Abstract
Transient Receptor Potential (TRP) proteins are cation channels composed of a transmembrane domain flanked by large N- and C-terminal cytoplasmic domains. All members of the vanilloid family of TRP channels (TRPV) possess an N-terminal ankyrin repeat domain (ARD). The ARD of mammalian TRPV6, an important regulator of calcium uptake and homeostasis, is essential for channel assembly and regulation. The 1.7 Å crystal structure of the TRPV6-ARD reveals conserved structural elements unique to the ARDs of TRPV proteins. First, a large twist between the fourth and fifth repeats is induced by residues conserved in all TRPV ARDs. Second, the third finger loop is the most variable region in sequence, length and conformation. In TRPV6, a number of putative regulatory phosphorylation sites map to the base of this third finger. Size exclusion chromatography and crystal packing indicate that the TRPV6-ARD does not assemble as a tetramer and is monomeric in solution. Adenosine triphosphate-agarose and calmodulin-agarose pull-down assays show that the TRPV6-ARD does not interact with either ligand, indicating a different functional role for the TRPV6-ARD than in the paralogous thermosensitive TRPV1 channel. Similar biochemical findings are also presented for the highly homologous mammalian TRPV5-ARD. The implications of the structural and biochemical data on the role of the ankyrin repeats in different TRPV channels are discussed.
The Transient Receptor Potential (TRP) proteins are a superfamily of cation channels with diverse functions. All TRP channels share a similar six-transmembrane-segment ion-transport domain flanked by intracellular N- and C-terminal domains, and are expected to function as tetramers. TRP channels are divided into seven subfamilies based on sequence similarity in their cytosolic domains, (TRPA – ANKTM1, TRPC – canonical, TRPM – melastatin, TRPN – NOMP-C, TRPV – vanilloid receptor, and the more distantly related TRPML – mucolipin and TRPP – polycystin) (1). The N-termini of the TRPA, TRPC, TRPN and TRPV channels contain between 3 and 31 ankyrin repeats (2), which are 33-amino acid residue motifs often involved in protein-protein interactions (3).
TRPV proteins have been identified in eukaryotes ranging from invertebrates to man and many are believed to play a role in sensing the environment (4–6). The mammalian TRPV subfamily is composed of six members that contain a six-repeat ankyrin repeat domain (ARD) in their N-terminal cytosolic domain (Figure 1A)(7–9). The TRPV channels can be further broken down into two subfamilies: TRPV1-4, all of which are thermosensitive, non-selective cation channels expressed primarily in sensory neurons and keratinocytes, and TRPV5 and TRPV6, both of which are highly selective for Ca2+ and are expressed primarily in epithelial tissue (10, 11). TRPV6 is the primary transporter of calcium in the intestinal epithelium, while TRPV5 is predominant in the kidney (12). Both TRPV5 and TRPV6 (previously known as CaT2 and CaT1, respectively (13)) are inward rectifying channels, constitutively active at low Ca2+ concentration and physiologic membrane potentials, and more than 100 times more selective for Ca2+ than Na+ (14).
Figure 1.
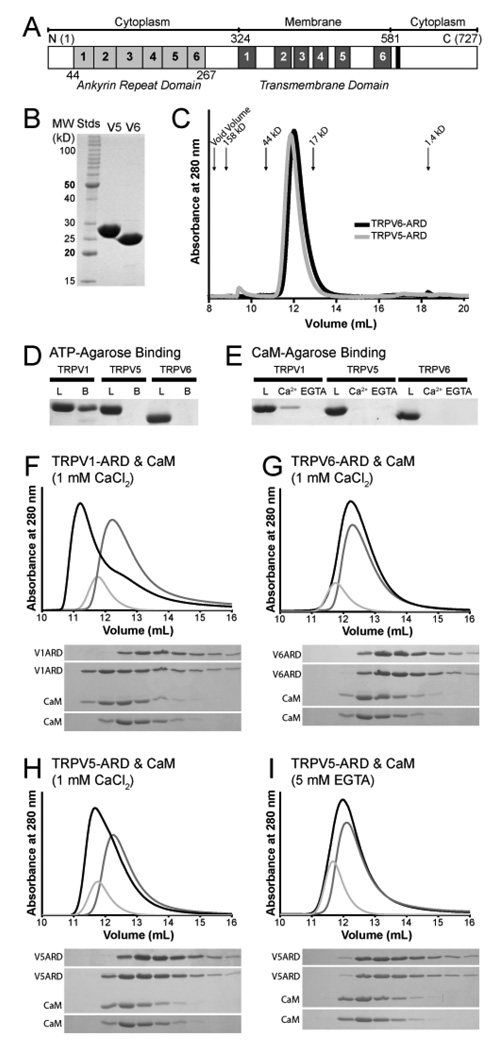
Biochemical properties of the TRPV5-ARD and TRPV6-ARD. (A) Schematic representation of a TRPV protein primary structure and the location of each domain. Individual ankyrin repeats and predicted transmembrane segments are numbered and colored light grey and dark grey, respectively. The conserved TRP-box in the C-terminus is colored black. The residue numbers correspond to those of mouse TRPV6. (B) 15% SDS-PAGE analysis of the TRPV5-ARD and TRPV6-ARD purified from E. coli. (C) Analytical Superdex 75 size exclusion chromatography of the TRPV5-ARD (grey) and TRPV6-ARD (black). Absorbance at 280 nm is plotted against elution volume. The void volume and elution volume of molecular weight standards are indicated. (D) ATP-agarose pull-down assay of TRPV ARDs, with 15% SDS-PAGE analysis of the loaded (L) and ATP-agarose-bound (B) protein. (E) CaM-agarose pull-down assay of TRPV ARDs, with 15% SDS-PAGE analysis of loaded protein (L) and protein bound in the presence (Ca2+) and absence (EGTA) of calcium. TRPV1-ARD (9) is included as a positive control in panels D and E. (F–I) Analysis of CaM/ARD complex formation by Superdex 75 size exclusion chromatography. Shown are representative traces from at least two experiments for TRPV1-ARD (F), TRPV6-ARD (G), and TRPV5-ARD with Ca2+ (H) or EGTA (I). Absorbance at 280 nm is plotted against elution volume with CaM alone in light grey, TRPV-ARD in grey and CaM plus TRPV-ARD in black. Shifts in retention time were confirmed by 20% SDS-PAGE analysis of 0.5 mL fractions covering the elution volumes between 10 and 15 mL for each injection.
TRPV5 and TRPV6 share approximately 75% sequence similarity and are capable of associating with each other; heterotetramers show intermediate electrophysiologic properties, depending on the ratio of TRPV5 to TRPV6 (15). Homo- and hetero-tetramerization is dependent on residues in all three regions of the protein, both the N- and C-terminal cytosolic domains and the transmembrane domain (16). Two ankyrin repeats of the TRPV6-ARD (repeats 3 and 5; residues 116–140 and 192–230, respectively) and ankyrin repeat 1 of TRPV5 (residues 64–76) have been implicated in channel assembly (17, 18). C-terminal residues 596–601 are essential for TRPV5 channel assembly (18). Furthermore, the C-terminal regions of both TRPV5 and TRPV6 are important for binding proteins involved in plasma membrane localization: the small GTPase Rab11a targets both TRPV5 and TRPV6 to the plasma membrane using residues 595–601 in TRPV5, and the corresponding residues (600–607) in TRPV6 (19); and residues 596–616 in TRPV5 (or 601–621 in TRPV6) are important for binding the PDZ domain-containing protein NHREF4, which may be involved in plasma membrane retention of the two channels (20). The C-terminal region also contains the TRP-domain, a short hydrophobic segment found in most TRP channels (21, 22).
The activity of TRPV6, but not TRPV5, is inhibited by direct binding of regulator of G-protein signaling 2 (RGS2) to the TRPV6 N-terminal cytosolic domain (23, 24). The Ca2+-sensing protein calmodulin (CaM) also inhibits TRPV6 (25). CaM binding to and inhibition of TRPV6 requires both the N- and C-terminal cytosolic domains and the transmembrane domain (26, 27). The activity of CaM on TRPV6 is opposed by Protein Kinase C (PKC), which is known to phosphorylate the C-terminal domain of TRPV6 (25).
In addition to its normal role as an epithelial calcium transporter, TRPV6 also plays a role in some types of prostate cancer (28, 29). The TRPV6-mediated increase in prostate cancer cell proliferation is mediated by stimulation of the Ca2+-activated transcription factor NFAT, which inhibits apoptosis (30). This can be replicated by over-expression of TRPV6 in HEK-293 cells, which increases intracellular Ca2+ concentrations, resulting in increased cell proliferation (31).
The TRPV6-ARD crystal structure was determined in order to better understand its role in channel assembly and regulation by other factors. Comparison with the crystal structures of the TRPV1 and TRPV2 ARDs and the sequences of other TRPV proteins leads to the identification of conserved structural features unique to the TRPV ARDs. We also show that the TRPV5 and TRPV6 ARDs are monomeric in solution, and unlike the TRPV1-ARD, they do not bind ATP or CaM to an appreciable extent. We discuss the implications of these structural and biochemical findings on the role of the ARD in the function of TRPV5 and TRPV6.
Materials and Methods
Cloning, expression and purification
The human TRPV5 and mouse TRPV6 cDNAs were obtained from ATCC. The TRPV6-ARD (residues 42–266) and TRPV5-ARD (residues 11–267) were cloned between the NdeI and NotI restriction sites of pET21-C6H (7). The ARDs were expressed in E. coli BL21(DE3) cells by induction with 0.4 mM IPTG (isopropyl-beta-D-thiogalactopyranoside) overnight at room temperature after the cells reached an OD600 of 0.4. Selenomethionine (Se-Met)-substituted TRPV6-ARD was expressed with feedback inhibition of methionine synthesis in M9 minimal medium supplemented with Se-Met under the same conditions. Cell pellets were resuspended in lysis buffer (20 mM TrisHCl (pH 8.0), 300 mM NaCl, 20 mM imidazole (pH 8.0) and 1 mM phenylmethylsulfonyl fluoride) supplemented with 0.1% Triton X-100, 0.2 mg/mL lysozyme, 50 µg/mL RNAse and 25 µg/mL DNAse, and lysed by sonication on ice. The cleared lysates were loaded onto Ni-NTA agarose (Qiagen) and eluted by a step gradient using lysis buffer containing 50, 100, 150 and 200 mM imidazole. Ten mM EDTA (pH 8.0) and 2.5 mM 2-mercaptoethanol were added to each fraction after elution. The ARDs were further purified on a Resource Q column (GE Healthcare) in 20 mM TrisHCl (pH 8.0), 2 mM EDTA (pH 8.0), 1 mM dithiothreitol (DTT) using a linear gradient of 0–1M NaCl. Size exclusion chromatography on a Superdex 200 column (GE Healthcare) in 10 mM TrisHCl (pH 8.0), 50 mM NaCl, 5% (v/v) glycerol and 1 mM DTT was used as a final purification step before concentration to ~30 mg/mL. The DTT concentration was raised to 10 mM in the anion exchange and size exclusion buffers during the purification of Se-Met-substituted TRPV6-ARD. Rat TRPV1-ARD and human CaM were purified using previously established protocols (9, 32).
Analytical size exclusion chromatography
All analyses were performed at 4 °C. TRPV5- and TRPV6-ARD were diluted in Tris buffered saline (TBS), pH 7.4 to 50 µM and injected on a Superdex 75 10/300 column (GE healthcare) in TBS (pH 7.4) with 1 mM DTT. Prior to injection the homogeneity of the samples was confirmed by 15% SDS-PAGE followed by Coomassie staining. Molecular weight standards (BioRad) were used to establish a standard curve to determine the apparent molecular weight of the eluted ARDs. The buffer used for CaM interaction assays was 20 mM TrisHCl, 150 mM NaCl, 1 mM DTT (pH 7.5) with either 1 mM CaCl2 or 5 mM EGTA. Samples containing 25 nmol of TRPV ARD, 25 nmol of CaM or both were incubated in the buffer for 45 minutes prior to injection.
ATP- and CaM-agarose pull-down assays
All binding assays were carried out at 4°C. For ATP-agarose pull-downs, 75 µL of a 50% slurry of agarose beads (11-atom spacer to ribose hydroxyls, Sigma) in 0.1 M TrisHCl (pH 7.5) and 0.5 M NaCl was added to 12.5 µg of protein in 0.9 mL binding buffer (10 mM TrisHCl (pH 7.5), 50 mM NaCl, 1 mM DTT and 0.15% n-decyl-β-D-maltopyranoside). The samples were incubated for 1.5 hours before washing three times with 0.9 mL binding buffer. The samples were eluted by incubating 5 min at 95°C in 75 µL 2× SDS sample buffer (100 mM TrisHCl (pH 6.8), 4% SDS, 0.2% bromophenol blue, 20% glycerol and 200 mM DTT) and analyzed by 15% SDS-PAGE. For CaM-agarose pull-downs, 60 µg of protein were combined with 50 µL of a 50% CaM-agarose slurry (Sigma) in a final volume of 0.3 mL binding buffer with either 2 mM CaCl2 or 5 mM EGTA (pH 7.5) and incubated for 2 hours before washing three times with 0.9 mL of the same buffer. Samples were eluted by incubating 5 min at 95°C in 50 µL of 2× SDS sample buffer and analyzed by 15% SDS-PAGE.
Crystallization of TRPV6-ARD
TRPV6-ARD crystals were grown by the hanging-drop vapor diffusion method at room temperature with a 1:1 protein to reservoir solution ratio, with 0.1 M NaHEPES (pH 7.5), 0.15 M K/Na Tartrate and 5% glycerol in the reservoir. Se-Met TRPV6-ARD crystals were grown from the same solution plus 10 mM DTT. Crystals were cryo-protected in 0.1 M NaHEPES (pH 7.5), 0.2 M K/Na Tartrate and 30% glycerol and flash frozen in liquid nitrogen.
Data collection and structure determination and analysis
X-ray data from native and Se-Met substituted crystals were collected at 100 K using an ADSC Q315 detector at the Advanced Photon Source ID24 beamline. Data were processed in HKL2000 (33) and data statistics are listed in Table 1. The TRPV6-ARD structure was determined to 1.7 Å by SIRAS (single isomorphous replacement with anomalous scattering). Initial phasing, solvent flattening and model building were carried out using autoSHARP (34, 35) and ARP/wARP (36–38). Model building was performed in COOT (39) and refinement with TLS (Translation/Libration/Screw) was carried out in REFMAC5 (40). Due to the lack of completeness in the low resolution data, only reflections between 8.0 and 1.7 Å were used in the final refinement. Final refinement statistics are listed in Table 1. The coordinates have been deposited in the Protein Data Bank with the entry code 2RFA. Figures were generated with PyMOL (41).
Table 1.
Data Collection and Refinement Statisticsa
| Data Collection | ||
|---|---|---|
| Space Group | P212121 | |
| Wavelength (Å) | 0.97921 | |
| Native | Se-Met | |
| Cell dimensions (a, b, c; Å) | 30.76, 63.05, 116.14 | 31.18, 62.51, 116.40 |
| Resolution (Å) | 30.0-1.7 (1.76-1.7) | 30.0-2.4 (2.49-2.4) |
| Rsym | 0.044 (0.445) | 0.094 (0.526) |
| I/σ(I) | 18.0 (3.4) | 11.8 (3.6) |
| Completeness (%) | 98.8 (95.8) | 100.0 (100.0) |
| Redundancy | 7.5 (6.4) | 5.9 (5.9) |
| Refinement | ||
| Resolution (Å) | 8.0 - 1.7 | |
| Number of Reflections | 25,172 | |
| Rwork / Rfree | 0.168 / 0.207 | |
| Molecules/Asymmetric Unit | 1 | |
| Residues in Model | 44–265 | |
| Number of Atoms | ||
| Protein | 1750 | |
| Water | 227 | |
| B-factors (Å2) | ||
| Protein | 35.4 | |
| Water | 56.9 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.014 | |
| Bond Angles (°) | 1.37 | |
Values from the highest resolution shell are in parentheses.
Results
The TRVP5 and TRPV6 ARDs are monomeric in solution
The ankyrin repeats of TRPV5 and TRPV6 play an important role in the tetramerization and assembly of their respective full-length channels (17, 18). Therefore, we investigated whether the isolated ARDs could themselves self-assemble as tetramers. The ARDs of TRPV5 (residues 11–267) and TRPV6 (42–266) were over-expressed and purified from E. coli (Figure 1A and B) and analyzed by size exclusion chromatography to determine their oligomerization state. Both ARDs eluted at volumes consistent with a monomer (Figure 1C). The molecular weights calculated from the amino acid compositions are 29.9 and 26.1 kD for TRPV5-ARD and TRPV6-ARD, respectively, and the apparent molecular weights determined from a standard curve from size exclusion were 29.7 and 27.4 kD, respectively. Furthermore, the TRPV6-ARD behaved as a monomer at all concentrations tested, up to 0.4 mM (data not shown).
The isolated TRPV5 and TRPV6 ARDs do not bind ATP
TRPV1-ARD is known to bind to both ATP and Ca2+-CaM (9), and it has been suggested that the ARD of TRPV6 binds CaM as well (27). We therefore tested whether the TRPV5 and TRPV6 ARDs could bind ATP or CaM in pull-down assays. Under conditions where the TRPV1-ARD was pulled down efficiently by ATP-agarose, neither the TRPV5-ARD nor TRPV6-ARD interacted with ATP-agarose (Figure 1D). This was not unexpected, as two of the positively-charged residues in the second ankyrin repeat of TRPV1 that contact the triphosphate moiety of ATP, K155 and K160, are substituted with negative or hydrophobic residues in TRPV5 and TRPV6 (E80 and I85 in both).
The ARD of TRPV5, but not TRPV6 binds CaM
We also identified conditions under which the TRPV1-ARD bound efficiently to CaM-agarose in a Ca2+-dependent manner, as expected from previous size-exclusion chromatography experiments showing that the two proteins form a 1:1 complex (9). Under those same conditions, the TRPV5-ARD and TRPV6-ARD were not pulled down by CaM-agarose in either the presence or absence of Ca2+ (Figure 1E).
We further investigated the interaction of CaM with TRPV5-ARD and TRPV6-ARD by size exclusion chromatography. As in the pull-down assays, TRPV1-ARD was used as a positive control (Figure 1F). Like the CaM-agarose pull-down assays, no binding was observed between TRPV6-ARD and CaM in the presence of calcium (Figure 1G). In light of previously published serial deletion analysis data showing an interaction between CaM and ankyrin repeat 2 of TRPV6 (27), it is surprising that TRPV6-ARD did not readily bind to CaM-agarose. However, analysis of the structure suggests an explanation for this behavior (see the Discussion below).
A small but significant shift in the elution volume of TRPV5-ARD was observed in the presence of CaM, indicating that the two interact (Figure 1H). Like TRPV1-ARD, the interaction of TRPV5-ARD with CaM could be blocked by removal of calcium with 5 mM EGTA and is therefore is calcium-dependent (Figure 1I). Like TRPV6, interactions between TRPV5 and CaM have been previously reported, but unlike TRPV6, Ca2+-CaM had no effect on TRPV5 activity (27).
Overall Structure of TRPV6-ARD
The crystal structure of TRPV6-ARD was determined using initial phases from single isomorphous replacement with anomalous scattering (SIRAS) and refined to a final Rwork/Rfree of 0.167/0.209 at a resolution of 1.7 Å (Table 1). In agreement with the monomeric state of the TRPV6-ARD observed by size exclusion chromatography, there was one monomer per asymmetric unit, with crystal packing incompatible with a tetrameric state.
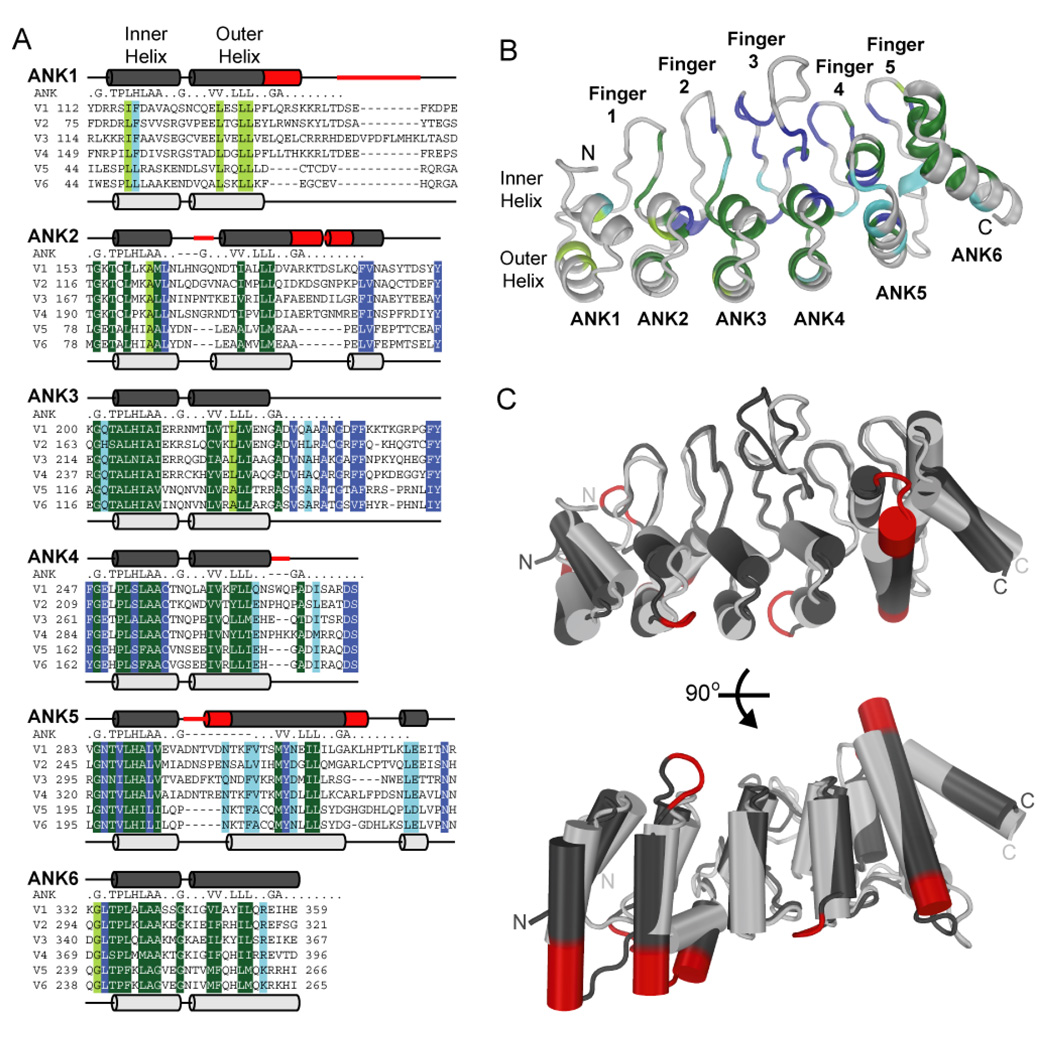
Like the ARDs of other mammalian TRPV proteins (7–9), the TRPV6-ARD is composed of six ankyrin repeats (Figure 2). These repeats have the characteristic anti-parallel inner and outer α-helices, with the helical layers linked together by finger loops (Figure 2). As in other ankyrin repeat-containing proteins, the hydrophobic core of the TRPV6-ARD is composed of conserved ankyrin-repeat consensus residues, while variable residues, which most likely specify binding partners, are found on the exposed faces of the repeat helices and at the tip of the finger loops. In addition, several residues conserved among TRPV ARDs line the base of the finger loops and form the short helices between repeat ANK2 and Finger 2 and ANK5 and Finger 5 (Figure 2B). As in the TRPV1 and TRPV2 ARDs, the TRPV6-ARD structure exhibits a large twist between ANK4 and ANK5. The molecular basis of this conserved twist is examined in detail below.
Figure 2.
Structure of the TRPV6 ARD. (A) Alignment of the ARDs of mammalian TRPV channels separated into individual repeats (ANK1-6). Residues that conform to the ankyrin repeat consensus (indicated above each repeat) in all species (from worms to man – see Supplementary Figure 1) are colored dark green and ankyrin repeat residues conserved in just mammalian TRPVs are colored light green. Residues that are conserved in TRPV ARDs and either are not part of or deviate from the ankyrin repeat consensus are colored dark blue for residues conserved in all species and light blue for residues conserved in mammalian TRPV ARDs only. For ARDs where a structure is yet to be determined (TRPV3, TRPV4 and TRPV5) the human sequences are used. For TRPV1, TRPV2 and TRPV6, the sequence of the crystallized species is used (rat TRPV1 and TRPV2, and mouse TRPV6). The secondary structure of TRPV1-ARD (dark grey; (9)) is indicated above each repeat and the secondary structure of TRPV6-ARD (light grey) is shown below. Insertions in TRPV1-4 compared to TRPV5 and TRPV6 are shown in red. (B) Sequence similarities mapped onto the overall structure of TRPV6-ARD. The backbone is colored according to the sequence similarity in the alignment show in A. (C) Location of the sequence insertions in TRPV1-4 relative to TRPV5 and TRPV6. The structures of the TRPV1 and TRPV6 ARDs were superimposed and colored according to the secondary structure in A. α-helices are shown as cylinders. The view at the bottom corresponds to a 90° rotation around the horizontal axis to highlight the fact that most of the insertions map to the ends of outer helices.
Comparison of the TRPV6-ARD structure to that of TRPV1-ARD and TRPV2-ARD
As stated in the introduction, the mammalian TRPV proteins can be subdivided into two subgroups, the TRPV1-4 channels involved in thermosensation and the TRPV5 and TRPV6 channels, critical components of calcium homeostasis. This subdivision is also reflected in phylogenetic analyses (42) and in sequence similarity across the entire sequence, including within the ARDs (43). Correspondingly, although the overall structure of the TRPV6-ARD is similar to the TRPV1 and TRPV2 ARDs, there are notable differences. There are six sequence insertions within the ARDs of TRPV1-4, compared to TRPV5 and TRPV6. A comparison of the TRPV6-ARD structure to those of TRPV1-ARD (9) and TRPV2-ARD (7, 8) reveals that four of the insertions result in extensions of the C-terminal end of outer helices and the other two extend the turn between helices in repeats ANK2 and ANK5 (Figures 2A and 2C). None of the insertions perturb the orientation or length of the finger loops, and the tip of the fingers are remarkably similar in the structure of the TRPV1-ARD and the TRPV6-ARD (Figure 2C). One notable exception is the tip of the longest finger, Finger 3.
The finger loops in TRPV6 and other TRPV proteins are longer than those in canonical ankyrin repeats with Finger 3 being the longest. Finger 3 is the only loop that adopts strikingly different conformations in the known TRPV ARD structures ((7–9); Figure 2C). The conformational variability is restricted to the tip of the finger, in the region corresponding to residues 153–159 in TRPV6. Notably, this region of Finger 3 is also the most variable in length in TRPV ARDs, varying from seven residues in TRPV2, TRPV5 and TRPV6 to 43 residues in the C. elegans TRPV OCR4 (Supplementary Figure 1).
Molecular basis for the unusual twist between ankyrin repeats 4 and 5 in the TRPV ARDs
As was previously observed in the TRPV1-ARD and TRPV2-ARD structures (7–9), there is a pronounced twist between TRPV6 repeats ANK1-4 and ANK5-6. This twist occurs in a region where the ARD of the TRPVs have conserved substitutions deviating from the ankyrin repeat consensus (blue residues in Figures 2A and 2B). It had been suggested that the twist between repeats in the TRPV2-ARD were a result of the extended outer helices (7), however TRPV6 shows the same twist despite having shorter outer helices (Figures 2A and 2C). Canonical ankyrin repeats, like repeats ANK2 through ANK4 in TRPV6, have a twist angle of approximately 6° between repeats and a distance of 12 Å between the final Cα of adjacent outer helices (see Supplementary Figure 2 for measurement details). In TRPV6, the twist angle between repeats ANK4 and ANK5 is 17° with a corresponding distance of 13 Å and the twist between repeats ANK5 and ANK6 is 22° with a distance of 17 Å. These increased twist angles are similar in the TRPV1- and TRPV2-ARDs (data not shown).
The twist between repeats ANK4 and ANK5 results from displacement of inner helix 5 from the regular packing of hydrophobic side chains observed between canonical ankyrin repeats. TRPV-conserved residues that deviate from ankyrin consensus in inner helix of repeat 5, V199 and L203 in TRPV6, replace the proline and alanine normally found in those positions (5 and 9, respectively; Figure 3A) (44, 45). Furthermore, I202 on inner helix 5 is rotated out of the hydrophobic core between repeats ANK4 and ANK5 and instead packs against the Y161 and Y162 side chains at the base of Finger 3 and C172 on the outer face of inner helix 4 (Figure 3B).
Figure 3.
Conserved TRPV residues induce the unusually pronounced twist in the ARD and define the orientation of the finger loops. In all panels, green dashed lines indicate hydrogen bonds typically observed in canonical ankyrin repeats and blue dashed lines represent hydrogen bonds between TRPV conserved residues. Distances normally observed in ankyrin repeats are shown as dashed black lines and significantly different distances in TRPV6-ARD are shown in red. All distances are in Å. All side chains are colored according to Figures 2A and 2B. Key residues are labeled, and numbers in parentheses correspond to the position of the residue within the ankyrin repeat consensus (see Figure 2A). (A) The canonical ankyrin repeat packing pattern can be observed between ANK3 and ANK4 (left), while V199 and L203 on the inner helix of ANK5 break the regular stacking of the helices (right). In panel A the regions illustrated are viewed approximately from the back of the orientation shown in Figure 3B. (B) A network of hydrophobic packing and hydrogen bonds between TRPV-conserved residues stabilize the base of Finger 3, the displaced inner helix of ANK5 and the displaced outer helix of ANK6, generating the pronounced twist between repeats 4 and 5 observed in all TRPV ARDs. Only ankyrin repeats 3 through 6 are shown as a Cα trace. Residues involved in the conserved hydrophobic network are shown as sticks and transparent spheres (with coloring conforming to Figure 2A). (C) TRPV substitutions disrupt the parallel orientation of the start and end of consecutive fingers. ANK3 (left) adopts a canonical ankyrin repeat fold with the threonine and histidine at positions 4 and 7, respectively, setting the orientation of the finger with respect to the helices. In contrast, the end of Finger 3 adopts a novel configuration as it lies against the inner helix of ANK4 (right).
Finally, a large twist within repeat ANK6 prevents a steric clash between the side chains of Y216 from repeat ANK5 and L258 from repeat ANK6 (Figure 3B). This results in a twist angle between the outer and inner helices of about 34°, approximately 15° more than seen in most ankyrin repeats. Meanwhile, Finger 5 and inner helix of repeat 6 are held in place by canonical ankyrin interactions.
Conserved TRPV residues determine the orientation of Finger 3 base
In all known TRPV ARD structures, including in TRPV6-ARD, Finger 3 breaks from the regular β-strand-like packing at the start and end of consecutive fingers. The position of an ankyrin repeat finger is usually fixed relative to the following inner helix by interactions between conserved threonine and histidine residues at positions 4 and 7, respectively, in the ankyrin repeat consensus, and the amide nitrogens of the first turn of the inner helix (Figure 3C left) (46). In repeat ANK4 of TRPV ARDs, position 4 is variable and position 7 is a conserved serine (Figure 2A). As a result, without the canonical constraint on backbone orientation, Finger 3 folds back along the face of inner helix 4 and the first ANK4 residue, Y162, packs against C172 (Figure 3C right). Instead of the canonical backbone β-strand hydrogen bonds observed between the end of Finger 2 and the start of Finger 3, the E164 side chain from Finger 3 bridges the gap and hydrogen bonds with S194 at the start of Finger 4 (Figure 3B & C). Y161 from Finger 3 also hydrogen bonds to D193 and N197 of Finger 4, further buttressing both loops in relation to each other. The base of Finger 3 is further stabilized by a conserved network of hydrophobic residues from repeats two through five that includes Y161, Y162 and C172 (Figure 3B).
Discussion
Biochemical and crystallographic analyses of the ARD of TRPV6 reveal that the six-repeat domain is monomeric. Like the TRPV1 and TRPV2 ARDs, the TRPV6-ARD shows a pronounced twist between repeats ANK1-4 and ANK5-6. The twist in the ARD is induced by conserved TRPV substitutions in the ankyrin repeat consensus and stabilized by conserved hydrophobic side chains at the bases of the unusually long Fingers 2 and 3. These hydrophobic residues also anchor the base of the most flexible and variable region in TRPV ARDs, the tip of Finger 3. The impact of these structural features on function and regulation of TRPV5 and TRPV6 are discussed below.
Role of the ARD in assembly of tetrameric TRPV5 and TRPV6 channels
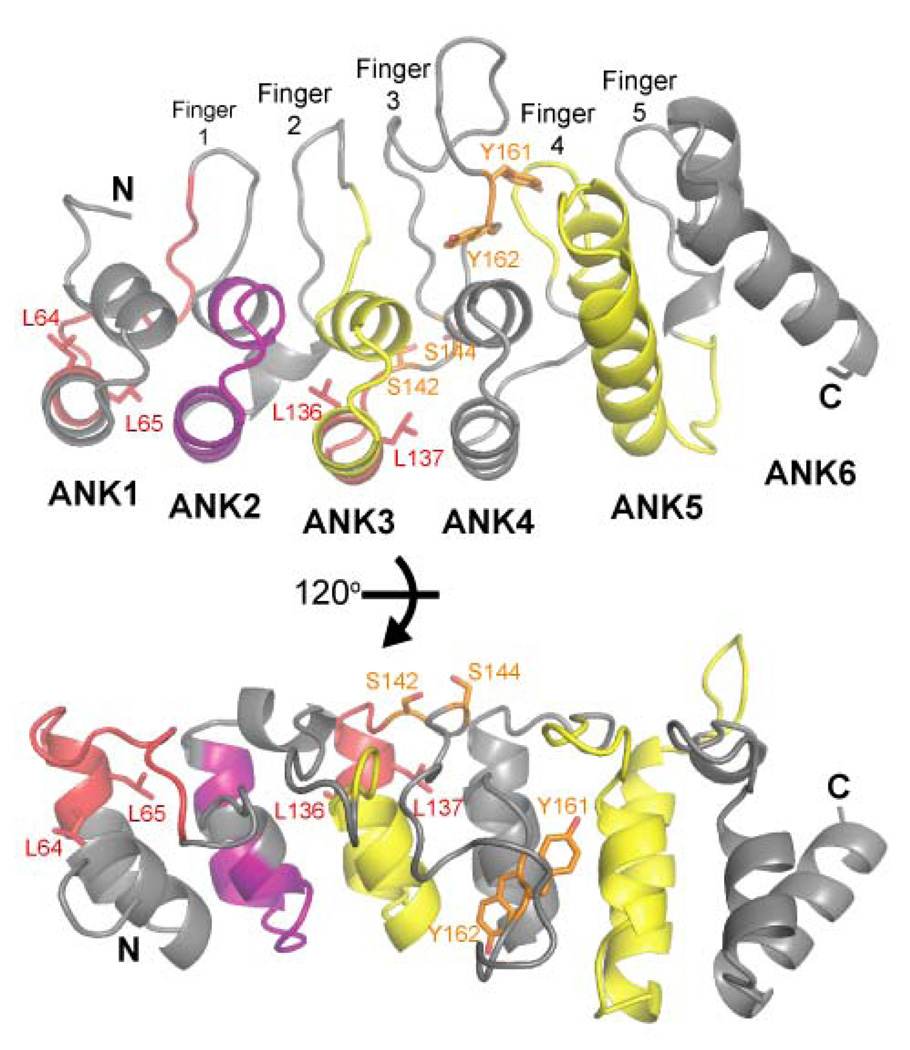
Previous reports have suggested that the ARDs of TRPV5 and TRPV6 are directly involved in tetramerization of the channels through interaction between the ARDs (17, 18). The locations are mapped onto the TRPV6 ARD structure in Figure 4. Our size exclusion data clearly indicate that the TRPV5-ARD and TRPV6-ARD are monomeric in solution. Furthermore, the packing and symmetry of in the TRPV6-ARD crystals are incompatible with tetrameric assembly of the ARD around a four-fold symmetry axis. Residues that had previously been identified as critical for tetramerization are also at important conserved ankyrin repeat positions. Residues that are critical for ankyrin repeat stability, particularly the paired leucines in the outer helices of repeats 1 and 3 (Figure 4) are either lost by deletion of ANK1 in TRPV5 or mutation of ANK3 in TRPV6 in the tetramerization-deficient mutants (17, 18). At positions in ankyrin repeats that are normally conserved, mutations to non-consensus residues typically decrease the stability of ARDs, while conversely for ARDs that have non-consensus residues at conserved positions, mutations back to ankyrin repeat consensus often increase ARD stability (45, 47, 48). Furthermore, the folding of ankyrin repeat structures is nucleated at specific repeats and propagated through the rest of the domain (49). Removal of ANK3 in TRPV6 (17) or the introduction of destabilizing mutations in TRPV5 (18) therefore likely disrupts the overall fold of the entire ARD, thereby preventing the assembly of tetrameric channels. Erler and colleagues also observed that two complementary segments of the TRPV6-ARD, ANK3-4 (residues 116–191) and ANK5-6 (residues 192–328), could interact in a two-hybrid assay (17). This finding could be explained by the two segments stacking to form a single ARD in trans. Furthermore, interactions between the N- and C-termini of both TRPV5 and TRPV6 have been observed (17, 18), and this may be the reason that interactions were also observed between the TRPV6 N-terminus and full-length protein, but not the N-terminus with itself in bacterial two-hybrid assays (17). It is also possible that TRPV5 and TRPV6 assembly is assisted by additional cellular factors which require the ARD and are unable to bind an ARD destabilized by mutation or partial deletion. In summary, while previous studies clearly demonstrate that the integrity of the ARD is important in channel assembly, our data indicate that it is not through self-tetramerization of the ARD. Rather, the TRPV ARDs likely regulate channel assembly through interactions with other factors or other regions within the channel.
Figure 4.
Previously identified functions of the TRPV6-ARD mapped onto the structure. Sections identified as critical for channel assembly/tetramerization (residues 64–76 in TRPV5 (18), and residues 136–140 in TRPV6 (17)) are colored red and the regions of ankyrin repeats 3 and 5 assigned a role in channel assembly (residues 116–163 and 192–230; (17)) are colored yellow. The region previously implicated in CaM binding (residues 85–100; (27)) is colored purple. Side chains whose phosphorylation may regulate TRPV6 activity are colored orange.
Regulation of TRPV5 and TRPV6 by CaM
Although in a previous study no effect of CaM on TRPV5 activity was observed (27), we observed an interaction between TRPV5-ARD and Ca2+-CaM in the size exclusion assay, but not the more stringent CaM-agarose pull-down assay, indicating that the interaction is weaker than that observed between TRPV1-ARD and CaM. It should also be noted that the TRPV5-ARD construct used contained an additional 30 residues at its N-terminus compared to the TRPV6-ARD construct used in this study. The TRPV5 construct was extended because the construct to most homologous to TRPV6-ARD could not be expressed in a soluble form. It is therefore possible that these extra residues contribute to the differences observed between TRPV5 and TRPV6 in the CaM-binding assays. The in vivo relevance of the weak interaction of CaM and TRPV5-ARD remains to be determined.
CaM is known to regulate TRPV6 and strong evidence exists for an interaction between CaM and the C-terminus of TRPV6 (25, 27). Others have also reported that CaM interacts with the TRPV6-ARD, in the same region important for CaM binding to TRPV1 ((9, 25, 27); Figure 4). However, in our assays we do not observe any binding of CaM to TRPV6-ARD under conditions where CaM does bind the TRPV1 and TRPV5 ARDs (Figure 1E–I). Like residues previously thought to be involved in tetramerization, the TRPV6-ARD residues implicated in CaM binding (residues 93–102; (27)) form part of the conserved ankyrin repeat core (Figure 4) and are unlikely to be available for interactions with other proteins. Recent cell-based studies, using a combination of electrophysiology and confocal microscopy, have also shown that the TRPV6 N-terminus does not play a significant role in its inhibition by CaM (50). Although both TRPV1 and TRPV6 are inhibited by CaM, the failure of the TRPV6-ARD to bind CaM (or ATP, a competitor for the CaM binding site and sensitizer in TRPV1) indicates that CaM regulates different members of the TRPV family through distinct mechanisms.
Regulation of TRPV6 by phosphorylation of ARD residues
It is possible that post-translational modifications are necessary for tetramerization of the TRPV6-ARD or for its association with CaM, and a number of confirmed and putative phosphorylation sites have been identified within the TRPV6-ARD. Phosphorylation of rat TRPV6 by a Src-like kinase leads to an increased Ca2+ influx through the channel, and both Y161 and Y162 were identified as putative phosphorylation sites through mutagenesis (51). The PTP1B phosphatase also interacts with the first 191 residues of TRPV6 and inhibits Ca2+ influx, likely through dephosphorylation of these tyrosine residues (51). Mutation of both tyrosines was necessary to prevent up-regulation of the channel by phosphorylation. In the TRPV6-ARD structure, the hydroxyl group of Y162 is exposed at the protein surface, while Y161 is buried and interacts with side chains from Finger 4 (Figures 3 and 4). If the observed tyrosine phosphorylation plays a significant role in regulating TRPV6 function in vivo, it is unlikely that Y162 is an important physiologic substrate of Src-like kinases and PTP1B, as it is not conserved in all TRPV6 proteins: it is a tyrosine in crayfish, zebrafish, mouse and rat; but a phenylalanine in human, chimpanzee, chicken and frog (Supplementary Figure 1). In the TRPV6-ARD structure, Y161 is buried and inaccessible to kinases, and therefore also seems an unlikely target for phosphorylation. As mentioned above, mutations away from ankyrin repeat consensus destabilize individual ankyrin repeats (47, 48). It is therefore possible that the presence of residues that deviate from the ankyrin repeat consensus in TRPVs makes this region dynamic enough to allow kinase access to Y161. The Y161 side chain has slightly above average B-factors for the structure (41.6 Å2 vs. 35.4 Å2) and is positioned where Finger 3 transitions from the stable region of conserved TRPV residues to the variable and flexible fingertip (Supplementary Figure 3). However, making Y161 accessible to a kinase still requires a rather large conformational change.
Two other putative phosphorylation sites have previously been identified in TRPV6 by sequence analysis (11). S142 and S144 are putative targets for CaM dependent kinase II (CaMKII) and protein kinase C (PKC), respectively. Both are surface-exposed at the base of Finger 3 and phosphorylation at one of the serines would disrupt recognition of the target site by the other kinase (Figure 4). It is noteworthy that neither serine is on the concave surface of the ARD that is typically involved in protein-ligand interactions in ankyrin repeat-containing proteins (46). Still, the possibility that these two putative phosphorylation sites play a role in the balance between PKC activity and increased intracellular Ca2+/CaM-binding in regulation of TRPV6 requires further investigation.
Conclusion
Comparison of the ARD structures from three TRPV proteins reveals conserved residues that produce the twist and stabilize the long fingers of these ARDs. Protein sequence analysis shows that the TPRV ARD “scaffold” is a conserved feature from worms to man. These conserved residues form a continuous network that stabilizes both the twists and turns of the fingers and helices. The reason for the conservation of this twisted ankyrin repeat structure and stabilization of the base of the elongated Finger 3 will require the elucidation of the structures of TRPV ARDs with some of their interacting partners.
TRPV6 and the closely-related TRPV5 channels are critical regulators of calcium homeostasis. Their activity, localization and assembly are all regulated in part through their N-terminal ARD. The crystal structure of the TRPV6 ARD reveals the location and environment of amino acid residues previously identified as mediators of these activities. Many of these residues actually form the conserved core of the TRPV6-ARD and provide the scaffolding that stabilizes the domain, shaping the surface accessible to interacting regulatory partners. The interactions that regulate TRPV6 are therefore likely to come from other residues, not yet identified, that are positioned by this scaffolding. The TRPV6-ARD structure provides a basis for further experiments such as mutagenesis of surface residues to disrupt potential ligand interfaces.
Supplementary Material
Acknowledgments
The authors would like to thank Albert Li, Shauna Stanton, and the other current and former members of the Gaudet lab for technical help, advice and discussion.
Abbreviations
- ANK
ankyrin repeat
- ARD
Ankyrin Repeat Domain
- ATP
adenosine triphosphate
- CaM
calmodulin
- CaMKII
calmodulin dependent kinase II
- DTT
dithiothreitol
- EDTA
ethylenediamine tetraacetic acid
- FRET
Förster resonance energy transfer
- GTP
guanosine triphosphate
- HEK
human embryonic kidney cells
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- NFAT
Nuclear Factor of Activated T-cells
- PAGE
polyacrylamide gel electrophoresis
- PDB
Protein Data Bank
- PKC
Protein Kinase C
- (RGS2)
regulator of G-protein signaling 2
- SIRAS
single isomorphous replacement with anomalous scattering
- SDS
sodium dodecyl sulfate
- Se-Met
Seleno-methionine
- TRP
Transient Receptor Potential
- TRPV
TRP Vanilloid
Footnotes
This work was supported by AHA SDG 0335134N, a McKnight Scholar Award and a Klingenstein Fellowship Award to RG. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The structure of the TRPV6 ankyrin repeat domain has been deposited in the Protein Data Bank as entry 2RFA.
Supporting Information Available
Supporting information consists of three supplemental figures. Supplementary Figure 1 is a sequence analysis of TRPV ARDs from worms to man. Supplementary Figure 2 illustrates the measurements of twists and distances between sequential ankyrin repeats. Supplementary Figure 3 shows the B-factor distribution of TRPV6-ARD mapped onto the structure as well as the electron density around Y161. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nilius B, Voets T. TRP channels: a TR(I)P through a world of multifunctional cation channels. Pflugers Arch. 2005;451:1–10. doi: 10.1007/s00424-005-1462-y. [DOI] [PubMed] [Google Scholar]
- 2.Owsianik G, D'Hoedt D, Voets T, Nilius B. Structure-function relationship of the TRP channel superfamily. Rev Physiol Biochem Pharmacol. 2006;156:61–90. [PubMed] [Google Scholar]
- 3.Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 4.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 5.Montell C. The venerable inveterate invertebrate TRP channels. Cell Calcium. 2003;33:409–417. doi: 10.1016/s0143-4160(03)00053-8. [DOI] [PubMed] [Google Scholar]
- 6.Vriens J, Owsianik G, Voets T, Droogmans G, Nilius B. Invertebrate TRP proteins as functional models for mammalian channels. Pflugers Arch. 2004;449:213–226. doi: 10.1007/s00424-004-1314-1. [DOI] [PubMed] [Google Scholar]
- 7.Jin X, Touhey J, Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J Biol Chem. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]
- 8.McCleverty CJ, Koesema E, Patapoutian A, Lesley SA, Kreusch A. Crystal structure of the human TRPV2 channel ankyrin repeat domain. Protein Sci. 2006;15:2201–2206. doi: 10.1110/ps.062357206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 10.Montell C, Birnbaumer L, Flockerzi V. The TRP channels, a remarkably functional family. Cell. 2002;108:595–598. doi: 10.1016/s0092-8674(02)00670-0. [DOI] [PubMed] [Google Scholar]
- 11.den Dekker E, Hoenderop JG, Nilius B, Bindels RJ. The epithelial calcium channels, TRPV5 & TRPV6: from identification towards regulation. Cell Calcium. 2003;33:497–507. doi: 10.1016/s0143-4160(03)00065-4. [DOI] [PubMed] [Google Scholar]
- 12.Hoenderop JG, Nilius B, Bindels RJ. Epithelial calcium channels: from identification to function and regulation. Pflugers Arch. 2003;446:304–308. doi: 10.1007/s00424-003-1045-8. [DOI] [PubMed] [Google Scholar]
- 13.Clapham DE, Julius D, Montell C, Schultz G. International Union of Pharmacology. XLIX. Nomenclature and structure-function relationships of transient receptor potential channels. Pharmacol Rev. 2005;57:427–450. doi: 10.1124/pr.57.4.6. [DOI] [PubMed] [Google Scholar]
- 14.Vennekens R, Hoenderop JG, Prenen J, Stuiver M, Willems PH, Droogmans G, Nilius B, Bindels RJ. Permeation and gating properties of the novel epithelial Ca(2+) channel. J Biol Chem. 2000;275:3963–3969. doi: 10.1074/jbc.275.6.3963. [DOI] [PubMed] [Google Scholar]
- 15.Hoenderop JG, Voets T, Hoefs S, Weidema F, Prenen J, Nilius B, Bindels RJ. Homo- and heterotetrameric architecture of the epithelial Ca2+ channels TRPV5 and TRPV6. Embo J. 2003;22:776–785. doi: 10.1093/emboj/cdg080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellwig N, Albrecht N, Harteneck C, Schultz G, Schaefer M. Homo- and heteromeric assembly of TRPV channel subunits. J Cell Sci. 2005;118:917–928. doi: 10.1242/jcs.01675. [DOI] [PubMed] [Google Scholar]
- 17.Erler I, Hirnet D, Wissenbach U, Flockerzi V, Niemeyer BA. Ca2+-selective transient receptor potential V channel architecture and function require a specific ankyrin repeat. J Biol Chem. 2004;279:34456–34463. doi: 10.1074/jbc.M404778200. [DOI] [PubMed] [Google Scholar]
- 18.Chang Q, Gyftogianni E, van de Graaf SF, Hoefs S, Weidema FA, Bindels RJ, Hoenderop JG. Molecular determinants in TRPV5 channel assembly. J Biol Chem. 2004;279:54304–54311. doi: 10.1074/jbc.M406222200. [DOI] [PubMed] [Google Scholar]
- 19.van de Graaf SF, Chang Q, Mensenkamp AR, Hoenderop JG, Bindels RJ. Direct interaction with Rab11a targets the epithelial Ca2+ channels TRPV5 and TRPV6 to the plasma membrane. Mol Cell Biol. 2006;26:303–312. doi: 10.1128/MCB.26.1.303-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Graaf SF, Hoenderop JG, van der Kemp AW, Gisler SM, Bindels RJ. Interaction of the epithelial Ca2+ channels TRPV5 and TRPV6 with the intestine- and kidney-enriched PDZ protein NHERF4. Pflugers Arch. 2006;452:407–417. doi: 10.1007/s00424-006-0051-z. [DOI] [PubMed] [Google Scholar]
- 21.Montell C. Physiology, phylogeny, and functions of the TRP superfamily of cation channels. Sci STKE. 2001;2001 doi: 10.1126/stke.2001.90.re1. RE1. [DOI] [PubMed] [Google Scholar]
- 22.Montell C. The TRP superfamily of cation channels. Sci STKE. 2005;2001 doi: 10.1126/stke.2722005re3. re3. [DOI] [PubMed] [Google Scholar]
- 23.Meyer MB, Watanuki M, Kim S, Shevde NK, Pike JW. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol Endocrinol. 2006;20:1447–1461. doi: 10.1210/me.2006-0031. [DOI] [PubMed] [Google Scholar]
- 24.Meyer MB, Zella LA, Nerenz RD, Pike JW. Characterizing early events associated with the activation of target genes by 1,25-dihydroxyvitamin D3 in mouse kidney and intestine in vivo. J Biol Chem. 2007;282:22344–22352. doi: 10.1074/jbc.M703475200. [DOI] [PubMed] [Google Scholar]
- 25.Niemeyer BA, Bergs C, Wissenbach U, Flockerzi V, Trost C. Competitive regulation of CaT-like-mediated Ca2+ entry by protein kinase C and calmodulin. Proc Natl Acad Sci U S A. 2001;98:3600–3605. doi: 10.1073/pnas.051511398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilius B, Weidema F, Prenen J, Hoenderop JG, Vennekens R, Hoefs S, Droogmans G, Bindels RJ. The carboxyl terminus of the epithelial Ca(2+) channel ECaC1 is involved in Ca(2+)-dependent inactivation. Pflugers Arch. 2003;445:584–588. doi: 10.1007/s00424-002-0923-9. [DOI] [PubMed] [Google Scholar]
- 27.Lambers TT, Weidema AF, Nilius B, Hoenderop JG, Bindels RJ. Regulation of the mouse epithelial Ca2(+) channel TRPV6 by the Ca(2+)-sensor calmodulin. J Biol Chem. 2004;279:28855–28861. doi: 10.1074/jbc.M313637200. [DOI] [PubMed] [Google Scholar]
- 28.Wissenbach U, Niemeyer BA, Fixemer T, Schneidewind A, Trost C, Cavalie A, Reus K, Meese E, Bonkhoff H, Flockerzi V. Expression of CaT-like, a novel calcium-selective channel, correlates with the malignancy of prostate cancer. J Biol Chem. 2001;276:19461–19468. doi: 10.1074/jbc.M009895200. [DOI] [PubMed] [Google Scholar]
- 29.Fixemer T, Wissenbach U, Flockerzi V, Bonkhoff H. Expression of the Ca2+-selective cation channel TRPV6 in human prostate cancer: a novel prognostic marker for tumor progression. Oncogene. 2003;22:7858–7861. doi: 10.1038/sj.onc.1206895. [DOI] [PubMed] [Google Scholar]
- 30.Lehen'kyi V, Flourakis M, Skryma R, Prevarskaya N. TRPV6 channel controls prostate cancer cell proliferation via Ca(2+)/NFAT-dependent pathways. Oncogene. 2007 doi: 10.1038/sj.onc.1210545. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz EC, Wissenbach U, Niemeyer BA, Strauss B, Philipp SE, Flockerzi V, Hoth M. TRPV6 potentiates calcium-dependent cell proliferation. Cell Calcium. 2006;39:163–173. doi: 10.1016/j.ceca.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Drum CL, Shen Y, Rice PA, Bohm A, Tang WJ. Crystallization and preliminary X-ray study of the edema factor exotoxin adenylyl cyclase domain from Bacillus anthracis in the presence of its activator, calmodulin. Acta Crystallogr D Biol Crystallogr. 2001;57:1881–1884. doi: 10.1107/s0907444901014937. [DOI] [PubMed] [Google Scholar]
- 33.Otwinnoski Z, Minor D. In: Methods in Enzymology. Carter CW, M S JR, editors. New York: Academic Press; 1997. pp. 307–326. [Google Scholar]
- 34.de La Fortelle E, Bricogne G. Methods in Enzymology. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 35.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods Mol Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 36.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 37.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 38.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. San Carlos, CA, USA: DeLano Scientific LLC; [Google Scholar]
- 42.Liedtke W, Kim C. Functionality of the TRPV subfamily of TRP ion channels: add mechano-TRP and osmo-TRP to the lexicon! Cell Mol Life Sci. 2005;62:2985–3001. doi: 10.1007/s00018-005-5181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phelps CB, Procko E, Lishko PV, Wang RR, Gaudet R. Insights into the Roles of Conserved and Divergent Residues in the Ankyrin Repeats of TRPV Ion Channels. Channels. 2007;1:148–151. doi: 10.4161/chan.4716. [DOI] [PubMed] [Google Scholar]
- 44.Mosavi LK, Minor DL, Jr, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci U S A. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 46.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zweifel ME, Leahy DJ, Hughson FM, Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12:2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreiro DU, Cervantes CF, Truhlar SM, Cho SS, Wolynes PG, Komives EA. Stabilizing IkappaBalpha by "consensus" design. J Mol Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe AR, Itzhaki LS. Rational redesign of the folding pathway of a modular protein. Proc Natl Acad Sci U S A. 2007;104:2679–2684. doi: 10.1073/pnas.0604653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derler I, Hofbauer M, Kahr H, Fritsch R, Muik M, Kepplinger K, Hack ME, Moritz S, Schindl R, Groschner K, Romanin C. Dynamic but not constitutive association of calmodulin with rat TRPV6 channels enables fine tuning of Ca2+-dependent inactivation. J Physiol. 2006;577:31–44. doi: 10.1113/jphysiol.2006.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sternfeld L, Anderie I, Schmid A, Al-Shaldi H, Krause E, Magg T, Schreiner D, Hofer HW, Schulz I. Identification of tyrosines in the putative regulatory site of the Ca2+ channel TRPV6. Cell Calcium. 2007;42:91–102. doi: 10.1016/j.ceca.2006.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.