Abstract
Collagen mimetic peptides (CMPs) have been used to elucidate the structure and stability of the triple helical conformation of collagen molecules. Although CMP homotrimers have been widely studied, very little work has been reported regarding CMP heterotrimers because of synthetic difficulties. Here we present the synthesis and characterization of homotrimers and ABB type heterotrimers comprising natural and synthetic CMP sequences that are covalently tethered to a template, a tris(2-aminoethyl) amine (TREN) succinic acid derivative. Various tethered heterotrimers comprising synthetic CMPs [(ProHypGly)6, (ProProGly)6] and CMPs representing specific domains of type I collagen were synthesized and characterized in terms of triple helical structure, thermal melting behavior and refolding kinetics. The results indicated that CMPs derived from natural type I collagen sequence can form stable heterotrimeric helical complexes with artificial CMPs and that the thermal stability and the folding rate increase with the increasing number of helical stabilizing amino acids (e.g. Hyp) in the peptide chains. Covalent tethering enhanced the thermal stability and refolding kinetics of all CMPs; however their relative values were not affected suggesting that the tethered system can be used for comparative study of heterotrimeric CMP's folding behavior in regards to chain composition and for characterization of thermally unstable CMPs.
Introduction
As a basic component of the extracellular matrix (ECM) and with more than 28 different types known to date, collagen is one of the most abundant and diversified proteins in mammals. It is a major structural component in most connective tissues such as skin, ligament, cartilage, bone, and tendon.1 The collagen molecule is characterized by the general Xaa-Yaa-Gly trimeric repeating motif in amino acid sequence where proline and hydroxyproline (Hyp, O) occur most often in the Xaa and Yaa positions, respectively.2 A hallmark structural feature of collagens is their unique triple helix in which three strands, each in a left-handed helix, intertwine with one another to form a right handed triple helix that is stabilized by inter-chain hydrogen bonds. Some collagen types are homotrimers (e.g. type II, III and VIII), while others are heterotrimers of ABB or ABC composition (e.g. type I, IV and V). The most abundant type I collagen, for example, exists as an ABB heterotrimer that consists of two α1(I) chains and one α2(I) chain which are genetically distinct but similar to each other in terms of amino acid sequence.3 The heterotrimeric nature of type I collagen plays an important role in collagen fiber packing as well as the mechanical properties and enzymatic susceptibility of collagen fibers.3-5 Mutations of heterotrimeric type I collagen (α1)2(α2)1 into homotrimeric type I collagen (α1)3 are reported to cause osteogenesis imperfecta in mice models.6,7
Collagen mimetic peptides (CMPs) have been used as a molecular model to elucidate both the three-dimensional structure of the collagen triple helix and the origin of its conformational stability.8-11 CMPs are generally 15 to 45 amino acid residues in length and are comprised of collagen-like Xaa-Yaa-Gly tri-residue repeats that have the inherent propensity to form the triple-helix structure. For decades, most research activities on CMPs have focused on homotrimeric CMPs where three identical polypeptide chains make up the triple helix. These CMPs are ideal models for the representation of homotrimeric collagens, such as type II and type III. It has proved more difficult to obtain and characterize heterotrimeric CMPs, which can serve as models for heterotrimeric collagens such as type I and IV, and only a few studies on heterotrimeric CMPs have been reported. Zagari and coworkers reported the melting behavior of CMP heterotrimers [(ProProGly)10]2(ProHypGly)10 and (ProProGly)10[(ProHypGly)10]2. These heterotrimers were identified from a mixture solution of (ProProGly)10 and (ProHypGly)10 after heating and cooling cycle.12 In addition to thermal transitions of the homotrimers, two additional transitions, which corresponded to the melting of the heterotrimeric CMPs, were observed by circular dichroism spectrometry (CD). The presence of heterotrimeric CMPs in the mixture solution indicated that CMPs with high Hyp content could form triple helices with CMP chains of low Hyp content. Hartgerink's research group recently reported the use of a similar melting/refolding strategy, combined with controlled charge-charge interactions, to prepare well-defined ABB and ABC types of CMP heterotrimers.13,14 When a mixture of charged single-stranded CMPs were gradually cooled down to below their melting temperatures, a stable heterotrimer was formed that contained CMP trimers with balanced electrostatic charges; upon cooling, this CMP mixture solution comprising anionic, cationic and neutral strands produced only ABC type heterotrimers,13,14 which allowed a folding kinetic study of a heterotrimer for the first time.15 However, this method is only suitable for a heterotrimeric system that contains well designed charge distributions and can not be applied to general heterotrimeric CMPs unless they are employed as part of a host-guest system. In addition, charge interactions that hold the heterotrimers together are easily disrupted by changes in pH and ionic strength, which limits the conditions for studying the trimers' folding behaviors.
Covalent bridges that link three peptide strands together have been used to enhance the stability of self-assembled triple helices and to produce heterotrimeric CMPs. Common methods involve cross-linking of side-chain functional groups16,17 or the use of trifunctional organic templates.18-20 Moroder and coworkers developed an effective method for heterotrimeric CMP synthesis by applying a cystine-knot strategy.17 The orthogonally protected thiol groups of cysteine residues were selectively de-protected and coupled to the thiols on other CMP strands which led to the formation of heterotrimeric CMPs. However, the order of chain alignment in these heterotrimeric CMPs had significant effects on triple helical stability and folding kinetics; two heterotrimers with the same composition but different chain register exhibited a 12°C difference in melting temperatures.21 This indicated that the structural constrain in one of the heterotrimers was very high which prevented an optimal conformation for triple helical structure. Goodman and coworkers reported the synthesis and biophysical analysis of template-assembled homotrimeric collagen structures using KTA (cis, cis-1, 3, 5-trimethylcyclohexane-1, 3, 5-tricarboxylic acid) or tris(2-aminoethyl) amine (TREN) succinic acid derivatives as templates.20,22 The conformation study showed that both KTA and TREN derivatives were relatively flexible templates which provided the one-residue shift necessary for the assembly of collagen-like triple helical structure, but the TREN template was reported to be more flexible than the KTA template and present less steric effects on the CMP's triple helical structure.22
Besides being a structural model of natural collagen, CMPs can also be used to produce various self-assembled biomaterials for potential applications in biomedicine.23-25 In particular, our group reported CMP binding to films and fibers of natural collagen, which is presumably mediated by the formation of hybrid complexes between CMPs and disentangled domains of the collagen molecules.26,27 These hybrid complexes can be modeled by template-tethered CMP heterotrimers, though currently all attempts at producing template-tethered CMPs have focused on homotrimeric CMPs, mainly due to the difficulties in selectively coupling one or two peptides to the templates. Here, we present a simple strategy for CMP heterotrimer synthesis using the TREN template, which involves serial solid phase and solution coupling of two different CMPs onto the template. We chose a well-established TREN template system to ensure optimal triple helix assembly of our target homo- and hetero-trimeric CMPs.22 Triple helical structures, thermal melting behaviors and refolding kinetics of various ABB type heterotrimers are presented which help us understand the heterotrimeric nature of collagens. In addition, the template-tethered heterotrimers can serve as an alternative to the host-guest system for studying thermally unstable CMPs derived from natural collagens and collagen-like sequences in non-collagenous proteins.
Materials and Methods
All reagents were obtained from Sigma-Aldrich (St. Louis, MO), Advanced Chemtech (Louisville, KY) or Novabiochem (La Jolla, CA) and used without further purification. Amino acids were purchased from Advanced Chemtech and Novabiochem. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry analyses were performed on an Applied Biosystems Voyager DE-STR spectrometer.
Peptide Synthesis
All peptides were synthesized using Fmoc mediated solid phase chemistry by automated or manual synthesis. Automated peptide synthesis was carried out on a peptide synthesizer (model 431) from Applied BioSystems (Foster City, CA). TentaGel R RAM resin (0.19 mmol of reactive sites/g; 0.1 mmol loading level; Peptides International, Louisville, KY) was loaded in the reaction vessel. Piperidine (20% by volume) in methylpyrrolidone (NMP) was used as the deprotection solution and effectiveness of the deprotection was monitored using a conductivity flow cell. The activation solution contained O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate (HBTU, 0.225 M) and N-hydroxybenzotriazole (HOBt, 0.225 M) in dimethylformamide (DMF). Five molar equivalents of Fmoc-protected amino acids were used. For side-chain protection, tert-butyl was used for Ser and Tyr, trityl for Asn and Gln, 4-{N-[1-(4,4-dimethyl-2,6-dioxocyclohexylidene)-3-methylbutyl]amino}benzyl ester (ODmab) for Glu and 1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl (Dde) was used for Lys. All amino acids were double-coupled followed by acetylation using a mixture of acetic anhydride (0.5 M), N-diisopropylethylamine (DIPEA, 0.125 M), and HOBt (0.015 M) in NMP to minimize the production of peptides with deleted sequence.
Manual peptide synthesis was carried out using 4 molar equiv of amino acids. Fmoc-deprotection was accomplished by treatment with 20% (v/v) piperidine in DMF for 40 min. The coupling reaction was performed using 4 molar equiv of HBTU and 6 molar equiv of DIPEA in NMP solution. All the coupling reactions were completed within 2∼4 hr and monitored by ninhydrin or chloranil tests. After the coupling of the last amino acid residue, the peptides were deprotected and the resins were dried under vacuum. The peptides were cleaved from the resin by treating the resin with a mixture solution of trifluoroacetic acid (TFA)/triisopropylsilane (TIS)/H2O (95:2.5:2.5 by volume) for at least 2 hr. The TFA solution was collected and ten-fold excess cold ether was added to precipitate the target peptides. The peptides were collected by centrifugation and dried under vacuum. The peptides were dissolved in deionized water and purified by reverse-phase HPLC with a C-18 column using a mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) as a mobile phase. The purified peptides were analyzed by MALDI-TOF MS (Supporting Information: Figure S1): m/z calculated 1745.0 [M] for P, found 1745.8 [M + H+]; m/z calculated 1841.0 [M] for O, found 1841.9 [M + H+]; m/z calculated 1790.9 [M] for α, found 1791.5 [M + H+]; m/z calculated 1804.9 [M] for T, found 1826.2 [M + Na+].
Template-Tethered Homotrimer Synthesis
The TREN template, TREN-(suc-OH)3, was synthesized according to the literature.22 DMF (0.1 mL) solution containing TREN-(suc-OH)3 (1 μmol, 0.45 mg) was mixed with HBTU (3.5 μmol in 7.0 μL of DMF solution) and DIPEA (82 μmol, 15 μL), and stirred at room temperature for 5 min. This mixture solution was added drop wise to 0.9 mL of DMF solution containing the peptide (4.0 μmol). The reaction was run for 24 hr at room temperature. The homotrimers were precipitated using 10 mL of cold ethyl ether and isolated by centrifugation.
Template-Tethered Heterotrimer Synthesis
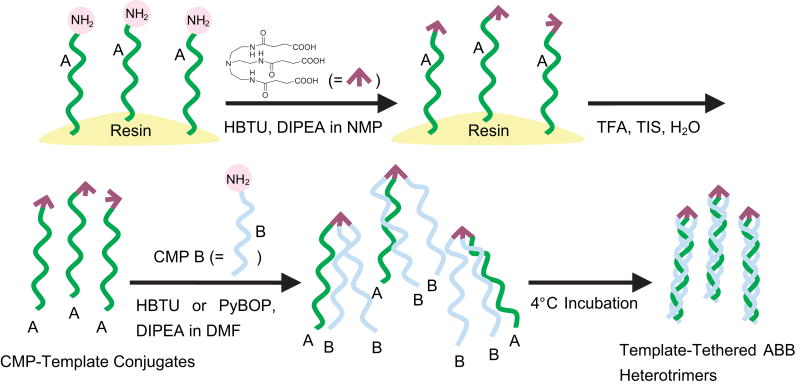
Heterotrimers were prepared by the route shown in Scheme 1, involving both solid phase and solution coupling. Conventional solid phase peptide synthesis was used to prepare CMP A on TentaGel R RAM resin. After final deprotection, 10 molar equiv of TREN-(suc-OH)3 was reacted to CMP A on the solid phase resin, using 40 molar equiv of HBTU (0.5 M in DMF solution). After 48 hr, the CMP-template conjugates were cleaved using a mixture of TFA/TIS/H2O=95/2.5/2.5 and purified by HPLC. (Supporting Information: Figure S2) CMP-template conjugates can also be synthesized by solution coupling. (See Supporting Information) The CMP-template conjugates (1 μmol) were dissolved in 0.1 mL of DMF to which were added HBTU (5 μmol in 10 μL of DMF solution) and DIPEA (82 μmol, 15 μL). The reaction mixture was stirred at room temperature for 10 min followed by addition of DMF solution (0.9 mL) containing 8 μmol of CMP B. After 48 hr of reaction time, the crude product was precipitated by addition of 10 mL of cold ethyl ether and isolated by centrifugation.
Scheme 1.
Synthetic strategy for template-tethered ABB type CMP heterotrimers.
For the synthesis of heterotrimers containing peptide α, the crude heterotrimer product isolated from centrifugation was re-dissolved in 2% hydrazine in DMF (1 mL) and stirred for 5 minutes to remove the side chain protective groups from Lys and Glu. The peptides were re-precipitated in 10 mL of cold ethyl ether and isolated by centrifugation.
HPLC Purification and MALDI Characterization of Template-Tethered CMPs
All peptides and template-tethered trimer products were purified by HPLC. HPLC purification was performed on a Varian Polaris 210 series equipped with Vydac C18 reverse phase column. Linear gradient mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) was used as a mobile phase. Semi-preparative column was used at a flow rate of 4 mL/min for peptide purification and analytical column was used at a flow rate of 1 mL/min for template-tethered CMP trimers. If the purity of HPLC purified products was less than 90%, the peptides were further purified by dialysis or by additional HPLC using a mixture of water (0.1% TFA) and methanol (0.1% TFA) as a mobile phase. MALDI-TOF MS was used to confirm the purity of the products (Supporting Information: Figure S1). MALDI MS m/z calculated 5627.4 [M] for P·P·P, found 5626.4 [M + H+]; m/z calculated 5723.4 [M] for O·P·P, found 5748.8 [M + Na+]; m/z calculated 5819.4 [M] for P·O·O, found 5820.1 [M + H+]; m/z calculated 5915.4 [M] for O·O·O, found 5914.1 [M + H+]; m/z calculated 5762.5 [M] for a·a·a, found 5765.2 [M + H+]; m/z calculated 5673.4 [M] for a·P·P, found 5668.3 [M + H+]; m/z calculated 5865.4 [M] for a·O·O, found 5865.0 [M + H+]; m/z calculated 5804.2 [M] for T·T·T, found 5804.4 [M + H+]; m/z calculated 5686.3 [M] for T·P·P, found 5684.9 [M + H+]; m/z calculated 5878.3 [M] for T·O·O, found 5879.2 [M + H+].
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra were collected on JASCO 715 spectrophotometer equipped with a JASCO PTC-348 WI temperature controller and a Hellma Cell (400 μL, 0.1 mm pathlength). Samples (0.2 mg/ml in 10 mM sodium phosphate buffer, pH 7.0) were stored at 4°C for at least 24 hr before the CD measurement and thermal unfolding studies. The thermal unfolding studies were performed by measuring the ellipticity maximum peak at 225 nm with 60 °C/hr heating rate for all CMPs. Mean residue ellipticity was calculated as follows: [θ] = (θ·m)/(c·l·nr), where θ is the observed ellipticity in mdeg, m is the molecular weight in g/mol, c is sample concentration in mg/mL, l is path length of the cuvette in cm, and nr is the number of amino acids in the peptide or in the template-tethered trimer molecule. Melting temperature (Tm) was determined by fitting the mean residue ellipticity to a two-state model.28 All melting experiments were repeated and Tms were found to be reproducible within ±1°C. To compare the effects of heating rate, compound T·O·O's melting experiment was conducted at both 60 °C/hr and 10 °C/hr heating rates (Figure S5). Tm determined from the slower heating rate was approximately 3°C lower than that determined from the fast heating rate.12 In addition, CD melting studies of heterotrimer a·O·O were carried out in both acidic (0.2 mg/ml in 50 mM H3PO4/NaH2PO4 buffer, pH 3.1) and basic (0.2 mg/ml in 50 mM sodium borate buffer, pH 9.3) solutions to investigate the effect of pH in the folding behavior of template-tethered heterotrimers.
Refolding Kinetics Studies
The refolding rate of CMP trimers were determined by first measuring the maximum mean residue ellipticity at 225 nm of the fully folded CMP trimers (0.2 mg/ml in 10 mM sodium phosphate buffer, pH 7.0) that had been incubated at 4°C for at least 24 hr ([θ]folded). The trimer molecules were thermally denatured by heating at a 1°C/min rate from 4°C to a temperature 20°C higher than their respective Tms (for O·O·O, 85°C was used due to the instrument limitation). CD signal was recorded ([θ]unfolded) after 5 min of incubation at that temperature. The solution was rapidly quenched to 4°C and the recovery of the ellipticity maximum was recorded over time ([θ]T). Fraction folded (FF) was calculated from the following equation: FF = ([θ]T − [θ]unfolded)/([θ]folded − [θ]unfolded) and the t1/2 is taken as the time that the melted CMP takes to achieve 50% fraction folded.
Results and Discussion
Design and Synthesis of CMPs
To demonstrate the advantage and versatility of the template tethering approach, we chose to study tethered trimers of various compositions comprising four CMPs: two conventional CMPs based on ProProGly (P) and ProHypGly (O) repeating units, and two natural collagen mimetic sequences, α and T, derived respectively from the middle domain (α2 chain, 269-286) and the triple helical C- terminal (α2 chain, 1091-1108) domain of rat tail type I collagen (Table 1). CMPs of ProProGly and ProHypGly triplet repeats have been extensively studied in the past by our group and others. 8,10,12,27 Their well-known triple helical stability and folding behavior allows for easy prediction and interpretation of the folding behavior of the template-tethered trimers. Previously, we reported CMP's structure-dependent binding affinity to type I collagen26 and hypothesized that Hyp-deficient domains or terminal domains could form loose structures that can be potential targets for CMP hybridization.27 Peptide α was selected from one of the thermally unstable domains of type I collagen, while the peptide T was selected from the triple helical C-terminal domain, which contains four Hyp residues.1,29 Although this sequence is rich in the helix-stabilizing amino acid Hyp, we thought that it could also form loose structures in the collagen triple helix because of its terminal location.
Table 1.
Sequences and CD melting temperatures of CMPs.
| Abbreviation | Peptide Sequence | Tm (°C) |
|---|---|---|
| P | G(PPG)6Y | -- |
| O | G(POG)6Y | 32 |
| α | GPKGELGPVGNPGPAGPAGY | -- |
| T | GSQGPAGPOGPOGPOGPOGY | 16 |
In designing the target peptides, we inserted an extra Gly at the N-terminus as a spacer to decouple direct interactions between the peptide and the template, and an extra Tyr at the C-terminus for convenient quantification of peptides using UV absorbance at 275 nm.
Conventional Fmoc chemistry was used to prepare the target peptides. In consideration of the subsequent template coupling reactions that occur between free amino termini of the CMPs and the carboxylic acid groups of the templates, all four peptides were synthesized on a TentaGel R RAM resin that liberates the target peptide with unreactive carbamide protecting the C-terminus. The side chains of reactive amino acids Lys and Glu were protected with Dde and ODmab, respectively, which are stable under peptide cleavage conditions (TFA treatment) and prevent undesired cross-links among various peptides during template coupling.
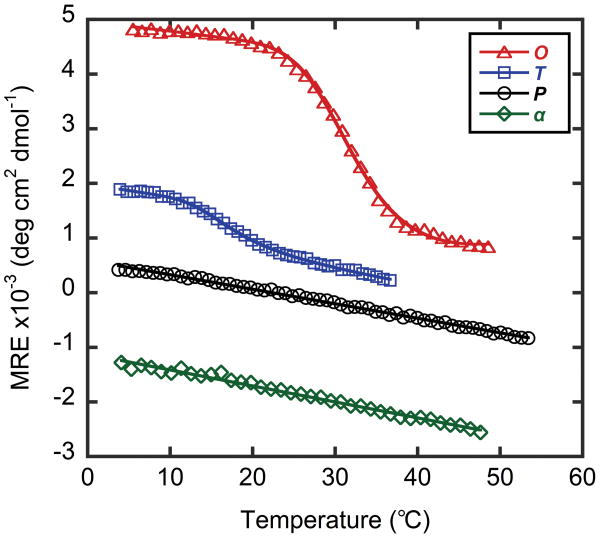
We conducted CD melting experiments to determine the thermal stability of the CMP triple helices (Table 1, Figure 1). Among the four peptides, O and T showed sharp sigmoidal thermal transitions; O melted at 32°C and T at 16°C. In contrast, a linear decrease in ellipticity was observed for P and α, suggesting little structure formation even at a temperature as low as 4°C.
Figure 1.
Thermal melting curves of untethered peptides O, T, P and α.
Synthesis of Template-Tethered Homo and Hetero CMP Trimers
Template-tethered homotrimers, designated as A·A·A, were synthesized by conjugating TREN-(suc-OH)3 templates directly onto purified CMPs in DMF solution at room temperature for 24 hr. According to recent report, ABB type heterotrimers can have 3 different chain registers with varying stability.30 Therefore, TREN template was chosen among other templates for its flexibility that could potentially accommodate all 3 chain registers and allow formation of the most stable triple helix.22 We examined three coupling reagents: 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), N,N′-diisopropyl-carbodiimide (DIC), and HBTU. The MALDI after the conjugation reaction showed that HBTU was most effective among the three tested. The homotrimer coupling yield of P·P·P after HPLC purification was 65% for HBTU activation but only 20% for EDC activation, while no significant product was obtained when DIC was used as the coupling reagent (Figure S3). When the same HBTU activation methods were applied to the synthesis of O·O·O, the yield after HPLC purification was only 32%. The reduction in reactivity for O may have been due to a small amount of water in the hydrophilic peptide O which can reduce the efficiency of HBTU as reported before.31 Because of small reaction scale, the exact yields of other homotrimers (α·α·α and T·T·T) after HPLC purification were not determined; however a comparative HPLC analyses suggested that they were in between those of P·P·P and O·O·O.
Template-tethered ABB type heterotrimers, designated as A·B·B, were synthesized by a two-step process. First, a single type A CMP was conjugated to the TREN-(suc-OH)3 template by reacting excess amount of template with the CMP directly on the resin. Due to the low density of reactive sites characteristic to the PEG-PS based Tentagel resin, only one of the template's three carboxylic acid moieties reacted with the resin-bound CMP. This process allowed easy synthesis of single CMP-template conjugate without any side products (e.g. double or triple CMP-template conjugates) to which full length type B CMP was added in solution to produce the template-tethered ABB type trimer. Compared to the homotrimer synthesis, the second coupling reaction proceeded with much lower reactivity as evidenced by the MALDI and HPLC profiles. We speculate that the CMP (chain A) conjugated to the template is sterically hindering the access of the next CMP (chain B) to the two carboxylic acids remaining on the template. Therefore, an excess amount of type B CMP was used for the reaction along with a long reaction time (48 hr) and the following adjustments were made to the coupling reagents of the second coupling step to achieve the satisfactory reaction yields. Similar to the homotrimer synthesis, P was found to be more reactive than O. For example, yields after HPLC purification was 15.6% for T·P·P and 8.8% for T·O·O.
During the 48 hr template conjugation reaction time, the HBTU had a tendency to react with the terminal amine of the CMP, forming the tetramethylguanidinium derivative.32 To avoid this side reaction, TREN-(suc-OH)3 was pre-activated with HBTU/DIPEA before addition to the peptides and the use of excess HBTU was avoided. In the case of coupling O to the template, we used benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP) instead of HBTU to avoid this side reaction.33 The PyBOP-mediated reaction was run at 60°C to compensate for its lower reactivity compared to HBTU.
Thermal Stability of Template-Tethered CMPs
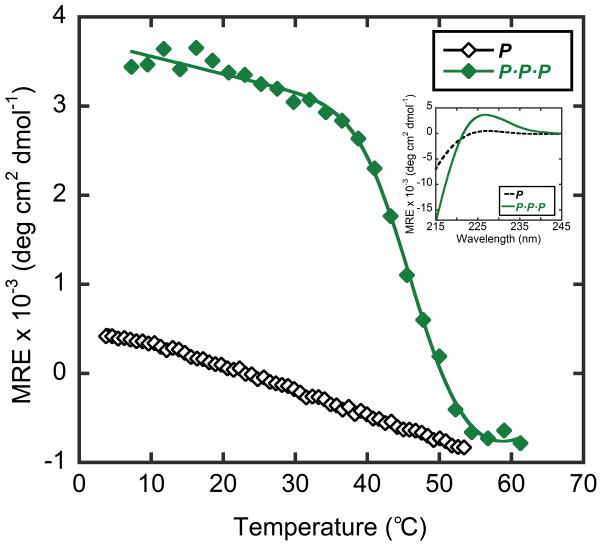
In order to demonstrate the validity of our approach, we first studied the melting and refolding properties of template-tethered homo and heterotrimers comprising only conventional CMPs, O and P. Our results revealed significant differences in structural stability between tethered and untethered CMPs. In drastic contrast to untethered P, when three strands of P were covalently tethered to the TREN-template, the homotrimer P·P·P displayed a CD spectrum characteristic of collagen triple helix (MREmax = 3,658 deg·cm2·dmol-1) and a well-defined sigmoidal melting curve with Tm at 47°C (Figure 2). Similarly, the tethered homotrimer O·O·O had a high melting temperature of 72°C, whereas the Tm of untethered O was 32°C (Figure 3). These drastic increases in Tm are mainly due to the entropic effect brought on by the template: folding and unfolding of tethered CMPs are intra-molecular processes while those of untethered CMPs are inter-molecular.22
Figure 2.
Thermal melting curves and CD spectra (inset) of untethered peptide P and template-tethered P·P·P. The high ellipticity at 226 nm and the sigmoidal melting curves for P·P·P indicate triple helix stabilization effect of the TREN template.
Figure 3.
CD melting studies of peptide O, and template-tethered CMP homotrimers and heterotrimers composed of P and O.
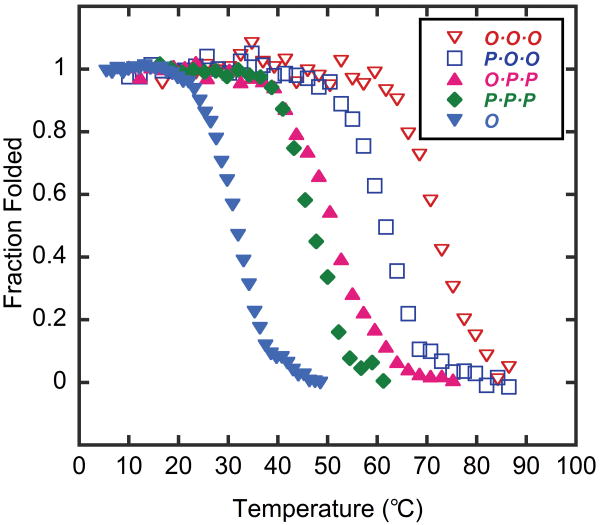
The CD melting study of the homo and heterotrimers revealed that the Tm of the tethered trimers rise with increasing numbers of O strands present in the triple helix due to the stabilizing effect of Hyp (Table 2, Figure 3).34,35 We observed a melting temperature elevation of 4°-11°C per replacement of one P strand with one O strand: P·P·P (47°C), O·P·P (51°C), P·O·O (62°C), and O·O·O (72°C). These results are in agreement with recent studies on untethered heterotrimeric CMPs [(PPG)10]2(POG)10 and (PPG)10[(POG)10]2,12,36 which were produced by cooling a mixture solution comprising the two melted CMPs. Zagari and coworkers observed Tm elevation of 6°-12°C per replacement of one (ProProGly)10 strand with (ProHypGly)1012 which is close to our template-tethered CMP system. Further examination of Tm values of tethered heterotrimers revealed cooperativity of O-O interactions in triple helix stabilization. The increase of Tm from homotrimer P·P·P to heterotrimer O·P·P was only 4°C, while the Tm difference between O·P·P (51°C) and P·O·O (62°C) was over 11°C. Considering that these jumps in melting temperatures are caused by the same substitution (single P→O), the marked differences in ΔTm are a clear indication of cooperativity in triple helix folding where O-O interactions are producing a synergistic stabilizing effect. Although the results showed a significant triple helix stabilization effect of the TREN-template, the template did not overwhelm the characteristics of each CMP strand, and the order of Tm remained the same as the untethered CMPs revealing the effect of the trimer's chain composition on the overall triple helical stability.
Table 2.
CD melting temperatures and refolding parameters of template-tethered trimers comprising synthetic CMPs.
| Compound | Tm (°C) | t1/2 (min) |
|---|---|---|
| P·P·P | 47 | 2.1 |
| O·P·P | 51 | 1.3 |
| P·O·O | 62 | 0.4 |
| O·O·O | 72 | -- |
Refolding Kinetics Studies of Template-Tethered CMPs
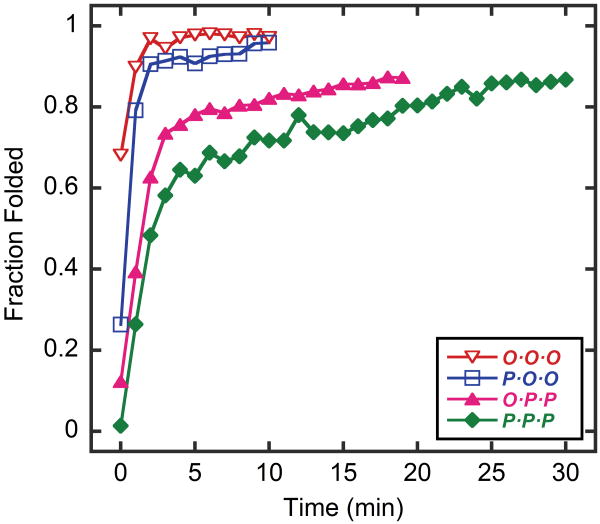
Using this template system, we were able to study the refolding behavior of CMP heterotrimers under various conditions. Such studies can only be performed with tethered heterotrimers since during the refolding process, untethered CMP chains can redistribute and form various heterotrimers with mixed compositions.12,36 The folding rates of the tethered homotrimers and heterotrimers were examined by rapidly quenching the thermally denatured homotrimers or heterotrimers to 4°C and observing the increase in CD signal at 225 nm over time. Untethered host-guest homotrimeric CMPs studied by Brodsky and coworkers had folding half times (t1/2) varying from tens of minutes to more than an hour.37 In contrast, our tethered CMP trimers showed a very fast refolding process with half times on the order of a few minutes (Table 2, Figure 4). Homotrimer O·O·O refolded so fast that we were unable to measure its t1/2; after the operation dead time, the first data point of O·O·O already corresponded to folding recovery of 68%. The t1/2 values of trimers P·O·O, O·P·P and P·P·P were 0.4 min, 1.3 min, and 2.1 min, respectively. Similar to the thermal melting studies, the rates of folding were heavily affected by the composition of the peptides: more O stands in the tethered trimer resulted in faster folding which also correlated with the melting temperatures as described above. The tethering of CMP strands increases the local concentration of unfolded CMPs. Given the high concentration dependency of the nucleation step in folding, the fast folding rates of these tethered CMPs were expected as others have reported similar fast refolding kinetics for FeII(bpy)3-tethered CMP homotrimers, which displayed a t1/2 value of less than one minute.38
Figure 4.
Refolding kinetics of thermally denatured template-tethered CMP homotrimers and heterotrimers at 4°C.
The template-tethered heterotrimeric CMPs can be considered a simplified model for heterotrimeric collagen molecules especially in the folding kinetic studies. Folding of collagens is nucleated by trimeric non-collagenous domains which are usually located at the C-terminus of a procollagen chain. While this suggests that the triple helix folding proceeds from the C- to the N-terminus, recent collagen sequence studies and folding kinetics studies of cross-linked CMP homotrimers suggest that collagen folding can also be nucleated at the N-terminus.39,40 The complex nucleation domain used in those reported folding studies could be replaced by the TREN-template which is structurally simple and readily available. For the TREN-template-tethered CMPs, we expect that the nucleation and follow-up helix propagation will preferentially occur at the N-terminal end at which the template is attached. In this regard, the TREN-(suc-OH)3 template CMP system provides not only heterotrimers of defined composition but also prominent nucleation sites for the helix formation that can be a suitable model for investigating the inter-strand interactions and the folding directionality of collagen.
Investigation of CMP-Collagen Interactions
Because of their common structural features and the nature of inter-strand interactions, synthetic CMPs and collagen are known to form hybrid complexes by heterogeneous triple helix assembly.41 Schiele and coworkers reported that the α1 chain of calf-skin collagen can form such hybrids with synthetic polypeptides including (ProAlaGly)n and (ProProGly)n.41 The hybridization was induced by cooling the melted mixture of α1 chains and synthetic polypeptides and the optical rotation data of the hybridized product indicated formation of the triple helix structure. Hybridization of type I collagen and synthetic CMPs have been extensively studied by our group where specific techniques were developed to allow CMPs to either adhere to or bury themselves within type I collagen fibers.26,27,42,43 The temperature-dependency of the collagen binding affinities that were determined for a series of CMPs with varying chain lengths indicated that the binding was mainly driven by triple helix propensity. By employment of CMP-functionalized gold nanoparticles, we identified periodic domains on collagen fibers that attract CMPs, indicating a high binding specificity.27,43,44 Although our TEM results were unable to identify the exact molecular location of these hybridization sites, we hypothesized that thermally unstable domains that lack Hyp or the termini of collagen molecule could induce microunfolding of triple helix and expose stretches of disentangled collagen chain that can hybridize with CMPs.
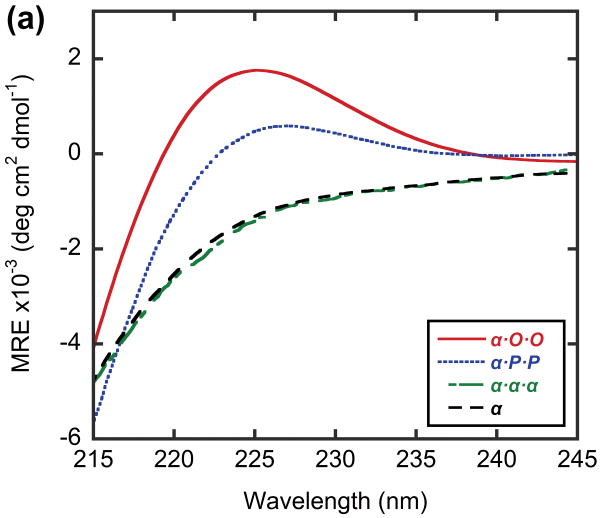
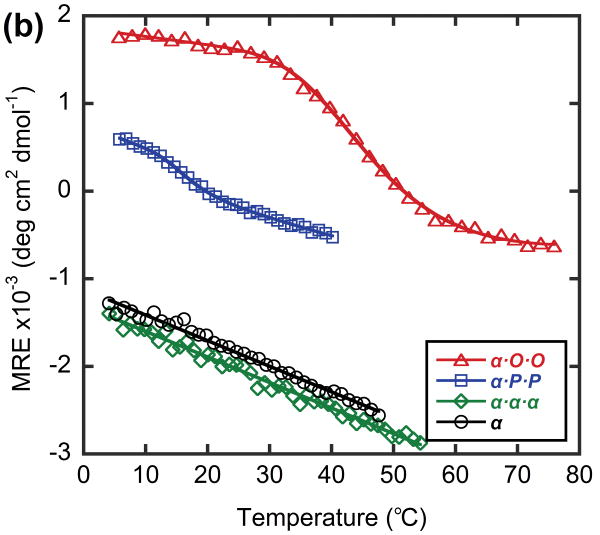
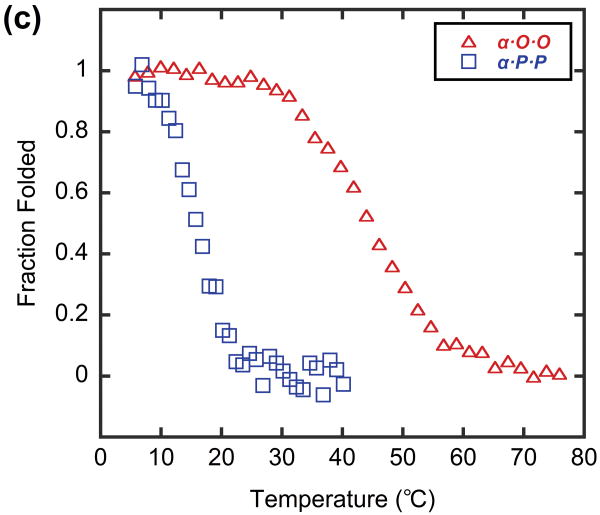
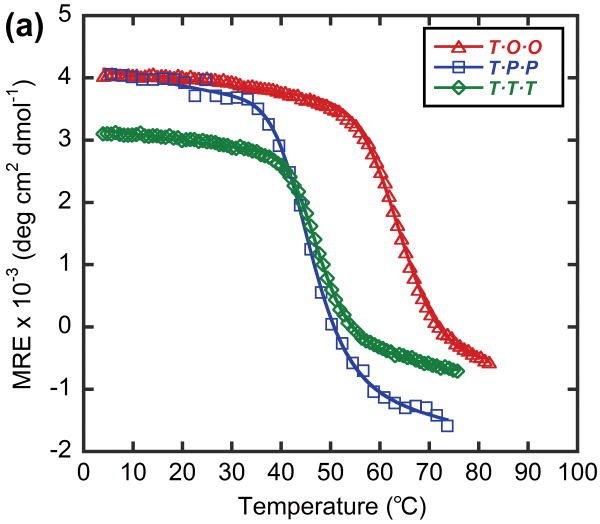
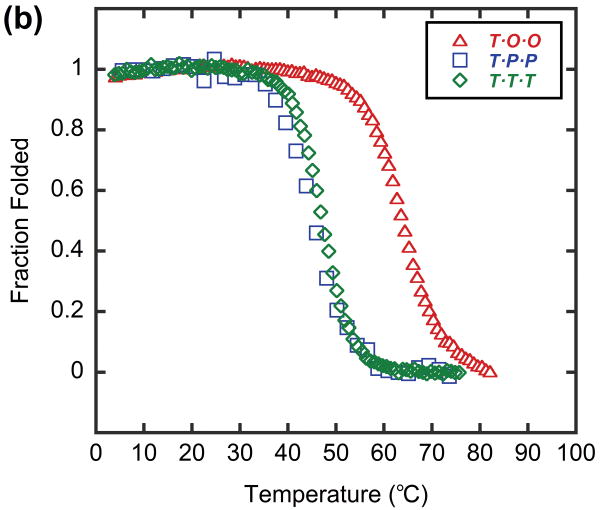
In order to model a potential hybrid complex that might form between CMP and collagen, we prepared a series of tethered heterotrimers comprising synthetic (O and P) and natural sequence (α and T) CMPs, which respectively represent the Hyp-deficient thermally labile domain and the triple helical C-terminal domain of type I collagen (Table 3). Heterotrimers α·O·O, α·P·P and T·O·O, T·P·P were produced to mimic hybrid complexes, and homotrimers α·α·α and T·T·T were produced to represent native collagen helices based on the similarity in amino acid sequence between the α1 and α2 chains of type I collagen. Peptide α, which had the lowest propensity for collagen triple helix among the four CMPs, did not form triple helices even when all three strands were covalently tethered to the template (α·α·α, Table 3), evidenced by i) no ellipticity maximum observed at 225 nm (MRE225 = -1415 deg·cm2·dmol-1, Figure 5a) and ii) an almost linear decrease in ellipticity during thermal melting studies (Figure 5b). In contrast, the CD spectrum of α·O·O showed positive maximum at 225.2 nm (MREmax = 1761 deg·cm2·dmol-1, Figure 5a) and the thermal melting experiment showed a clear sigmoidal curve with Tm at 44°C (Figure 5b, 5c), which indicated the characteristic triple helical structure with relatively high thermal stability. The Tm of T·T·T, was 47°C, but the T·O·O heterotrimer exhibited an even higher Tm of 64°C (Figure 6, Table 3). These results clearly demonstrate that the synthetic/natural CMP hybrid complexes do fold into stable triple helices, and that unstable sequences derived from natural collagen can form much more stable helices by hybridization with the peptide O. In addition, heterotrimers α·P·P (Tm: 16°C) and T·P·P (Tm: 45°C) folded into triple helical structure with lower thermal stability (Table 3, Figure 5, 6) which verifies the strong triple-helical stabilization effect of Hyp even in these heterotrimeric systems. However, these P containing hybrids still showed enhanced or similar triple helical stability when compared to their natural homotrimer counterparts (Table 3), suggesting that polypeptide (ProProGly)n is well capable of forming triple helical hybrids with natural collagen sequence.41
Table 3.
CD melting temperatures of template-tethered homo/heterotrimers comprising natural collagen sequence α and T.
| Compound | Tm (°C) |
|---|---|
| α·α·α | -- |
| α·P·P | 16 |
| α·O·O | 44 |
| α·O·O (pH 3.1) | 40 |
| α·O·O (pH 9.3) | 43 |
| T·T·T | 47 |
| T·P·P | 45 |
| T·O·O | 64 |
Figure 5.
CD spectra (a) and thermal melting curves (b) of α and α-containing template-tethered homotrimers and heterotrimers, shown as mean residue ellipticity. (c) Thermal melting curves of heterotrimer α·P·P and α·O·O, shown as fraction folded. Peptide α and homotrimer α·α·α do not self assemble into triple helices, but heterotrimers α·P·P and α·O·O form stable triple helices with defined Tm values.
Figure 6.
Thermal melting curves of T·T·T, T·P·P and T·O·O, shown as mean residue ellipticity (a), and fraction folded (b).
We admit that our template-tethered heterotrimer is an oversimplified model of the CMP-collagen hybrid complex that does not consider any effect from kinetic competition by homotrimer formation during CMP invasion or a neighboring effect during hybridization, both of which are critical to CMP-collagen interactions. However, this was our first attempt at reproducing the potential CMP-collagen interaction at a molecular level and will serve as the foundation for further studies aimed at improving this system to better mimic CMP-collagen complexes. Using the template-tethered heterotrimer system, we were able to verify the gain in thermal stability that was expected when unstable natural collagen sequences form a hybridized complex with synthetic CMP with high triple helical propensity.
The unstable triple helical domains in a collagen chain play important roles in biological activities including ligand binding and enzymatic susceptibility.29 Mutations that lead to structural instability in the triple helix have been related to debilitating diseases such as osteogenesis imperfecta.45 CMPs derived from these thermally unstable domains are often too unstable to be studied directly by CD. Therefore, Brodsky's research group pioneered the host-guest system.2,45-47 In this system, the unstable target peptide of interest is flanked at one or both termini by three to five ProHypGly triplets that act as external triple helix stabilizers, elevating the Tm of the unstable peptide to where it can be easily determined by CD. Recently, Hartgerink and coworkers have extended this host-guest approach to include heterotrimers by applying charge-charge interactions that mediate controlled non-templated heterotrimer assembly.13-15
The template-tethered system, we believe, is an alternative approach that allows thermal stability studies of unstable domains that are too long or too unstable to be studied in the host-guest system. In contrast to adding synthetic peptide flanks, the peptide of interest can be forced to assemble with CMPs of high helical propensity such as (ProHypGly)n by conjugating them to a template to form a tethered heterotrimer. The CD melting temperatures for α·O·O (44°C), P·O·O (62°C) indicate that the peptide α is thermally less stable than P even though both peptides do not form homotrimeric helices on their own. The peptides P (no melting) and T (Tm=16°C), despite the big difference in their melting behaviors, seem to show similar triple helical thermal stabilities when they are conjugated to a template in a homotrimeric form (P·P·P, 47°C; T·T·T, 47°C) or in an ABB type heterotrimeric form with the same B composition (P·O·O, 62°C; T·O·O, 64°C). This suggests that stand-alone peptide P may be thermally unstable but as part of a collagen protein, this sequence could be almost as stable as sequence T.
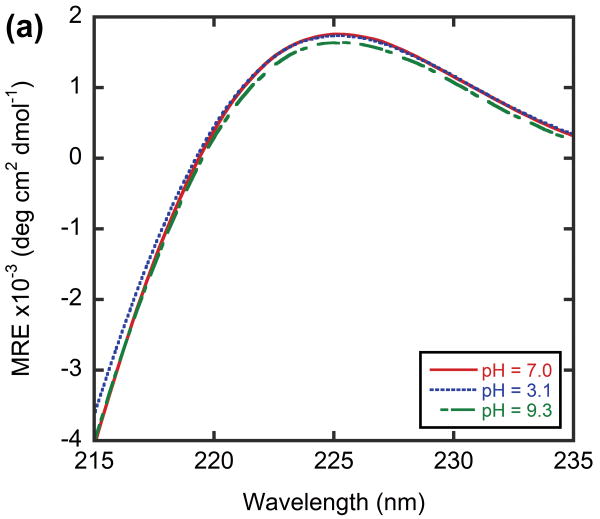
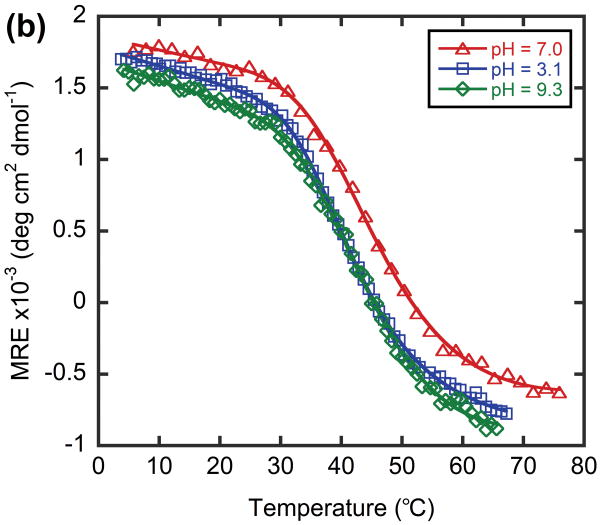
To the best of our knowledge, there are only two other reported methods for the preparation of heterotrimeric CMPs of defined composition: i) covalent cysteine crosslinking methods developed by Moroder and coworkers17 and ii) the host-guest peptide system that make use of electrostatic interactions developed by Hartgerink and coworkers.15 The first method involves cumbersome chemistry, and the registry of CMP chain alignment is known to affect the triple helical stability and folding kinetics.21 Our TREN-template-tethered heterotrimers are easy to synthesize, and their folding behaviors are not limited by the crosslinking/tethering process due to the C3 symmetry and the flexibility of the TREN template. The second method can be applied to prepare ABB and even ABC types of CMP heterotrimers as long as the sequence of interest is short enough to be included in the host-guest system. However, the ionic interactions that dictate the heterotrimer composition are easily disrupted by ionic strength and pH as evidenced by melting of such compounds under acidic condition.13 We performed the CD studies on the heterotrimer α·O·O under both acidic and basic conditions to showcase the versatility of our system in allowing heterotrimer folding studies under various pH conditions. The heterotrimer displayed a triple helical conformation with similar CD spectra and melting behaviors under both pH 3.1 and 9.3 (Figure 7). However, Tm values obtained under these conditions were 1∼4°C lower than Tm obtained under pH 7.0 (Table 3) suggesting the existence of small non-intermolecular interactions from the KGE triplet unit within the single α strand.48 These results suggest that our covalent tethered system can not only be used for studying the thermal stabilities of heterotrimers under various pH conditions, but can also be used to separate intra-chain factors from inter-chain factors that contribute to triple helix stabilization. Charge-charge interactions are essential for the stabilization of various collagen like triple helices found in bacteria.49,50 Considering the complementary nature of the electrostatic interaction and its pH dependence, the TREN-template CMP heterotrimer will be an important platform for investigation of the folding behavior of bacterial collagen-like proteins.
Figure 7.
CD spectra (a) and thermal melting curves (b) of α·O·O under acidic, basic and neutral conditions.
Conclusion
Since its introduction, collagen mimic peptide has been a successful model for elucidating the structural and biophysical characteristics of collagen.11,29,51 Even though natural collagens are either homotrimeric or heterotrimeric, most CMPs studied to date are limited to homotrimers due to challenges associated with heterotrimer synthesis.13,15,17,21 In this article, we introduced a novel method to assemble ABB type heterotrimeric CMPs by stepwise covalent conjugation of two different peptides to a TREN-template. We presented the design, synthesis and characterization of a series of tethered heterotrimers comprising CMPs of both natural and synthetic sequences. Systematic comparison of their CD melting curves revealed stabilization of the triple-helical structure by Hyp and the cooperativity of Hyp-rich chains in the trimer stabilization. In addition, the template-tethered system allowed facile study of the melting and folding behavior of heterotrimers under varying pH conditions by providing fixed CMP composition and a well-defined nucleation site for folding. Using the TREN template tethering methodology, we prepared CMP heterotrimers that model potential hybrid complexes that might form during the CMP-collagen binding process.26,27,43 Great gain in triple helical stability was observed when CMPs of natural sequence were forced to form hybridized trimers with CMPs of high triple helical propensity such as (ProHypGly)6. We believe that the two-step tethering approach described in this article provides one of the most versatile means to prepare template-tethered heterotrimeric CMPs of defined composition, both in terms of the straightforward synthetic procedure and the variety of CMP composition. We also believe that the TREN-tethered CMP system can be useful for studying the thermal stability and folding behavior of heterotrimeric collagens and collagen-like proteins, especially the ones that are composed of highly charged amino acids, such as those found in bacteria, which are likely to be sensitive to chain composition and common environmental conditions (e.g. pH).
Acknowledgments
We are grateful to Professor Yuan Chuan Lee (Department of Biology, Johns Hopkins University) and Dr. Xuesong Jiang for informative discussions. This work was supported by grant from the NSF (DMR-0645411) and NIH (R21GM-74812).
References
- 1.Weiss JB, Jayson MIV. Collagen in Health and Disease. Churchill Livingstone; 1982. [Google Scholar]
- 2.Ramshaw JAM, Shah NK, Brodsky B. J Struct Biol. 1998;122:86–91. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- 3.Miles CA, Sims TJ, Camacho NP, Bailey AJ. J Mol Biol. 2002;321:797–805. doi: 10.1016/s0022-2836(02)00703-9. [DOI] [PubMed] [Google Scholar]
- 4.Pfeiffer BJ, Franklin CL, Hsieh Fh, Bank RA, Phillips CL. Matrix Biol. 2005;24:451–458. doi: 10.1016/j.matbio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 5.McBride DJ, Choe V, Shapiro JR, Brodsky B. J Mol Biol. 1997;270:275–284. doi: 10.1006/jmbi.1997.1106. [DOI] [PubMed] [Google Scholar]
- 6.Chipman S, Sweet H, McBride DJ, Davisson M, Marks SJ, Shuldiner A, Wenstrup R, Rowe D, Shapiro J. Proc Natl Acad Sci U S A. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro JR, McBride DJ, Fedarko NS. Connect Tissue Res. 1995;31:265–268. doi: 10.3109/03008209509010820. [DOI] [PubMed] [Google Scholar]
- 8.Yuji K, Rume S, Kinji K, Toshizo I. Biopolymers. 1970;9:415–425. [Google Scholar]
- 9.Miles CA, Bailey AJ. J Mol Biol. 2004;337:917–931. doi: 10.1016/j.jmb.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Li MH, Fan P, Brodsky B, Baum J. Biochemistry. 2002;32:7377–7387. doi: 10.1021/bi00080a007. [DOI] [PubMed] [Google Scholar]
- 11.Bella J, Eaton M, Brodsky B, Berman HM. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 12.Berisio R, Granata V, Vitagliano L, Zagari A. Biopolymers. 2004;73:682–688. doi: 10.1002/bip.20017. [DOI] [PubMed] [Google Scholar]
- 13.Gauba V, Hartgerink JD. J Am Chem Soc. 2007;129:2683–2690. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- 14.Gauba V, Hartgerink JD. J Am Chem Soc. 2007;129:15034–15041. doi: 10.1021/ja075854z. [DOI] [PubMed] [Google Scholar]
- 15.Gauba V, Hartgerink JD. J Am Chem Soc. 2008;130:7509–7515. doi: 10.1021/ja801670v. [DOI] [PubMed] [Google Scholar]
- 16.Fields CG, Mickelson DJ, Drake SL, McCarthy JB, Fields GB. J Biol Chem. 1993;268:14153–14160. [PubMed] [Google Scholar]
- 17.Ottl J, Moroder L. J Am Chem Soc. 1999;121:653–661. [Google Scholar]
- 18.Kemp DS, Petrakis KS. J Org Chem. 1981;46:5140–5143. [Google Scholar]
- 19.Erik, T. R.; Dirk, T. S. R.; Hans, W. H.; Philip, G. d. G.; Rob, M. J. L., 2002, p 4613-4621.
- 20.Feng Y, Melacini G, Taulane JP, Goodman M. J Am Chem Soc. 1996;118:10351–10358. [Google Scholar]
- 21.Sacc B, Renner C, Moroder L. J Mol Biol. 2002;324:309–318. doi: 10.1016/s0022-2836(02)01065-3. [DOI] [PubMed] [Google Scholar]
- 22.Kwak J, Capua AD, Locardi E, Goodman M. J Am Chem Soc. 2002;124:14085–14091. doi: 10.1021/ja0209621. [DOI] [PubMed] [Google Scholar]
- 23.Kotch FW, Raines RT. Proc Natl Acad Sci U S A. 2006;103:3028–3033. doi: 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koide T, Homma DL, Asada S, Kitagawa K. Bioorg Med Chem Lett. 2005;15:5230–5233. doi: 10.1016/j.bmcl.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Fields GB, Lauer JL, Dori Y, Forns P, Yu YC, Tirrell M. Biopolymers. 1998;47:143–151. doi: 10.1002/(SICI)1097-0282(1998)47:2<143::AID-BIP3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Wang AY, Mo X, Chen CS, Yu SM. J Am Chem Soc. 2005;127:4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 27.Wang AY, Foss CA, Leong S, Mo X, Pomper MG, Yu SM. Biomacromolecules. 2008;9:1755–1763. doi: 10.1021/bm701378k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horng JC, Hawk AJ, Zhao Q, Benedict ES, Burke SD, Raines RT. Org Lett. 2006;8:4735–4738. doi: 10.1021/ol061771w. [DOI] [PubMed] [Google Scholar]
- 29.Brodsky B, Persikov AV. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 30.Russell LE, Fallas JA, Hartgerink JD. J Am Chem Soc. 2010;132:3242–3243. doi: 10.1021/ja909720g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakakibara S, Inouye K, Shudo K, Kishida Y, Kobayashi Y, Prockop D. Biochim Biophys Acta. 1973;303:198–202. doi: 10.1016/0005-2795(73)90164-5. [DOI] [PubMed] [Google Scholar]
- 32.Story SC, Aldrich JV. Int J Pept Protein Res. 1994;43:292–296. doi: 10.1111/j.1399-3011.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 33.Albericio F, Bofill JM, El-Faham A, Kates SA. J Org Chem. 1998;63:9678–9683. [Google Scholar]
- 34.Burjanadze TV. Biopolymers. 1979;18:931–938. doi: 10.1002/bip.1979.360180413. [DOI] [PubMed] [Google Scholar]
- 35.Jenkins CL, Raines RT. Nat Prod Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 36.Slatter DA, Miles CA, Bailey AJ. Journal of Molecular Biology. 2003;329:175–183. doi: 10.1016/s0022-2836(03)00380-2. [DOI] [PubMed] [Google Scholar]
- 37.Ackerman MS, Bhate M, Shenoy N, Beck K, Ramshaw JAM, Brodsky B. J Biol Chem. 1999;274:7668–7673. doi: 10.1074/jbc.274.12.7668. [DOI] [PubMed] [Google Scholar]
- 38.Koide T, Yuguchi M, Kawakita M, Konno H. J Am Chem Soc. 2002;124:9388–9389. doi: 10.1021/ja026182+. [DOI] [PubMed] [Google Scholar]
- 39.Snellman A, Tu H, Vaisanen T, Kvist AP, Huhtala P, Pihlajaniemi T. EMBO J. 2000;19:5051–5059. doi: 10.1093/emboj/19.19.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank S, Boudko S, Mizuno K, Schulthess T, Engel J, Bächinger HP. J Biol Chem. 2003;278:7747–7750. doi: 10.1074/jbc.C200698200. [DOI] [PubMed] [Google Scholar]
- 41.Heidemann ER, Harrap BS, Schiele HD. Biochemistry. 1973;12:2958–2963. doi: 10.1021/bi00740a002. [DOI] [PubMed] [Google Scholar]
- 42.Wang AY, Leong S, Liang YC, Huang RCC, Chen CS, Yu SM. Biomacromolecules. 2008;9:2929–2936. doi: 10.1021/bm800727z. [DOI] [PubMed] [Google Scholar]
- 43.Xiao M, Yoojin A, Chang-Soo Y, Seungju MY. Angew Chem. 2006;45:2267–2270. doi: 10.1002/anie.200504529. [DOI] [PubMed] [Google Scholar]
- 44.Miles CA, Bailey AJ. Micron. 2001;32:325–332. doi: 10.1016/s0968-4328(00)00034-2. [DOI] [PubMed] [Google Scholar]
- 45.Beck K, Chan VC, Shenoy N, Kirkpatrick A, Ramshaw JAM, Brodsky B. Proc Natl Acad Sci U S A. 2000;97:4273–4278. doi: 10.1073/pnas.070050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Biochemistry. 2000;39:14960–14967. doi: 10.1021/bi001560d. [DOI] [PubMed] [Google Scholar]
- 47.Persikov AV, Ramshaw JAM, Brodsky B. J Biol Chem. 2005;280:19343–19349. doi: 10.1074/jbc.M501657200. [DOI] [PubMed] [Google Scholar]
- 48.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Biochemistry. 2005;44:1414–1422. doi: 10.1021/bi048216r. [DOI] [PubMed] [Google Scholar]
- 49.Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. J Biol Chem. 2007;282:29757–29765. doi: 10.1074/jbc.M703991200. [DOI] [PubMed] [Google Scholar]
- 50.Xu C, Yu Z, Inouye M, Brodsky B, Mirochnitchenko O. Biomacromolecules. 2010;11:348–356. doi: 10.1021/bm900894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]