have the ability to deliver macromolecular cargos, e.g., polypeptides [9,10], oligonucleotides [11], nanoparticles [12], and even liposomes [13] into cells. Importantly, the delivered cargos have produced biological activities in vivo [11,14,15].
Despite the promise of polycationic CPP as carriers, the mechanisms for their internalization are mostly unknown. Several endocytic pathways including clathrin-dependent endocytosis, caveolae/lipid raft-mediated endocytosis and macropinocytosis, have been reported to be involved in the uptake of CPPs [16–18]. The interaction between the polycationic CPPs and the negatively charged heparin sulfate proteoglycans (HSPGs) of plasma membrane has been proposed to be a shared internalization mechanism [19,20].
We are seeking to develop systems to deliver therapeutic proteins to motor neurons. To identify neuronal CPPs, peptides were displayed on phage and assessed for internalization by cultured neuronal cells. Here, we report the identification and characterization of a conformationally constrained 13mer peptide that is internalized by cells through lipid raft-dependent endocytosis.
Materials and methods
Reagents
Heparinase III, methyl-β-cyclodextrin (MβCD) and propidium iodide (PI) were obtained from Sigma–Aldrich (St. Louis, MO). Peptides labeled with FITC (fluorescein isothiocyanate) at the N-terminus were synthesized as C-terminal peptide amides by the Tufts University Core Facility (Boston, MA). Peptides were stored frozen at −80 °C until further use. The peptide solutions were freshly prepared by dissolving peptides in distilled water (pH 8.0, adjusted with analytical pure NH3·H2O). The cyclic 13mer phage library displayed different peptides having the structure of CX11C (C = cysteine, X = any amino acid residue).
Cell culture
N2A cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air and cultured in Eagle’s Minimum Essential Medium (MEM) supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. For fluorescence assays, cells were grown in 35 mm tissue culture dishes or 6-well plates (with or without coverslips) at a cell density of 4 × 105 cells/5 ml medium/dish or 2 × 105 cells/3 ml medium/well overnight. CGNs were isolated and cultured from 8 day old Wistar rats as described previously [21]. Culture medium was renewed 0.5 h prior to experiments.
Peptide phage library amplification and selection
Phage work was performed as previously described [22]. The procedure for selection of cell-internalized phage was adapted from Gao et al [23]. Briefly, 4 × 1010 cfu of peptide phage library was added into 4 × 106 N2A cells in 100 mm tissue culture dishes and incubated at 37 °C for 3 h. Cell screening of phage was stopped by immersion of dishes in ice for 10 min. The cells were stringently washed with Dulbecco’s phosphate buffered saline (DPBS), glycine buffer (50 mM glycine, 0.5 M NaCl, pH 2.5) and PBS (pH 7.2), sequentially. Cells were lysed with 1 ml of 0.1 M triethylamine (TEA) and neutralized with 0.5 ml of 1 M Tris–HCl (pH 7.4). One milliliter of the cell lysate was used to amplify internalized peptide-phage as described above and used in the next round of selection. DNAs of 10 phage colonies from the fourth round of selection were prepared using the Wizard Plus SV Miniprep DNA Purification System (Promega) for sequence analysis.
Site-directed mutagenesis
S41 phage mutants were generated using the QuikChange® XL Site-Directed Mutagenesis Kit (Strata-gene) according to the manufacturer’s instruction. The primer pairs for each mutant are as follows. S41 mutant1 (M1): Pm1 5′-ccg gccatggccgcaatggtgcagtggagcttg-3′, Pm1a 5′-caagctccactgcaccattgc ggccatggccgg-3′; S41 mutant2 (M2): Pm2 5′-gccgcatgcgtgcaggctag cttgctccggggg-3′, Pm2a 5′-cccccggagcaagctagcctgcacgca tgcggc-3′; S41 mutant3 (M3): Pm3 5′-cgtgcagtggagattgctccggggg-3′, Pm3a 5′-cccccggagcaatctccactgcacg-3′. Successful mutagenesis was confirmed in each case by DNA sequencing. The mutated DNAs were transformed into E coli ER2738 and phage displaying the modified S41 peptides was amplified as described above.
Immunofluorescence assay
N2A cells were incubated with 2 × 1010 cfu phage at 37 °C for 2 h. Following washes with DPBS, cells were treated with trypsin–EDTA at 37 °C for 15 min, washed, replated into 35 mm dishes (with coverslip) and grown overnight. After washing, cells were fixed and permeabilized with 100% methanol for 10 min, washed once with phosphate buffered saline (PBS) and blocked with 10% normal goat serum (Rockland Immunochemicals) at room temperature (RT) for 2 h. Cells were then incubated at 4 °C overnight with mouse M13 mAb (1:100 dilution, Amersham) and stained with FITC-conjugated anti-mouse IgG (1:150 dilution, ZYMED) at RT for 1h. 1μg/ml propidium iodide (PI) was added for nucleus staining. After washing with PBS and distilled water, cells were mounted with Vectashield mounting medium (Vector Laboratories) and observed under a fluorescence microscope. CGNs were treated as described above except that the trypsinization step was omitted. Images were taken by a digital camera connected to the microscope.
FACS analysis on cellular uptake of S41
N2A cells were preincubated (in 35 mm dishes) at 4 °C or pretreated (in 6-well plate) at 37 °C for 0.5 h in the presence of 8 mM MβCD or 20 mU/ml heparinase III. Cells were then incubated with 7.5 μM FITC-labeled S41 peptide at 4 °C or at 37 °C for 1 h in the absence or presence of above chemicals. Cells were treated with trypsin–EDTA at 37 °C for 15 min, collected by centrifugation (1000g for 3 min), resuspended in 800 μl of DPBS and immediately analyzed (10,000 events/sample) by fluorescence activated cell sorter (FACS) (FACScalibur, Becton–Dickinson). For data analysis, WINMDI software (Joe Trotter, The Scripps Institute) was used. The statistical significance was evaluated by t-test at the 95% confidence level.
Confocal laser scanning microscope analysis on cellular uptake of S41
FITC-labeled S41 (7.5 μM) was added into N2A cells and incubated at 37 °C for 1 h. For lipid rafts labeling, the cells were washed six times with DPBS, 2 ml of chilled growth medium and 2 μL of 1 mg/ml Alexa Fluor 555-labeled cholera toxin B subunit (CT-B) conjugate stock solution (Molecular Probes) were added and the cells incubated at 4 °C for 15 min. After washing three times with DPBS, 2 ml of growth medium containing 10 μL of the anti-CT-B rabbit serum (Molecular Probes) were added and incubated with the cells at RT for 1 h. Following washes with DPBS, 2 ml of medium was added into cells and observed by confocal laser scanning microscope.
Results
Selection of S41 as a cell-penetrating peptide
A conformationally constrained peptide display library of phage-displaying random 11mer peptides flanked by cysteines (CX11C) was used to select cell-penetrating peptides (CPPs) in N2A cells. After four rounds of selection for cell internalization, the titer ratio of phage in the recovered cell lysate vs. input phage increased 50-fold, suggesting enrichment of phage that become internalized. Sequencing of 10 random clones following the fourth round of selection revealed that all phage displayed the peptide CVQWSLLRGYQPC, designated S41. Database search in the NCBI GenBank showed that S41 has homology to a motif in the N-termini of MHC class I molecules (mouse, rat, and human) (Table. 1A). To validate that phage displaying S41 were selectively internalized into cells, these phage were mixed with a 1000-fold excess of phage displaying random CX11C peptides and subjected to two rounds of internalization selection in N2A cells. Twelve random phagemid clones were picked from the cell lysate and DNA sequencing revealed all to encode the S41 peptide, demonstrating that S41 phage are selectively internalized in these cells.
Table 1.
Amino acid sequences of the novel cell-penetrating peptide S41, homologues and mutations. One-letter amino acid codes are used. (A) Sequence of S41 and alignment with a region of MHCI from human (aa 128–140), rat (aa 105–117), and mouse (aa 125–137). (B) Amino acid sequences of S41 mutants created for this study.
| A | |||||||||||||
| S41 | C | V | Q | W | S | L | L | R | G | Y | Q | P | C |
| MHC Glass I Human | G | P | D | G | R | L | L | R | G | Y | N | Q | F |
| MHC class I Rat | G | T | D | W | S | L | L | R | G | Y | R | Q | D |
| MHC class I Mouse | G | S | D | W | R | L | L | R | G | Y | Q | Q | F |
| B | |||||||||||||
| S41 (wild-type) | C | V | Q | W | S | L | L | R | G | Y | Q | P | C |
| Mutant l (Ml) (C1M) | M | V | Q | W | S | L | L | R | G | Y | Q | p | C |
| Mutant 2 (M2) (W4A) | C | V | Q | A | S | L | L | R | G | Y | Q | P | C |
| Mutant 3 (M3) (S5R) | C | V | Q | W | R | L | L | R | G | Y | Q | p | C |
Immunofluorescence assay on the cellular uptake of S41 phage
Uptake and intracellular localization of phage displaying S41 peptide was next examined by immunofluorescence assay. Following incubation of N2A cells with S41 phage or control phage, phage was detected with a mouse anti-M13 mAb and/or a FITC-labeled anti-mouse IgG secondary antibody. The N2A cells incubated with S41 phage in the presence of the M13 primary antibody showed strong intracellular staining localizing to the perinuclear region (Fig. 1A). No staining was observed in the absence of the M13 primary antibody (Fig. 1B), while cells incubated with control phage showed weak non-specific staining (Fig. 1C). A similar result was observed in CGNs immunostaining (Fig. 1E and F) showing that S41 phage is actively endocytosed in primary neurons in addition to neuroblastoma cells.
Fig. 1.
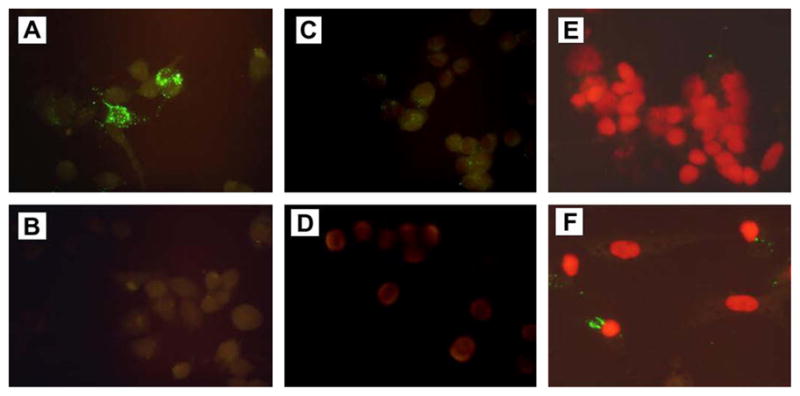
Uptake of S41 phage into mouse neuroblastoma N2A cells and rat cerebellar granule neurons (CGNs) detected by immunofluorescence. Panels show immunofluorescence (IFA) detection of internalized phage in N2A cells (A–D) and CGNs (E,F). Cells were incubated with S41 phage (A, B and F), control phage (C) or remained untreated (D,E). IFA was performed as described in “Materials and methods”. Cells were stained with FITC-labeled secondary antibody in the presence (A, C, D, E, and F) or absence (B) of mouse anti-M13 mAb. All cells were stained with FITC-labeled anti-mouse IgG and counterstained with propidium iodide (PI) to show the nuclei prior to microscopic examination.
S41 Mutations impair the cellular uptake of S41 phage
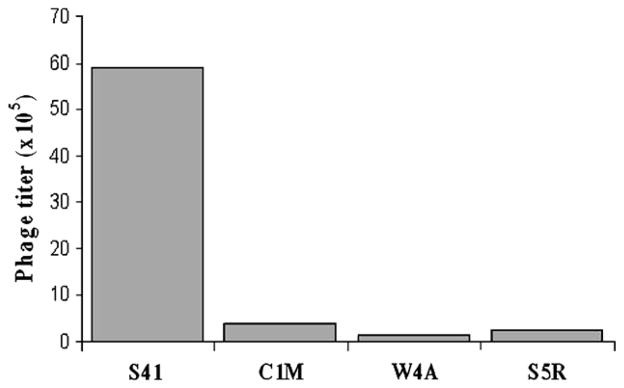
Three single-amino acid mutations of the S41 peptide sequence were created (Table 1B) to test the sensitivity of S41 internalization to specific changes in amino acid sequence. Cyclic peptides often have increased biological activity over their linear analogues, presumably because of their conformationally constrained structure [24,25]. To test if the cyclic structure is important in S41 function, we created a cysteine to methionine mutation (C1M) that eliminates one of the flanking cysteines that permits intrapeptide linkage and cyclization. The significance of the homology of S41 to MHC I (Table 1A) was tested by creating two mutations. The W4A mutation reduces homology to MHC I, while the S5R mutation increases homology to the MHC I sequence WRLLRGYQ. Phage displaying the S41 mutant peptides were amplified and compared to phage displaying wild type S41 to test the ability of the various displayed peptides to promote phage internalization into N2A cells. Input phage numbers were constant and the amount of phage recovered from cell lysates provided a measure of the efficiency of phage internalization (Fig. 2). Uptake of phage displaying the mutant S41 peptides was also assessed by IFA in N2A cells and corroborate the phage titer results showing that the phage are not significantly internalized (data not shown). These experiments indicate that the cyclic loop in S41 is necessary to its internalization and that the cell internalization property of the S41 peptide is vulnerable to at least some single amino acid changes. The fact that increasing the MHC I homology of S41 did not improve uptake does not support the hypothesis that this homology is related to the internalization function.
Fig. 2.
Single amino acid changes to S41 have a major effect on its ability to promote phage internalization by N2A cells. 2 × 1010 cfu of phage displaying the S41 peptide, or the mutant versions of S41 shown in Table 1B, were incubated with N2A cells. The cells were then stringently washed to strip bound phage as described in “Materials and methods” and the titer of phage remaining in cell pellet extract was determined. Internalized phage titers are shown for S41 and mutants C1M, W4A, and S5R. Shown is a representative of six different experiments.
FACS analysis on cellular uptake of synthetic S41 peptide
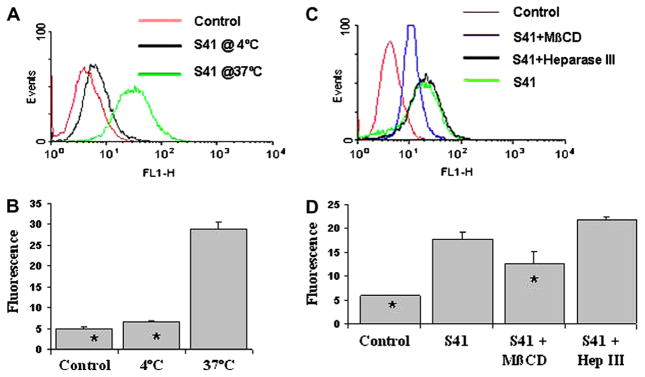
Recently, it has been postulated that apparent CPP internalization can be caused by fixation artifacts [26]. To exclude this for S41, we obtained a FITC-labeled synthetic S41 peptide and examined its internalization ability in living N2A cells by flow cytometry. Low temperature causes the lipid membrane to become more rigid and minimizes active processes of cellular transport and should inhibit uptake of true CPPs. As shown on Fig. 3A and B, S41 uptake into cells was greatly inhibited when performed at 4 °C indicating that uptake of S41 is temperature-dependent and that internalization involves active processes such as endocytosis.
Fig. 3.
The influence of temperature, MβCD and heparinase III (Hep III) on the internalization of FITC labeled S41 into N2A cells. FITC-labeled S41 were incubated with N2A cells at different temperatures or with MβCD and Hep III, and then subjected to flow cytometry. A and C show fluorescence scans while B and D show the average fluorescence. Means ± SD are indicated. Each sample was analyzed in duplicate or triplicate. Values significantly (p < 0.05) different from S41 treated cells at 37 °C were marked with asterisk.
To further explore the mechanisms involved in S41 internalization, synthetic S41 peptide uptake was measured by fluorometry in the presence of caveolin/lipid rafts-mediated endocytosis inhibitor MβCD. This experiment showed that the presence of 8mM MβCD led to a 30% inhibition of S41 uptake (Fig. 3C and D), consistent with the results of phage titer comparison experiments (data not shown). This observation suggests the involvement of lipid rafts in the cellular uptake of S41.
The heparin sulfate (HS) chains of cell membrane heparin sulfate proteoglycans (HSPGs) play an important role in the cellular uptake of polycationic CPPs. To evaluate whether heparin sulfates affect the uptake of S41, N2A cells were treated with 20 mU/ml of heparinase III (an enzyme that degrades the HS chains [27]) for 1 h. As shown in Fig. 3C and D, the uptake of S41 in heparinase III treated cells was not significantly affected, compared with the uptake of S41 in untreated cells.
Co-localization of S41 peptide with lipid rafts
Substantial evidence has been reported that lipid rafts play a key role in a variety of cellular processes, for example, signal transduction, receptor-mediated internalization, as well as in the infectious cycles of several viruses and bacterial toxins, such as cholera toxin (reviewed by [28,29]). Cholera toxin subunit B (CT-B) binds to the ganglioside GM1, which selectively partitions into lipid rafts [30]. To seek further evidence that S41 enters cells through lipid rafts-mediated endocytosis, we examined whether S41 co-localizes with fluorescein-labeled CT-B. As observed under confocal microscope, S41 co-localizes, at least in part, with Alexa555-labeled CT-B in living N2A cells (Fig. 4).
Fig. 4.
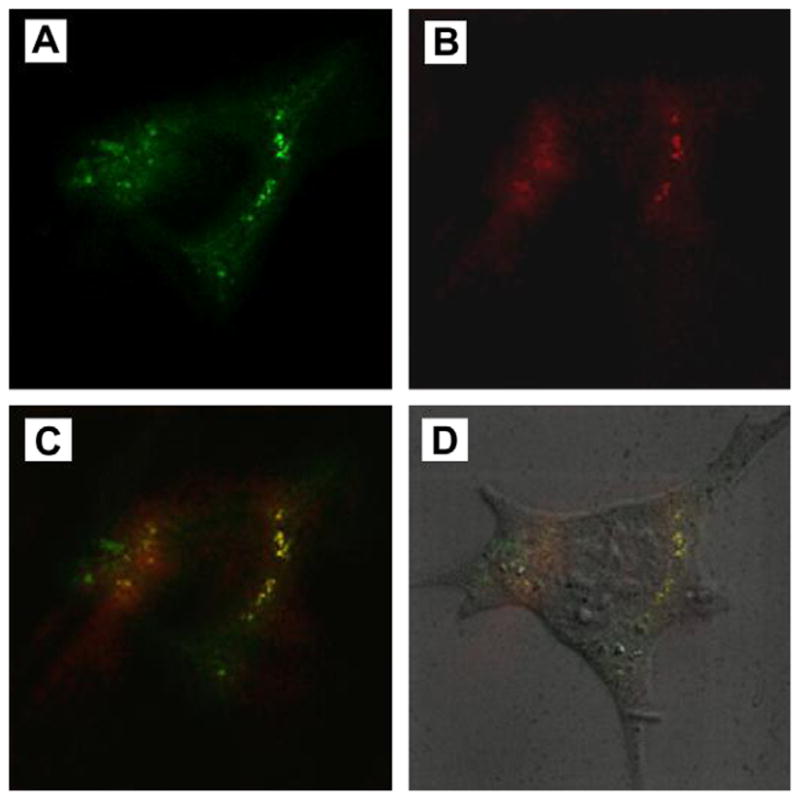
S41 peptide co-localizes with lipid rafts in living N2A cells. N2A cells were incubated with FITC-labeled S41 peptide, Alexa555-labeled CT-B, and anti-CT-B antibody, then subjected to confocal laser scanning microscopy. Images were obtained of a stained single cell filtering for FITC (A) or Alexa555 (B), merged (C) or an overlay of panel C with a DIC (differential interference contrast) microscopy image (D).
Discussion
In this study, we report the identification and characterization of a novel cell-penetrating peptide-S41. The data presented here indicate that S41 is actively taken by cells through a lipid rafts-dependent endocytic pathway. This conclusion is based on (i) clear intracellular staining of S41 phage following incubation with N2A cells and primary rat cerebellar granule neurons, (ii) sequence- and temperature-dependent uptake of the S41 peptide, (iii) inhibition of S41 internalization by MβCD, and (iv) S41 peptide co-localization with lipid rafts in living cells.
The low permeability of cell plasma membrane to polar molecules is a major obstacle to the growing needs for protein drug delivery requiring the translocation into cells. CPPs have attracted increasing interest for their small size and, more importantly, for their capability to efficiently deliver membrane-impermeable molecules into the cells in vivo and in whole organisms [11,31,32]. The well-studied CPPs, such as AntP, VP22, nano-arginine peptide(R9) and TAT, are enriched in basic amino acid residues (Arg and Lys) resulting in their interaction with the ubiquitously occurring HSPGs in cell plasma membrane and non-selective internalization property. S41 is distinct from the classical polycationic CPPs in that this 13mer peptide has only one basic amino acid (Arg). As shown in our FACS analysis results, heparinase III pretreatment did not affect FITC-labeled S41 uptake. In another assay, the cellular uptake of S41 phage was not inhibited by the addition of exogenous heparin, too (data not shown), indicating that HSPGs are not required for S41 internalization.
Our data showed that the cellular uptake of S41 is temperature-dependent and inhibited by MβCD. Furthermore, it co-localizes, at least in part, with lipid rafts in living cells. These results demonstrated that S41 enters cells through lipid rafts-dependent endocytosis. Further testing of S41 cell specificity and cargo carrying capacity will be important to evaluating the application of S41 as a novel cell-penetrating peptide.
Acknowledgments
We thank Ms. Jacque Tremblay for excellent technical assistance and Drs. Hanping Feng and Jong-Beak Park for helpful discussions. We also thank Dr. Saul Tzipori, PI of the Tufts Microbiology and Botulism Research Unit of the Food and Waterborne Diseases Integrated Research Network, for his support of this project. This work was supported by contract NO1-AI30050 from the National Institutes of Health (NIH) and the National Institute of Allergy and Infectious Diseases (NIAID).
References
- 1.Lindgren M, Hällbrink M, Prochiantz A, Langel U. Cell-penetrating peptides. Trends Pharmacol Sci. 2003;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 2.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 3.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat transactivator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 4.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 5.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 6.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 7.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 8.Kown MH, Yamaguchi A, Jahncke CL, Miniati D, Murata S, Grunenfelder J, Koransky ML, Rothbard JB, Robbins RC. L-arginine polymers inhibit the development of vein graft neointimal hyperplasia. J Thorac Cardiovasc Surg. 2001;121:971–980. doi: 10.1067/mtc.2001.112532. [DOI] [PubMed] [Google Scholar]
- 9.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 11.Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 12.Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT, Weissleder R. Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- 13.Torchilin VP, Rammohan R, Weissig V, Levchenko TS. TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors. Proc Natl Acad Sci USA. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi SK, Hashimoto K, Koni PA. Induced DNA recombination by Cre recombinase protein transduction. Genesis. 2002;33:48–54. doi: 10.1002/gene.10089. [DOI] [PubMed] [Google Scholar]
- 15.Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc Natl Acad Sci USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Caveolae-mediated internalization of extracellular HIV-1 tat fusion proteins visualized in real time. Mol Ther. 2003;8:284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 17.Fittipaldi, Ferrari A, Zoppe M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J Biol Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 18.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 19.Silhol M, Tyagi M, Giacca M, Lebleu B, Vives E. Different mechanisms for cellular internalization of the HIV-1 Tat derived cell penetrating peptide and recombinant proteins fused to Tat. Eur J Biochem. 2002;269:494–501. doi: 10.1046/j.0014-2956.2001.02671.x. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. Possible existence of common internalization mechanisms among arginine-rich peptides. J Biol Chem. 2002;277:2437–2443. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 21.Schousboe A, Meier E, Drejer J, Hertz L. Preparation of primary cultures of mouse (rat) cerebellar granule cells. In: Shahar A, de Vellis J, Vernadakis A, Haber S, editors. A Dissection and Tissue Culture Manual for The Nervous System. Alan R Liss; New York: 1989. pp. 203–206. [Google Scholar]
- 22.Sepulveda J, Shoemaker CB. Design and testing of PCR primers for the construction of scFv libraries representing the immunoglobulin repertoire of rats. J Immunol Methods. 2008;332:92–102. doi: 10.1016/j.jim.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao C, Mao S, Ditzel HJ, Farnaes L, Wirsching P, Lerner RA, Janda KD. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. Bioorg Med Chem. 2002;10:4057–4065. doi: 10.1016/s0968-0896(02)00340-1. [DOI] [PubMed] [Google Scholar]
- 24.Aumailley M, Gurrath M, Müller G, Calvete J, Timpl R, Kessler H. Arg-Gly-Asp constrained within cyclic pentapeptides Strong and selective inhibitors of cell adhesion to vitronectin and laminin fragment P1. FEBS Lett. 1991;291:50–54. doi: 10.1016/0014-5793(91)81101-d. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki N, Nakatsuka H, Mochizuki M, Nishi N, Kadoya Y, Utani A, Oishi S, Fujii N, Kleinman HK, Nomizu M. Biological activities of homologous loop regions in the laminin alpha chain G domains. J Biol Chem. 2003;278:45697–45705. doi: 10.1074/jbc.M304667200. [DOI] [PubMed] [Google Scholar]
- 26.Lundberg M, Wikström S, Johansson M. Cell surface adherence and endocytosis of protein transduction domains. Mol Ther. 2003;8:143–150. doi: 10.1016/s1525-0016(03)00135-7. [DOI] [PubMed] [Google Scholar]
- 27.Linhardt RJ, Turnbull JE, Wang HM, Loganathan D, Gallagher JT. Examination of the substrate specificity of heparin and heparan sulfate lyases. Biochemistry. 1990;29:2611–2617. doi: 10.1021/bi00462a026. [DOI] [PubMed] [Google Scholar]
- 28.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 29.Nicholsa BJ, Kenworthya AK, Polishchuka RS, Lodgea R, Robertsa TH, Hirschberga K, Phairb RD, Lippincott-Schwartza J. Rapid cycling of lipid raft markers between the cell surface and golgi complex. J Cell Biol. 2001;153:529–542. doi: 10.1083/jcb.153.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merritt EA, Sixma TK, Kalk KH, van Zanten BA, Hol WGJ. Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT) Mol Microbiol. 1994;13:745–753. doi: 10.1111/j.1365-2958.1994.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 31.Foerg, Merkle HP. On the biomedical promise of cell penetrating peptides: limits versus prospects. J Pharm Sci. 2008;97:144–162. doi: 10.1002/jps.21117. [DOI] [PubMed] [Google Scholar]
- 32.Morris MC, Deshayes S, Heitz F, Divita G. Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]