Abstract
Peptide detectability is defined as the probability that a peptide is identified in an LC-MS/MS experiment and has been useful in providing solutions to protein inference and label-free quantification. Previously, predictors for peptide detectability trained on standard or complex samples were proposed. Although the models trained on complex samples may benefit from the large training data sets, it is unclear to what extent they are affected by the unequal abundances of identified proteins. To address this challenge and improve detectability prediction, we present a new algorithm for the iterative learning of peptide detectability from complex mixtures. We provide evidence that the new method approximates detectability with useful accuracy and, based on its design, can be used to interpret the outcome of other learning strategies. We studied the properties of peptides from the bacterium Deinococcus radiodurans and found that at standard quantities, its tryptic peptides can be roughly classified as either detectable or undetectable, with a relatively small fraction having medium detectability. We extend the concept of detectability from peptides to proteins and apply the model to predict the behavior of a replicate LC-MS/MS experiment from a single analysis. Finally, our study summarizes a theoretical framework for peptide/protein identification and label-free quantification.
Introduction
For a specific shotgun proteomics platform, collectively including the steps from the sample preparation protocol to peptide fragmentation and database search, peptide detectability is defined as the probability of observing (or identifying) a peptide from a standard sample mixture.1 Peptide detectability can be approximately computed using machine learning models given the peptide sequence and its parent protein, where the training is typically performed using a set of model proteins mixed at equal abundances. The predicted peptide detectability has been used to address several problems in shotgun proteomics, including protein inference2–4 and label-free quantification.1,5
In practice, however, several issues emerge if one relies on standard protein mixtures for detectability prediction. First, peptide detectabilities trained for standard conditions, referred to as standard detectabilities, cannot be directly used to predict peptides that are likely to be observed in non-standard samples under diverse cellular conditions because the peptides in non-standard samples typically have unequal abundances. In other words, detectability of a peptide in a complex sample, referred to as the effective detectability,3,4 is not an intrinsic property of the peptide, since it depends on both the physicochemical properties of the peptide/protein and its abundance. Second, it may be impractical to provide a standard sample mixture for every proteomics platform in order to predict peptide detectabilities for this platform. Finally, standard protein mixtures are relatively simple, consisting of 10–50 proteins, and are often non-representative of complex mixtures. As a result, the detectability predictor trained using these data may not be able to achieve high accuracy when applied to complex biological samples.
In addition to the use of standard samples, several groups have proposed to learn peptide detectabilities using identified peptides from single or pooled biological samples.5–11 Although these predictors benefit from the large data sets employed in the training step, they may be affected by the unequal abundances of identified peptides as protein and peptide quantities can vary by orders of magnitude in biological samples.12 For example, peptides with high standard detectability and low abundance as well as those peptides with relatively low standard detectability that are detected owing to their high abundance may confuse the model during training. Such predictors may appear to be designed to learn effective detectability, but because effective detectability is not an intrinsic property of a peptide, and proteins with a small number of identified peptides may be removed during training, it is not immediately obvious whether their output is closer to either the standard or the effective detectability.
To address these concerns, the goal of this work is to deepen our understanding of both standard and effective detectability as well as the properties of computational models used to learn them. In addition, we seek to expand the use of detectability in biological experiments. While standard detectability has been a useful concept in protein inference and quantification, the notion of effective detectability is inherently related to two important concepts in proteomics: peptide repeatability and peptide reproducibility.13 Peptide repeatability is related to the technical replicates of the sample, when the same sample is analyzed by the same operator using the same instrument in consecutive runs. On the other hand, the reproducibility analysis implies that the operator and/or the instrument, but not the analytical platform or the protocols, are varied. Because effective detectability is tied to both sample and proteomics experiment, including sample preparation, MS platform, operator, and peptide identification software, it provides the asymptotic limit of peptide repeatability as the number of technical replicates approaches infinity. Thus, accurate prediction of the effective detectability from complex mixtures can directly estimate repeatability of the peptide and provide an upper limit on peptide reproducibility using a single run. As such it may be incorporated into experimental design in proteomics in order to improve confidence for those proteins identified from a small number of peptide hits.
In practical terms, we present a novel iterative algorithm to simultaneously learn standard and effective peptide detectabilities from complex samples. We provide evidence that this algorithm is accurate, but more importantly, it improves protein inference algorithms and enables estimation of the number of protein identifications in replicated experiments. It also facilitates estimation of the number of proteins in an organism that may not be identifiable under realistic assumptions of their relative quantities. Finally, we analyze the practical limits on the prediction of peptide detectability.
Materials and Methods
Iterative learning of peptide detectabilities
We consider a set of proteins reliably detected in a tandem mass spectrometry experiment. For training purposes, a set of (tryptic) peptides from these proteins is partitioned into the positive (identified) and negative (not identified) sets. For simplicity, miscleaved, truncated, degenerate or post-translationally modified peptides are not considered. We then train a computational model to approximate both the standard and effective detectability of any given peptide as well as protein relative quantities (abundances).
The training method consists of two components: (i) a modular neural network that incorporates estimated protein quantities as well as peptide sequence properties as input to predict the standard and effective peptide detectabilities simultaneously (Figure 1); and (ii) a procedure to estimate protein abundances based on the predicted effective peptide detectabilities and the confidence of the identified peptides. The model training starts by assuming equal protein quantities (i.e. qi = 1 for each protein i) and then iterating the above-mentioned steps until the model converges to a solution. To train a network, we utilize protein quantities estimated in the previous iteration, whereas to estimate protein quantities we utilize the neural networks trained in the previous step to approximate the effective peptide detectabilities. The pseudo code is shown in Algorithm 1.
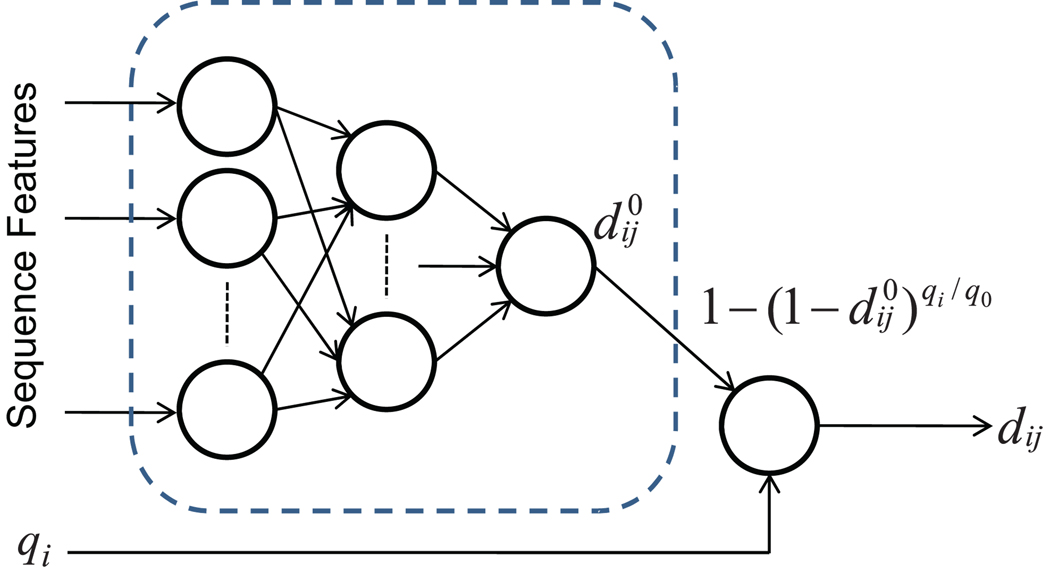
Figure 1.
Neural network architecture used for simultaneously predicting standard and effective peptide detectabilities. Indices i and j represent proteins and peptides, respectively. Peptide/protein sequence features and protein quantities qi are presented to the model. The deterministic transformation in the rightmost node ensures that the subnetwork in the dashed rectangle estimates the standard detectability , whereas the final output estimates the effective detectability dij.
Neural network architecture
We use two groups of features as input to the network (Figure 1). The first set of 292 features were computed solely from peptide sequences and the neighboring residues in the proteins from which they were digested. Similar to the model we described previously,1 these features include amino acid compositions, peptide length, peptide mass, N- and C-terminal residues, sequence complexity, and several derived or predicted peptide properties such as aromatic content, hydrophobicity, flexibility, hydrophobic moment and intrinsic disorder. The second group of features includes only the estimated quantity of the protein of origin for the peptide.
For a specific peptide j from protein i, the prediction process is carried out as follows: (i) all sequence-based features are derived from its primary structure (including sequence of its parent protein) and are input to the neural network together with the quantity measure of protein i; (ii) its standard detectability is approximated by using the subnetwork involving only sequence-based features (as shown in the dashed rectangle in Figure 1); (iii) the effective detectability dij is calculated in the rightmost node in Figure 1 from the standard detectability and estimated quantity qi. The last step is referred to as the quantity adjustment.
It is worth emphasizing that the standard detectability is predicted by the subnetwork involving only peptide sequence features (Figure 1). However, during the training process, only effective detectabilities can be compared with the known identification results and can be used to optimize the model when the proteins in the sample are not at the same abundance. Thus, both detectabilities are learned simultaneously.
Algorithm 1.
The proposed algorithm for learning standard and effective peptide detectabilities. Indices i and j represent proteins and peptides, respectively; pij = peptide j from protein i; 𝒫i = the set of tryptic peptides from protein i; ANN = the artificial neural network as depicted in Figure 1; = standard detectability estimate for pij; dij = effective detectability estimate for pij; qi = relative quantity estimate for protein i.
|
Estimation of peptide quantities
We propose here a simple approach to estimate protein quantity based on peptide identification information only. The approach requires minimum information from an LC-MS/MS experiment, i.e. neither spectral counts nor ion intensity information is needed. Hence, it can be applied to any type of input. Assuming that the quantity of protein i in the sample is qi, the effective detectability dij of peptide j from this protein can be modeled as
| (1) |
where q0 is the peptide quantity at the standard condition and is the standard detectability of peptide j. Clearly, .
Because we ignore the influence of degenerate, miscleaved, truncated, and post-translationally modified peptides, all identified peptides from protein i can be considered to have the same abundance. Then, the prior expectation for the number of identified peptides from protein i at quantity qi can be expressed as
| (2) |
where 𝒫i is the set of peptides in protein i (in a somewhat abused notation where 𝒫i represents both peptides and their indices). On the other hand, we can compute the posterior expectation of the number of identified peptides directly from the peptide identification results using peptide detectability dij as a prior probability.3,4 Therefore, we also have
| (3) |
where LRij is the identification likelihood ratio of peptide j, defined as the ratio of the likelihood of a peptide being identified and the likelihood of it not being identified. We note that LRij can be computed from the peptide identification scores (as reported by search engines) based on probabilistic models such as PeptideProphet.14 When a peptide is identified multiple times, the maximum value can be taken as its identification score (likelihood). A derivation of Eq. 3 is provided in Supplementary Materials. In the extreme case, if we consider all identified peptides as true positives, we can assign LRij = ∞ for identified peptides, and LRij = 0 for non-identified peptides.3,4 In this case, corresponds to a count of confidently identified peptides. If we can approximate standard peptide detectabilities of peptides in 𝒫i using a neural network, then we can estimate the abundance of protein i by combining the prior and posterior expectations from Eqs. 2 and 3 as
| (4) |
The new quantity qi in Eq. 4 can be obtained by a simple bisection search algorithm until the difference between the left- and right-hand side of the equation falls below a small prespecified threshold. This approach is similar to the method of moments commonly used in statistical parameter estimation.15 An alternative strategy is the maximun likelihood (ML) parameter estimation. We describe the ML approach in the Supplementary Materials and show that it provides similar solutions as Eq. 4.
Quantities qi are normalized by q0, which itself is selected such that the average standard detectability in the set of identified proteins equals 0.5. More specifically, the relative quantity of the standard sample (q0) is computed by solving
| (5) |
where 𝒫 = ∪i𝒫i is the set of all peptides and |𝒫| is the cardinality of 𝒫. Then, the new value of qi is computed as
| (6) |
We emphasize that the normalization step is critical for the convergence of the iterative learning algorithm.
Protein detectability
Here we extend the concept of peptide detectability to protein detectability. Since peptide detectability is defined as
| (7) |
we similarly define detectability of protein i as
| (8) |
where xi = 1 indicates that protein i exists in the sample, qi is the quantity of the protein, and yij = 1 indicates that peptide j from protein i is correctly identified based on the MS/MS spectra (formally, xi and yij are indicator variables). Thus, ∑j yij ≥ c means that at least c peptides from protein i are correctly identified. In this work, we consider c = 1, and use di(q) to represent di,1(q). Generally, correct peptide identifications cannot be guaranteed, thus c > 1 or protein dependent values may be necessary.
Assuming conditional independence of peptide identifications given that the protein is present in the sample, we can compute protein detectability di based on peptide detectabilities dij. This assumption is reasonable because peptides from the same protein typically do not elute at the same time and hence do not compete for ionization and fragmentation. Given the conditional independence condition, we have
| (9) |
We can further define protein standard detectability , and obtain . This detectability has the same form as peptide detectability shown in Eq. 1.
Neural network training
The neural network was optimized towards the maximum log-likelihood (cross-entropy) measure
| (10) |
where 𝒫 is the set of all peptides, dij is the approximated effective detectability of peptide j from protein i, and tj is the target variable (tj = 1 for identified peptides and tj = 0 for non-identified peptides). Prior to the network training, we applied a t-test feature selection, as described previously.1 Each model was trained with 20 random initializations of neural network weights to alleviate the problem of the local minimum convergence and up to 10 iterations were allowed for protein quantity estimation (usually less than 5 iterations were sufficient for convergence). Finally, to avoid overfitting and obtain better generalization performance we used the Bayesian regularization algorithm to train the network.16
Model comparison
The new Iterative Learning (IL) algorithm was trained using proteins with 2 or more confidently identified peptides. We refer to the model predicting effective detectabilities as IL2, whereas its variant predicting standard detectabilities (rightmost node in the dashed box in Figure 1) is referred to as IL20. For comparison purposes, we also implemented several simple methods for peptide detectability prediction. The first method (M2) used all proteins with 2 or more identified peptides for training without the protein quantity adjustment. This was equivalent to assuming that all proteins were mixed at equal abundances in the sample. The second and third methods, similar to the one implemented in APEX,5,10 excluded low abundance proteins from the training set by selecting proteins with a large number identified peptides (i.e. ≥5 in model M5 and ≥10 in model M10). These methods were aimed at exploiting smaller but more homogeneous training sets with respect to the quantities of identified proteins. Technically, the learning algorithm for models M2, M5, and M10 uses the sub-neural network inside the dashed box in Figure 1, whereas IL2 utilizes the full neural network. The final method used for the comparisons was one we constructed previously.1 This method, referred to as the Standard Sample Predictor (SSP), was trained on a standard sample of 12 model proteins mixed at similar abundances. Although the training set of the SSP model was guaranteed to be homogeneous with respect to quantity, the performance of the method may suffer due to the small size of the training data.
Data sets
The data set used in this study includes 20 replicate LC-MS/MS analyses of a biological sample from the bacterium Deinococcus radiodurans. D. radiodurans proteins were extracted by four passes through a French press at 16000psi, cleared by centrifugation at 13000g for 45 minutes. Duplicate protein extracts were trypsin digested overnight in the presence of 0.05% RapiGest SF (Waters, Milford, MA) acid-labile surfactant and 25mM ammonium bicarbonate after reduction and alkylation with dithiothreitol and iodoacetamide. Trypsin was deactivated and the acid-labile surfactant was cleaved with the addition of 5µL of 90% formic acid followed by incubation at 37°C for 2 hours and centrifugation at 13000g for 10 minutes. The peptide samples were cleaned by solid phase extraction using a Waters OASIS HLB cartridge and the manufacturer’s protocol. After removing the solvent by speed-vac at 45°C for 2 hours, the digest was suspended in 200µL of solvent.
The 20 replicates comprised 2 × 10 experiments, i.e. the original sample was split in two separate vials and digested with trypsin. Peptides corresponding to at most 1.3µg of protein were loaded and separated using a 120-minute gradient from 3% to 40% acetonitrile at 250nL/min using a nano 2DLC (Eksigent Technologies, Dublin, CA) on a 15cm, 75µm capillary column packed in-house with 5µm C-18AQ particles (Michrom Bioresources, Auburn, CA). The eluting peptides were electrosprayed into the source of a ThermoFinnigan (San Jose, CA) LCQ Deca XP ion-trap mass spectrometer. A dynamic exlusion protocol was used to limit the acquisition of each precursor mass to twice over a 45-second window.
The peptides were identified using MASCOT,17 allowing for variable oxidation of methionine and fixed carbamidomethylation at cysteine, with the false discovery rate (FDR) of 0.01 using the decoy D. radiodurans database. In total, the Deinococcus data included 1301 identified peptides (126 of charge +1, 1022 of charge +2, and 277 of charge +3) at 1% peptide-spectrum match (PSM) level FDR. At a 1% peptide (with charge) level FDR, the data consisted of 1013 identified peptides (48 of charge +1, 857 of charge +2, and 210 of charge +3). For the training set used in the machine learning step, only proteins with 2 or more unique and confidently identified (at 1% PSM FDR) peptides were included. For these proteins, all peptides were assumed to be unique to them. Note that the 20 replicates of the the Deinococcus sample are not technical replicates in the strict sense, because the sample was split prior to digestion and analyzed using different LC columns. The data set is available upon request.
Results
Comparison of prediction models
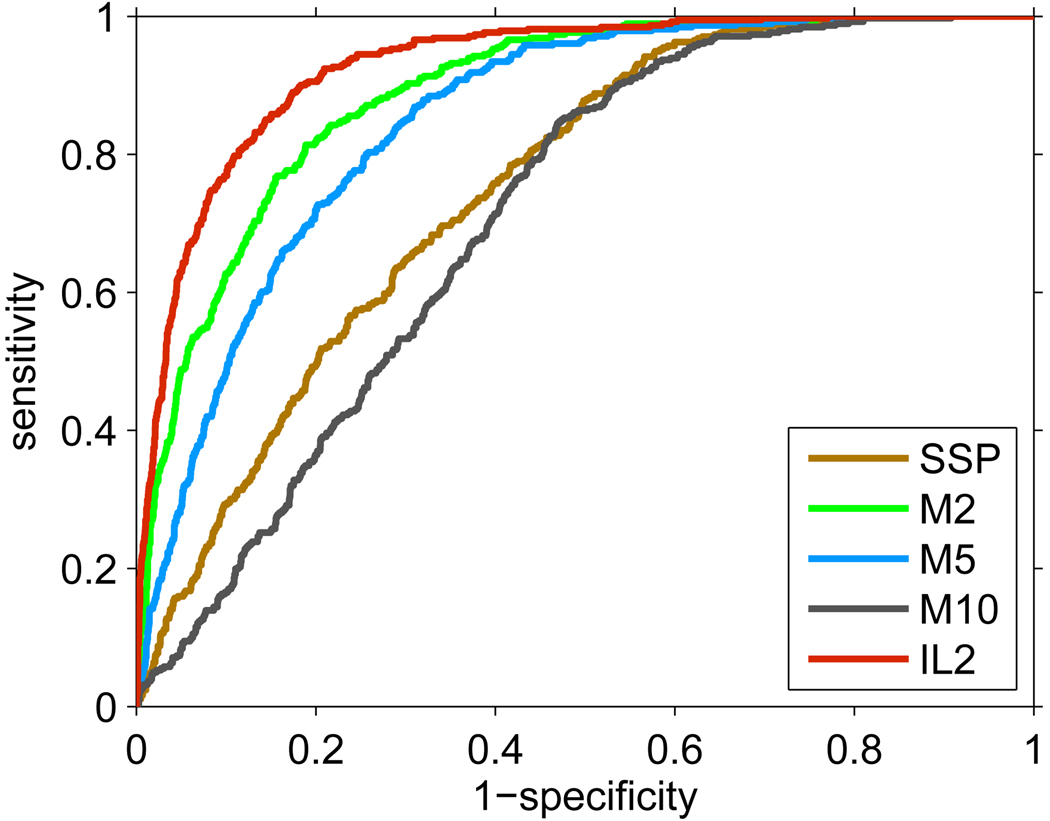
We assessed the performance of several computational models in the task of predicting proteotypic peptides7,18,19 in a complex mixture. In this experiment, the first of the twenty Deinococcus runs was used for training and quantity estimation, while the remaining 19 runs were used for defining which peptides were proteotypic (peptides unique to their parent protein, identified in ≥50% of the experiments in which their parent protein is identified). Four different detectability predictors were compared: (i) SSP model for the detectability prediction of peptides of charge +2 trained on a standard sample;1 (ii) predictor M2 trained on the proteins with ≥2 peptide hits in a complex sample; (iii) predictors M5 and M10 trained on the highly abundant proteins (i.e. proteins with ≥5 and ≥10 peptide hits, respectively) from a complex sample; and (iv) the new predictor (IL2) trained using the iterative learning procedure with quantity adjustment using proteins with ≥2 peptide hits from a complex sample. Only peptides whose m/z was within the instrument range were used for training and accuracy estimation. Note that models IL2 and M2 were trained on exactly the same data, whereas models M5 and M10 exploited only a subset of proteins. Approximately 40% of the proteins had ≥5 identified peptides, while only about 10% had ≥10 identified peptides.
The Receiver Operating Characteristic (ROC) curves for the five predictors on the Deinococcus data set are shown in Figure 2. ROC curves are commonly used to visualize the true positive rate (or sensitivity) against the false positive rate (or 1 − specificity) for a binary classifier.20 The series of true/false positive rates are obtained by varying the decision threshold on the raw prediction outputs. The performance accuracies, measured by the Area Under the ROC Curve (AUC), of the predictors were ranked as IL2 > M2 > M5 > SSP > M10 (Table 1). The SSP model was inferior to most algorithms since it was trained on a small sample that also had a different sample preparation procedure and instrument (Thermo Electron LTQ linear ion-trap mass spectrometer) from the samples used in this work. Surprisingly, M2 outperformed M5 and M10 models, which indicates that it may not be necessary to restrict the training set to highly abundant proteins as in previous work.5,10 The iterative training method (IL2) further improved the proteotypic peptide prediction over M2 (IL2 and M2 models used exactly the same training set, which indicates the usefulness of the quantity adjustment).
Figure 2.
ROC curves of predictions of proteotypic peptides for the five detectability predictors. The detectability and the protein quantities were estimated on the first replicate. The proteotypic peptides were defined by the remaining 19 Deinococcus analyses. The corresponding AUC values for each curve are shown in Table 1.
Table 1.
Performance comparisons among different detectability predictors in predicting proteotypic peptides. Models M2, M5, M10 and IL2 were trained on the Deinococcus data generated in this study, while the standard sample predictor (SSP) was developed in previous work.1 For all models, the first replicate was used for training and the remaining 19 experiments to define proteotypic peptides. The performance was evaluated based on the Area Under the ROC curve (AUC), cross-entropy (ce), and mean square error (mse). The difference between models M2 and IL2 is significant with the p-value P = 9.8 · 10−13, based on the likelihood ratio test with 111 degrees of freedom (111 being the number of extra parameters used in IL2 but not M2 i.e., the number of proteins included in training). The p-values for the pairwise comparisons of the AUC values are shown in Table S1, Supplementary Materials.
| Performance measures |
Models | ||||
|---|---|---|---|---|---|
| SSP | M10 | M5 | M2 | IL2 | |
| AUC | 0.750 | 0.708 | 0.849 | 0.889 | 0.928 |
| ce | 1980.2 | 1227.8 | 1012.1 | 779.1 | 654.1 |
| mse | 0.240 | 0.124 | 0.102 | 0.078 | 0.065 |
In addition to providing an accurate effective detectability predictor, our goal was to understand the nature of the predictions of the remaining models trained on complex samples. To achieve this, we compared the outputs of models M2, M5 and M10 to the standard detectability predictor IL20 and effective detectability predictor IL2. Table 2 shows the Pearson correlation coefficients between the outputs of these models on the first replicate of the Deinococcus data.
Table 2.
Relationships among different models measured by the Pearson correlation coefficient. Models M2, M5, M10 and IL2 were trained on the first replicate of the Deinococcus data; the standard sample predictor (SSP) was developed in previous work on a somewhat different platform.1 IL20 represents the standard detectability output from the IL2 model (Figure 1). All models except IL2 were normalized to have the mean value of 0.5 using Eq. 5. Rep stands for the Repeatability model where detactabilities were approximated by the fraction of times a peptide (from an identified protein in the first replicate) was identified in the remaining 19 analyses. For the smallest correlation coefficient 0.27, the p-value for it not being zero is 4.0 · 10−54 (t = 15.80, two-tail t-test with degree of freedom 3108, where 3110 is the sample size or number of peptides). The p-values for correlation between M2 and IL2 (0.69) being smaller than the correlation between M2 and IL20 (0.9) is << 1.0 · 10−10 (z = 34.16, one-tailed Steiger’s Z-test for correlated correlations, computed using FZT calculator (http://psych.unl.edu/psycrs/statpage/comp.html)). The correlation between M10 and SSP (0.86) vs. correlation between M5 and SSP (0.68) are different with p-value << 1.0 ·10−10 (Steiger’s z = 24.40).
| Models | SSP | M2 | M5 | M10 | IL20 | IL2 | Rep |
|---|---|---|---|---|---|---|---|
| SSP | 1.00 | 0.67 | 0.68 | 0.86 | 0.63 | 0.45 | 0.28 |
| M2 | 1.00 | 0.88 | 0.66 | 0.91 | 0.69 | 0.43 | |
| M5 | 1.00 | 0.72 | 0.83 | 0.63 | 0.40 | ||
| M10 | 1.00 | 0.60 | 0.44 | 0.27 | |||
| IL20 | 1.00 | 0.68 | 0.41 | ||||
| IL2 | 1.00 | 0.65 | |||||
| Rep | 1.00 |
The high correlation between the outputs of M2 and IL20 together with the modest correlation between M2 and IL2, suggest that M2 model learns standard peptide detectability. This result is not completely unexpected given that effective detectability cannot be predicted from sequence alone. Somewhat surprisingly, however, we find that the standard sample predictor (SSP) was highly correlated with M10 model, despite somewhat different analytical platforms. We believe this may be caused by the effects of competition which are more pronounced in the learning of IL2 and M2 models. These models may be capturing the relationship between sequence and retention times at which the competition for ionization is the strongest for a particular group of peptides. In such a way, the detectability of peptides in these high-competition regions may be reduced. Thus, we believe that both SSP and IL20 models learn peptide detectability, but that they may be capturing different properties of peptides and analytical platforms. In such a case, SSP models would be more appropriate for the application in simple protein mixtures, whereas the IL20 model would more effectively capture properties of a system with significant competition.
Effective detectability as a probability estimate
While AUC is a useful measure of classification performance, it only tests for a correct ranking of predictions. This is unsatisfactory here since detectability must also represent the prior probability of peptide identification.3,4 Such applications require not only the correct ranking but also that the predicted detectability be an accurate estimate of the probability that the peptide will be identified.
To assess this property, we tested whether the predicted effective detectability correlates with the repeated identification of the same peptide in multiple LC-MS/MS runs. In this experiment, we used the first of the Deinococcus replicates (PQ21) to train the detectability predictors, whereas the remaining 19 analyses were used to calculate the relative frequency of peptide identifications for all tryptic peptides (both identified and non-identified) from the identified proteins in the first replicate. Figure 3 shows that the quantity-adjusted detectability model (IL2) provides significantly better approximation (mean squared error mse = 0.047; correlation coefficient ρ = 0.65) of the technical replicability of a peptide than the standard detectability models (IL20: mse = 0.29, ρ = 0.41; M2: mse = 0.059, ρ = 0.43; SSP: mse = 0.22, ρ = 0.28). Note that the ideal detectability predictions should appear on the dashed diagonal line in this diagram.
Figure 3.
Experimental peptide repeatability as a function of peptide detectability. For each model, peptides are binned by predicted detectability (bin size = 0.1) and in each bin the repeatability is averaged. Predictors were trained on the first LC-MS/MS replicate (PQ21), and the repeatability is estimated based on the remaining 19 analyses of the Deinococcus sample. The mean square errors of each fit are shown in Table 1.
Better detectability prediction improves protein inference
An important application of peptide detectability is in protein identification. Because effective detectability is the conditional probability that a peptide is identified in an LC-MS/MS experiment given that the protein(s) containing the peptides exist at some known quantities, it can be naturally incorporated into a Bayesian network model reflecting the peptide/protein identification process.3,4
A straightforward question is the extent to which the improved accuracy of detectability prediction influences protein inference. To address this question, we compared three standard detectability models (M2, M5, and IL20) coupled with the MSBayesPro algorithm3,4 on the first replicate of the sample. In order to carry out probabilistic inference, we converted the MASCOT scores of peptides to probabilities using PeptideProphet.14 Peptides identified multiple times (at same or different charge states), were represented by the maximum probability values. For the training of detectability models (M2, M5, IL20), only proteins with 2 or more identified peptides (at 1% FDR) were used, but all proteins with one or more peptides identified with probability >0.05 were included in protein inference.
We used the maximum a posteriori (MAP) decoding solution and the marginal posterior probabilities3,4 for protein identification. By both criteria we observed that the new algorithm led to an improved number of protein identifications compared to M2 and M5 models (Figure 4). Protein level FDR curves showed similar results (Figure 4(b)). Together, these results suggest that the improvements in detectability prediction by including low quantity proteins and the use of quantity adjustment during training are beneficial to protein inference models.
Figure 4.
The effect of standard detectability predictions on protein identifications using two iterations of the MSBayesPro protein inference algorithm.3,4 Two criteria were used: (a) the number of identified proteins through the MAP decoding of protein configuration, (b) FDR curves using protein marginal posterior probability P as a score for each protein. Decoy Deinococcus database was used to estimate FDR. Note that no decoy proteins were identified by the MAP decoding.
Standard vs. effective detectability
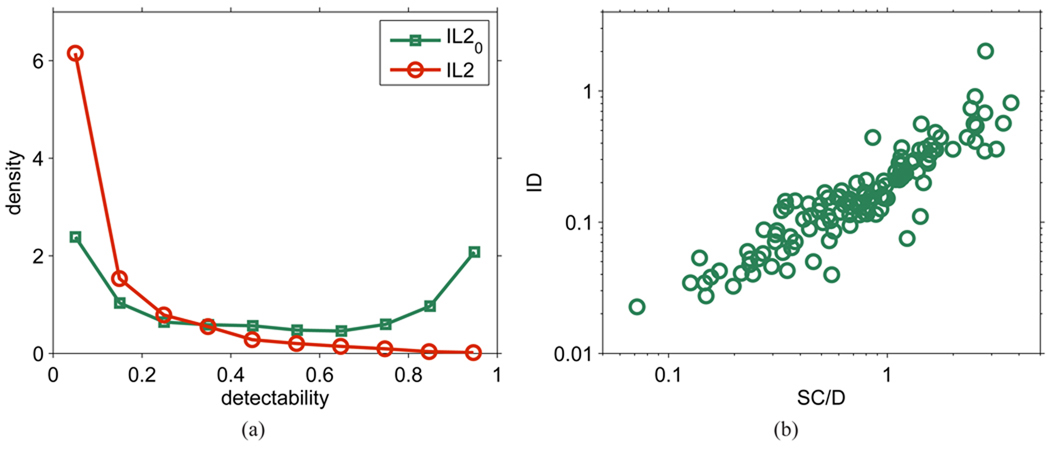
Figure 5(a) shows the distribution of predicted standard and effective detectabilities for all peptides in a protein database using the IL2 and IL20 models. We observe a U-shaped distribution for standard detectability, indicating that a majority of peptides have either high detectability (close to 1) or low detectability (close to 0), whereas a relatively small fraction of peptides have medium detectability. In contrast, the distribution of effective detectability shows a high peak close to 0 and a steep decrease in density as the detectability increases. The distribution of effective detectability can be explained by the distribution of standard detectability combined with the distribution of protein quantities in biological samples.21
Figure 5.
(a) Distribution of values of predicted detectability based on the first LC-MS/MS analysis of the Deinococcus sample. (b) Quantity estimation by peptide identification (ID), according to Eqs. 5–6, compared to the spectral counting SC/D method (in-house reimplementation of the APEX method5,10).
As indicated in the peptide quantity-detectability model (Eq. 1), we propose that quantity and standard detectability together determine the detection probability of a peptide. To model this effective detectability, or peptide repeatability, we employed a simple approach for estimating protein quantity (hence peptide quantity, because we ignored degenerate peptides or post-translational modifications). Estimated protein quantities from Eq. 3, however, correlate well (ρ = 0.77) with those of normalized spectral counts used in the APEX approach,5,10 with a linear trend across the entire range of values (Figure 5(b)). This result suggests that our simple quantity estimation, which corresponds to a detectability-weighted peptide count, is reasonably accurate.
Note that the five most abundant proteins identified in the Deinococcus sample were 50S ribosomal protein L28 (NP_296244.1), FraH-related protein (NP_285656), 30S ribosomal protein S13 (NP_295848), co-chaperonin GroES (NP_294329.2), and 30S ribosomal protein S16 (NP_295018).
Empirical limits of detectability prediction
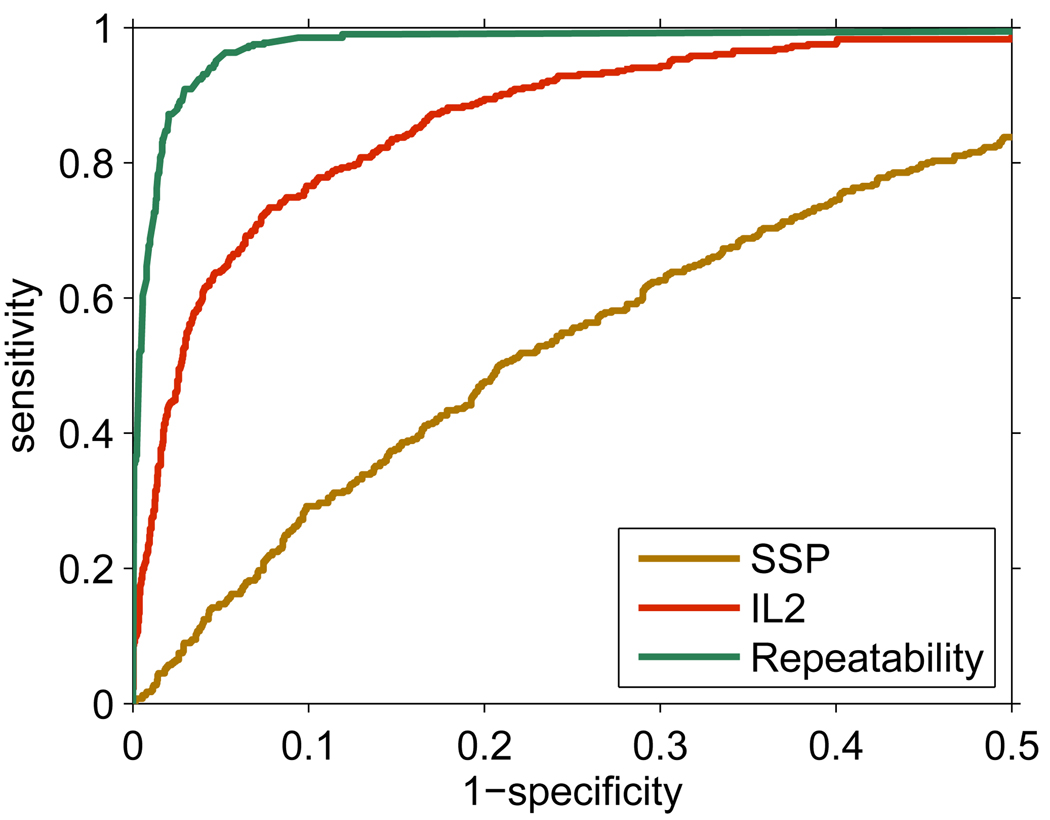
The model of peptide detectability is limited by several factors, including the particular peptide-spectrum matching algorithm, data representation, selection of the machine learning model, and the assumption that all peptides in the sample are independent. Here, we evaluated the empirical upper limits of the performance of the peptide detectability predictors. A detectability predictor (IL2) trained on the first Deinococcus analysis (PQ21) was compared to another predictor (’Repeatability’) obtained by averaging the identifications over the remaining 19 replicates in the task of predicting identified peptides (Figure 6). Observe that the Repeatability model is not a predictor in the traditional sense, it uses the empirical repeatability values (the fraction of times a peptide was identified over the 19 experiments) directly as predictive score, and serves as an indicator of an uppper limit for the effective detectability prediction. Both predictors were also compared with the SSP model.
Figure 6.
The upper limit of detectability prediction, demonstrated by the ROC curves of the models predicting effective detectability on the first analysis of the Deinococcus sample (false positive range 0 to 0.5). Three models are compared: the standard detectability predictor (SSP), the effective detectability predictor (IL2), and the model computing the repeatability of peptide identifications by the proportion of the remaining 19 replicate experiments in which a peptide was identified. The corresponding AUC values are: 0.984 (Repeatability), 0.925 (IL2) and 0.740 (SSP).
The AUCs of the three models suggest that the iterative learning with quantity adjustment significantly improved prediction of peptide detectability over standard detectability. However, this model was significantly outperformed by the empirical repeatability of peptide identifications. This suggests that some of the assumptions used in our model may be violated. For example, we have not explicitly modeled competition among co-eluting peptides for ionization and fragmentation in a complex proteome (eliminating this limitation was beyond the scope of this study). In addition, a design of more sophisticated machine learning models could further improve the prediction.
Estimating protein identifications in replicate analyses
In order to predict the cumulative protein identifications in replicated LC-MS/MS experiments from a single analysis, it is necessary to estimate the distribution of protein quantities in the Deinococcus sample. However, because many proteins do not have any identified peptides, estimating quantities of individual proteins based on a single experiment cannot be accomplished. On the other hand, simply estimating the fraction of unidentified proteins at each quantity level is straightforward once the distribution of protein quantities is estimated.
To accomplish this, let us first define the average effective detectability of all proteins in D. radiodurans at quantity q and calculate it as , where N is the number of proteins in D. radiodurans database (N = 3167 proteins from 2 chromosomes and 2 plasmids, retrieved from GenBank on 08/27/2009). Using the definition of protein detectability from Eq. 8, if the sample contains k different proteins (randomly selected from the proteome) at quantity q, we expect k · μd(q) of these proteins to be identified in any single experiment. Thus, given all the identified proteins at quantity level q, the number of unidentified proteins at that same quantity level can be easily estimated.
Based on previous work,5,22–24 we fit the observed protein quantities {qi} from the first experiment to a log-normal probability distribution
| (11) |
where parameters μ and σ were to be determined (note that the log-normal distribution is similar to the power-law distribution21).
The parameters of the distribution were estimated using the maximum likelihood approach with two modifications. First, each protein at quantity qi was assigned weight 1/μd(qi) to compensate for the tendency of low quantity proteins to remain unidentified (we refer to this weighting procedure as the bias correction step). Second, due to the properties of analytical platforms, proteins at quantities lower than the minimum observed quantity qm will not be identified at all, resulting in a left-truncated log-normal quantity distribution. Accordingly, parameters μ and σ were estimated (μ̂, σ̂) by maximizing the following log-likelihood function
| (12) |
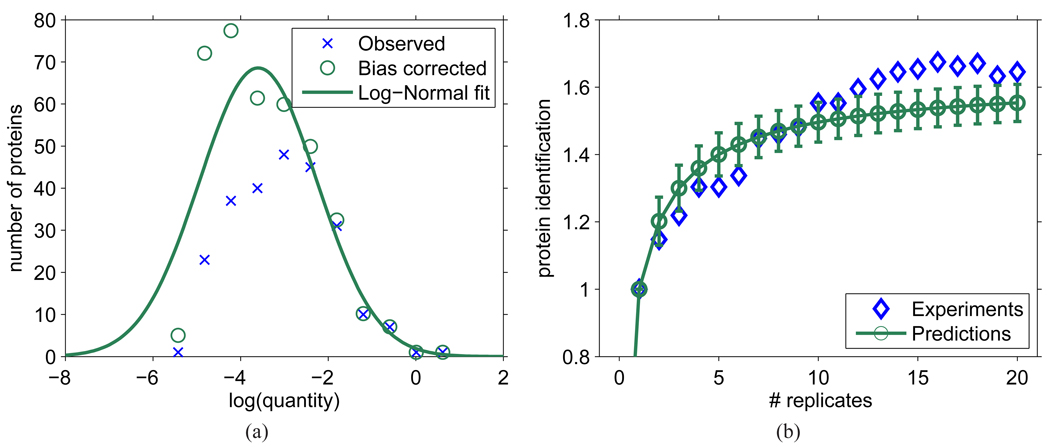
where Φ (·) is the cumulative density function of a normally distributed random variable. Figure 7(a) shows the distribution of original protein quantities, the bias-corrected distribution, and the fitted distribution that maximizes logL.
Figure 7.
Predicting cumulative protein identifications in replicated LC-MS/MS experiments based on one survey experiment. (a) Estimated quantity distribution of all proteins in the sample (identified or not) based on the first experiment PQ21. The bias corrected values were obtained by weighting the obverved values by the inverse of the average protein detectability 1/μd(q), while the log-normal fit curve was obtained by maximum likelihood fitting using Eq. 12. (b) Estimated number of proteins identified (relative to number of identifications in the first experiment, n1) after replicated LC-MS/MS experiments. Protein quantities were sampled from the log-normal distribution estimated in (a) and the standard protein detectabilities were derived from standard peptide detectabilities predicted using the IL20 predictor trained on experiment PQ21.
In order to estimate the number of experimentally identified proteins after multiple runs, we used a 5% peptide-level FDR to obtain a list of identified peptides, which were then mapped to the proteins from D. radiodurans. The number of correct protein identifications is then estimated as the difference between the number of proteins identified from the target database and the number of proteins from the decoy database. Obviously, a number of proteins identified by single peptides are false identifications, while many other proteins identified by correct peptide-spectrum matches were missing because those peptides were not in the list of confident peptide identifications.
Given the standard protein detectability predictions for all proteins in D. radiodurans and estimated quantity distribution f (q, μ̂, σ̂2), the number of proteins identified after r replicate experiments, n̂r, can be estimated as follows: (i) sample quantities from the distribution f (q, μ̂, σ̂2) for all proteins in D. radiodurans proteome; (ii) compute the effective detectability for each protein i; (iii) denoting n1 as the number of proteins identified in the first LC-MS/MS run, calculate the expected number of proteins (identified or not) in the sample as ; (iv) finally, calculate the expected number of protein identifications after r replicate experiments n̂r as
| (13) |
In Figure 7(a) we show the estimated log-normal distribution of protein quantities, while in Figure 7(b) we compare the predicted and observed number of protein identifications in 20 replicate analyses. Our primary concern was not whether a particular protein was identified, but rather the number of identified proteins. Good agreement between the expected and observed accumulation of identified proteins provides evidence that the theoretical framework and assumptions used for estimating peptide/protein detectabilities were reasonable and can be effectively used in practice. Note that for the replicate experiments, the peptides were first pooled together and then a 5% peptide-level FDR cutoff was estimated and applied. By doing this, the number of false identifications was prevented from accumulating. This approach, however, may result in a decreased number of protein identifications with a number of pooled experiments.
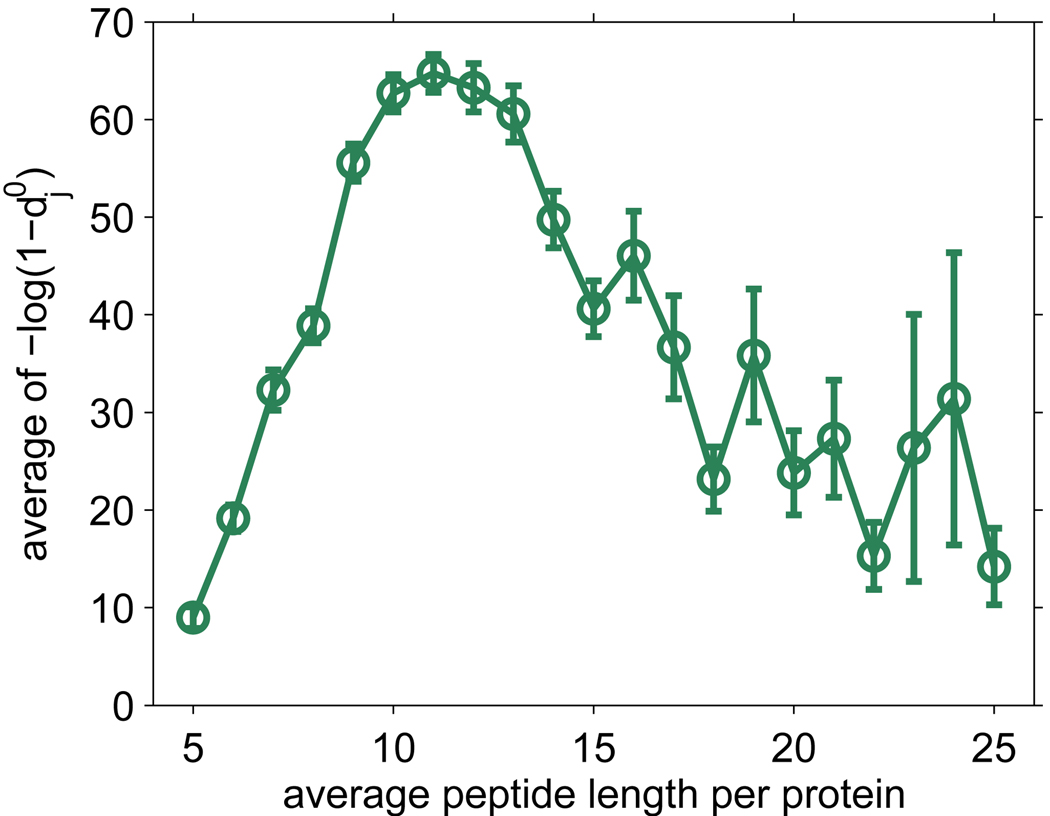
The estimated quantity of proteins in the Deinococcus sample can also be used to gain insight into the number of proteins from D. radiodurans that can be identified by LC-MS/MS if at sufficiently high quantity qM. We used the estimated quantity distribution from the first run of the Deinococcus sample and examined how many proteins would be identifiable in a single run with the criteria that the effective detectability di(qM) ≥ 0.5. Surprisingly, 99.7% of proteins would be identifiable if qM were set to be the maximum estimated quantity among identified proteins, while 94.8% of proteins would be identifiable with qM equal to an average of the observed quantities for the identified ribosomal proteins (ribosomal proteins were used to represent housekeeping proteins). This result is consistent with the good proteome coverage already achieved for D. radiodurans25 and a consensus that the dynamic range of the cellular proteome and analytical platform are major challenges in a proteomics experiment.26 Finally, Figure 8 shows the average protein detectability as a function of proteins’ average (tryptic) peptide length in D. radiodurans. Observe that proteins with average peptide length of around 11 are most detectable.
Figure 8.
Average protein detectability in D. radiodurans proteome as a function of the average tryptic peptide length in proteins. Note that is a monotonic transformation of standard protein detectability , where j is the index for proteins.
Discussion
The goal of this study was to investigate the properties and expand the use of peptide detectability in shotgun proteomics, both with respect to statistical inference of peptide and protein presence (and quantities), and experiment design. We developed a novel computational model for predicting standard and effective detectabilities in a proteomics experiment. This iterative algorithm is based on simultaneous learning of standard and effective detectabilities coupled with a label-free quantity estimation. The experiments performed in this work provide evidence that the new algorithm provides practically useful detectability estimates. The results shown were obtained using the first replicate analysis (PQ21) as the training set, however, similar results were achieved when a different run was used instead of PQ21 (Supplementary Materials; we note that the variability of estimates is significantly influenced by the outcome of the first replicate analysis). We have also experimented with several other data sets from different proteome samples and different platforms and observed similar trends among the models (data not shown).
Although we provided evidence that the proposed detectability learning algorithm and proteomics modeling approaches are useful, there are several limitations worth futher inverstigation. First, only fully tryptic peptides were considered throughout this study. Extending the approach to semi/non-tryptic peptides and peptides with missed cleavages should generally improve the applicability of peptide detectability. Second, degenerate peptides that are present in multiple proteins (e.g. splice isoforms, protein families) are not formally addressed in this work. However, we believe that this is primarily a problem of estimating protein/peptide quantity, thus, if coupled with more powerful quantity estimation algorithms, the effective detectability should be readily applicable to the proteomes with extensive homology. Finally, we emphasize that the machine learning framework proposed here for detectability learning should generally work with data from sub-proteome samples. However, estimation of protein identifications in replicate analyses should be carried out with caution due to the lack of evidence that the log-normal protein quantity distribution holds in sub-proteomes.
An important part of this work was to investigate the behavior of previously proposed detectability models.1,5,7–11 We hypothesized that detectability models M2, M5 and M10, that are trained on complex samples, may not be able to infer standard detectabilities as a result of unequal protein abundances in the training data. However, since the effective detectability is not an intrinsic property of a peptide, such models cannot be expected to accurately approximate peptide repeatability either. Our experiments show that detectability models trained on complex biological mixtures, in fact, are able to approximate standard detectability. Such argument is supported by the fact that the outputs of IL20 and M2 models were significantly correlated (Table 2), but also by the ability of the M2 model to be quantity-adjusted (one step application of Eq. 4, post-learning) to achieve only slightly lower performance accuracy than the IL2 model (data not shown). If the data set is large enough, noise present in the training data will not significantly influence models M2, M5, and M10. Their final output can be seen as standard detectability for peptides when the proteins are present at one fixed quantity, probably corresponding to the average over all identified proteins. These detectability models thus benefit from the ability of machine learning models (neural networks in this work) to infer posterior probabilities even in the presence of large amounts of class-label noise.27
We also suggest that due to inherent differences in distributions of identified peptides over organisms and experimental conditions, one can expect the best performance if the detectability predictor is trained in-sample, that is, after the first set of peptide identifications are made.10 Such predictors are likely to outperform pre-trained models (compare SSP model with M2, M5, and M10). However, if the number of identified proteins in an experiment is not sufficiently large, the pre-trained models can still be valuable and are also easier to use by experimental scientists. We suggest that while using the pre-trained approach, only the standard detectability predictions be transferred. Protein quantities, which are sample specific, should be estimated from the sample of interest (e.g. using a one-step quantity calculation from Eq. 4), thus enabling the estimation of effective detectabilities.
Accurate estimation of standard detectability was shown to have important applications in computational MS/MS proteomics. Previously, we used standard detectability to predict peptides likely to be truncated28 and improve protein inference.2–4 Here, we showed that an improvement in detectability prediction directly results in increased number of protein identifications. However, an accurate estimation of detectability can also result in improved label-free protein quantification,1,5 better identification of post-translational modifications or more effective experiment design. Our work demonstrates that it is possible to accurately estimate, from a single analysis, the behavior of a replicated experiment with respect to achieving a saturation point in protein identifications. Such estimates were previously possible only for organisms for which protein quantities were roughly known and under relatively standard cellular conditions.29
Interestingly, we observed a U-shaped distribution of the standard detectability and L-shaped distribution of the effective detectability in a complex sample. This indicates that peptides in a protein at standard quantity can be roughly divided into detectable and non-detectable, with a relatively small fraction of peptides with medium detectability. Also, we suggest that a transformation from a U-shaped to the L-shaped distribution is the result of large variability in protein quantity, and potentially competition. This has implications for proteotypic peptide identification which are typically defined as peptides frequently observed (i.e. >50% of the time) in the experiments where their parent proteins were identified.7,18,19
We showed that a peptide detectability predictor can accurately approximate peptide repeatability, and thus can be used for the prediction of proteotypic peptides from the biological samples of species that have not been extensively studied. We note that the symmetric shape of the standard detectability distribution is influenced by an assumption that the average detectability over all peptides is 0.5. In our experience (data not shown) it is reasonable to estimate that the average standard peptide detectability was close to 0.5. Other mean values may skew this distribution to the left (if average detectability is lower than 0.5) or to the right, but it always results in some peptides being detectable and others undetectable.
Finally, in this work the concept of peptide detectability was extended to proteins, in order to study detectability of cellular proteomes and also guide MS/MS proteomics experiments. As shotgun proteomics typically requires multiple replicated experiments, our work can provide informatics support that is necessary for such approaches from the increased power in protein identification and quantification to understanding the behavior of replicate analyses. We believe that the concept of peptide detectability is key to a Bayesian formalism in the analysis of tandem mass spectrometry data. Thus, better understanding of its definitions, strategies of learning as well as its use, are important for experimental and computational communities.
Supplementary Material
Acknowledgement
We thank three anonymous reviewers for their comments and suggestions that improved the quality of this work. This project was funded by the NIH grant R01 RR024236-01A1 and NCI grant U24 CA126480-01.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tang H, Arnold RJ, Alves P, Xun Z, Clemmer DE, Novotny MV, Reilly JP, Radivojac P. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 2.Alves P, Arnold RJ, Novotny MV, Radivojac P, Reilly JP, Tang H. Advancement in protein inference from shotgun proteomics using peptide detectability. Pac Symp Biocomput. 2007:409–420. [PubMed] [Google Scholar]
- 3.Li YF, Arnold RJ, Li Y, Radivojac P, Sheng Q, Tang H. A Bayesian approach to protein inference problem in shotgun proteomics. Research in Computational Molecular Biology (RECOMB’08), LNCS 4955. 2008:167–180. doi: 10.1089/cmb.2009.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li YF, Arnold RJ, Li Y, Radivojac P, Sheng Q, Tang H. A Bayesian approach to protein inference problem in shotgun proteomics. J Comput Biol. 2009;16:1183–1193. doi: 10.1089/cmb.2009.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 6.Le Bihan T, Robinson M, Stewart I, Figeyss D. Definition and characterization of a "trypsinosome" from specific peptide characteristics by nano-HPLC-MS/MS and in silico analysis of complex protein mixtures. J. Proteome Res. 2004;3:1138–1148. doi: 10.1021/pr049909x. [DOI] [PubMed] [Google Scholar]
- 7.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 8.Wedge DC, Gaskell SJ, Hubbard SJ, Kell DB, Lau KW, Eyers C. Peptide detectability following ESI mass spectrometry: prediction using genetic programming; GECCO ’07: Proceedings of the 9th annual conference on Genetic and evolutionary computation; New York, NY, USA. 2007. pp. 2219–2225. [Google Scholar]
- 9.Sanders W, Bridges S, McCarthy F, Nanduri B, Burgess S. Prediction of peptides observable by mass spectrometry applied at the experimental set level. BMC Bioinformatics. 2007;8 Suppl 7:S23. doi: 10.1186/1471-2105-8-S7-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel C, Marcotte E. Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat Protoc. 2008;3:1444–1451. doi: 10.1038/nport.2008.132. [DOI] [PubMed] [Google Scholar]
- 11.Webb-Robertson B-JM, Cannon WR, Oehmen CS, Shah AR, Gurumoorthi V, Lipton MS, Waters KM. A support vector machine model for the prediction of proteotypic peptides for accurate mass and time proteomics. Bioinformatics. 2008;24:1503–1509. doi: 10.1093/bioinformatics/btn218. [DOI] [PubMed] [Google Scholar]
- 12.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Tabb DL, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2009 doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate 27 the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 15.Lindgren B. Statistical theory. Chapman & Hall/CRC; 1993. [Google Scholar]
- 16.Mackay DJC. Bayesian interpolation. Neural Computation. 1992;4:415–447. [Google Scholar]
- 17.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 19.Blonder J, Veenstra T. Computational prediction of proteotypic peptides. Expert Rev Proteomics. 2007;4:351–354. doi: 10.1586/14789450.4.3.351. [DOI] [PubMed] [Google Scholar]
- 20.Friedman J, Tibshirani R, Hastie T. The elements of statistical learning: data mining, inference and prediction. New York: Springer-Verlag; 2001. [Google Scholar]
- 21.Mitzenmacher M. A brief history of generative models for power law and lognormal distributions. Internet mathematics. 2004;1:226–251. [Google Scholar]
- 22.Bengtsson M, Ståhlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghaemmaghami S, Huh W, Bower K, Howson R, Belle A, Dephoure N, O’Shea E, Weissman J. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 24.Ueda HR, Hayashi S, Matsuyama S, Yomo T, Hashimoto S, Kay SA, Hogenesch JB, Iino M. Universality and flexibility in gene expression from bacteria to human. Proc Natl Acad Sci U S A. 2004;101:3765–3769. doi: 10.1073/pnas.0306244101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipton MS, et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci U S A. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picotti P, Bodenmiller B, Mueller L, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magdon-Ismail M, Nicholson A, Abu-Mostafa Y. Financial markets: very noisy information processing. Proc. IEEE. 1998;86:2184–2195. [Google Scholar]
- 28.Alves P, Arnold RJ, Clemmer DE, Li Y, Reilly JP, Sheng Q, Tang H, Xun Z, Zeng R, Radivojac P. Fast and accurate identification of semi-tryptic peptides in shotgun proteomics. Bioinformatics. 2008;24:102–109. doi: 10.1093/bioinformatics/btm545. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Sadygov R, Yates J., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.