Abstract
Although cellular behaviors are dynamic, the networks that govern these behaviors have been mapped primarily as static snapshots. Using an approach called differential epistasis mapping, we have discovered widespread changes in genetic interaction among yeast kinases, phosphatases, and transcription factors as the cell responds to DNA damage. Differential interactions uncover many gene functions that go undetected in static conditions. They are very effective at identifying DNA repair pathways, highlighting new damage-dependent roles for the Slt2 kinase, Pph3 phosphatase, and histone variant Htz1. The data also reveal that protein complexes are generally stable in response to perturbation, but the functional relations between these complexes are substantially reorganized. Differential networks chart a new type of genetic landscape that is invaluable for mapping cellular responses to stimuli.
One of the most basic approaches to understanding gene function relies on the identification of genetic interactions, which occur when the phenotypic effects of one gene depend on the presence of a second. Recently, a number of technologies have been developed to systematically map genetic interaction networks over large sets of genes in budding yeast (1–3) and other model organisms (4, 5). Thus far, these networks have been constructed only under normal laboratory conditions. However, cells are constantly bombarded by signals and stresses, such as ligands, drugs, hormones, toxins, or other environmental conditions. Although it is clear that some genetic interactions are condition-dependent (6, 7), to what extent environmental stresses can affect genetic interaction networks, and the pathways they represent, is still unknown.
To gain insight into how genetic networks are altered by stress, we assembled a large genetic interactome with and without perturbation by the DNA-damaging agent methyl methane-sulfonate (MMS). Using the technique of epistatic miniarray profiles (E-MAP) (8), genetic interactions were interrogated among a set of 418 yeast genes selected to provide broad coverage of the cellular signaling and transcriptional machinery, including nearly all yeast kinases, phosphatases, and transcription factors, as well as known DNA repair factors (fig. S1 and table S1). About 80,000 double-mutant strains were generated from all pairwise mutant combinations of the 418 genes, in which mutations were complete gene deletions (nonessential genes) or hypomorphic alleles (essential genes) as appropriate. Double-mutant combinations were grown with or without 0.02% MMS, and their colony sizes were analyzed statistically to compute a genetic interaction score (S score) in each condition (9), which indicates whether the strain was healthier or sicker than expected (positive or negative S, respectively) (10).
From established score thresholds for positive and negative interactions (S ≥ +2.0, S ≤ −2.5) (9) we identified two genetic networks: a set of 1905 interactions for the untreated condition, and a set of 2297 interactions under MMS. Analysis of these “static” genetic maps showed strong associations with physical interaction networks of various kinds. For example, gene pairs with either positive or negative genetic interactions were highly enriched for proteins known to physically interact. In addition, both maps were enriched for known kinase- and phosphatase-substrate pairs, as well as transcription factor-target pairs (fig. S2). The correspondence to physical and functional associations reflects the predictive power of this genetic interaction data set.
Comparison of the genetic networks across conditions revealed large differences, with more interactions unique to each map than in common (Fig. 1A). For example, more than 70% of positive interactions identified under MMS were not identified in the untreated sample, which reflects widespread DNA damage–induced epistasis (Fig. 1B). To assess these changes in interaction, each gene pair was associated with its difference in S score across conditions (Fig. 1C). A P value for this difference was calculated using the null distribution of score differences observed when comparing replicate interaction measurements from the same condition (fig. S3) (10). This method identified 873 differential genetic interactions at P ≤ 0.001, with a corresponding false-discovery rate of ~9% (fig. S4 and table S2). We term this approach differential epistasis mapping (dE-MAP), as it is based on the difference of two static networks generated using the E-MAP methodology. A total of 379 interactions were “negative differential,” which indicated DNA damage–induced lethality or sickness, whereas 494 were “positive differential,” which indicated inducible epistasis or suppression (Fig. 1D). The majority (62%) of differential interactions were not detectable in either static condition, most likely because they are too weak to detect in any single condition yet display a substantial change in interaction between conditions.
Fig. 1.
An epistasis map for DNA damage signaling. (A) Comparison of genetic interactions (positive, S ≥ +2; negative, S ≤ −2.5) uncovered in untreated or DNA damage treated (+MMS) conditions. Control represents interactions from two independent experiments in untreated conditions. (B) Percentage of interactions (positive or negative) identified in each condition that are specific to that condition. (C) Differences between the untreated and treated maps identify differential interactions. (D) Scatter of S scores between untreated and treated maps and identification of positive differential (green, P ≤ 0.001) and negative differential interactions (red, P ≤ 0.001). (E) Enrichment of interactions involving known DNA repair genes, shown for static and differential networks and including both positive and negative interactions. For the right-most bar, all differential interactions also identified in the static network are removed. (F) Enrichment of interactions involving genes with various functions (10).
To determine whether static untreated, static treated, or differential genetic networks best uncover DNA damage-response pathways, we examined a reference set of 31 known DNA repair genes (table S1). We noted that static networks were no more likely than random to include interactions with genes in this reference set (Fig. 1E). This lack of enrichment was observed in the untreated genetic network, as well as, surprisingly, the static network obtained under MMS. In contrast, the differential network—obtained through the quantitative difference in interaction across conditions—was highly enriched for interactions involving DNA damage-response genes such as RAD52, TEL1, and DUN1 (Fig. 1E) (10).
We noted that both the static treated and untreated networks were dominated by interactions involving genes that function in chromatin organization (Fig. 1F). This strong chromatin signal has been previously reported in budding and fission yeasts and C. elegans (2, 4, 5, 11). Through network subtraction, however, the “housekeeping interactions” due to chromatin are removed, which allows sensitive detection of differentially represented pathways. Thus, network comparison reveals a landscape of genetic interactions particularly tailored to the cellular response of interest.
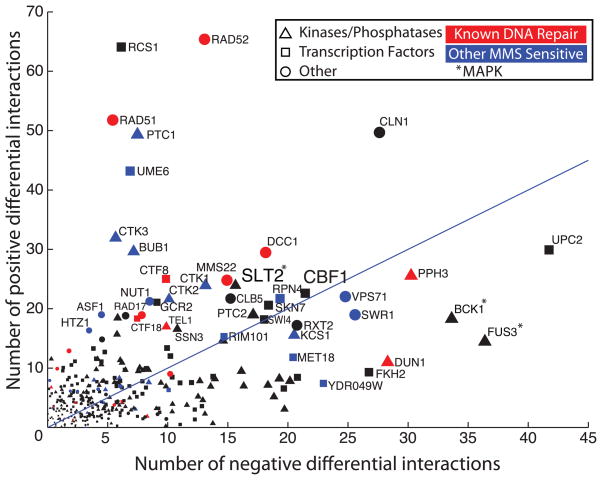
Analysis of static networks has found that network “hubs”, i.e., genes with many interactions, modulate a variety of cellular functions and are more likely to be essential for viability (12). For the differential network, we found that the number of interactions per gene was correlated with the sensitivity to MMS of the corresponding gene deletion strain (r = 0.35, P < 10−5) (fig. S5, A and B). Differential interaction hubs were also more likely to be essential for growth under a variety of drug treatments and stresses (fig. S5, C and D), consistent with previous observations for static hubs (13).
Further investigation showed that many differential interaction hubs were already well known to function as key components of DNA repair pathways (Fig. 2), which led us to predict that the remaining hubs might encode this role. Two such differential interaction hubs encode Slt2 and Bck1, mitogen-activated protein kinases (MAPKs) that have been implicated in the maintenance of cell wall integrity but not yet linked to DNA repair (Fig. 2). We found that Slt2 is both up-regulated and translocated to the nucleus upon MMS treatment, and it is required for appropriate regulation of ribonucleotide reductase genes in response to DNA damage (fig. S6, A to D) (14). Furthermore, both MAPKs show strong genetic interactions with DNA damage checkpoint genes (fig. S6, E and F), which suggests that they may function in a parallel signaling pathway.
Fig. 2.
Identification of differential genetic interaction hubs. The scatterplot shows the number of positive and negative differential interactions associated with each gene in this study. The 30 genes whose deletions are the most sensitive to MMS are indicated (blue) (24), excluding those already known to function in DNA repair (red) (table S1).
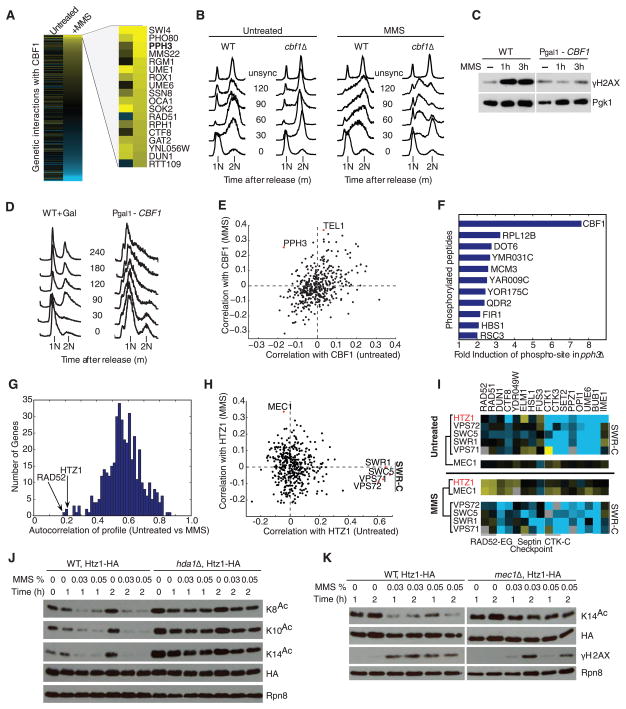
Another differential interaction hub not previously linked to DNA repair was centromere binding factor 1 (CBF1) (Fig. 2), a sequence-specific transcription factor and component of the inner kinetochore (15). CBF1 gained strong genetic interactions upon MMS treatment (Fig. 3A), which suggested an additional role in DNA repair. We found that Cbf1 is required for appropriate activation of cell cycle checkpoints (Fig. 3B) and that CBF1 overexpression interferes with induction of the damage-dependent histone modification γH2AX (Fig. 3C) and leads to cell cycle arrest in G1 (Fig. 3D). Furthermore, MMS treatment causes the CBF1 profile of genetic interaction scores (across all genes on the E-MAP) to become correlated with the profiles of TEL1 kinase and PPH3 phosphatase, which encode key proteins regulating the DNA damage checkpoint (Fig. 3E) (16). Mass spectrometry–based phosphoproteomics showed that Cbf1 is hyperphosphorylated at a conserved serine-glutamine motif (SQ145-146) in pph3Δ cells (Fig. 3F and table S3). As the checkpoint kinases Mec1 and Tel1 have been shown to target Cbf1 at the same SQ site (17), it is likely that Pph3 is the protein phosphatase that counteracts the effect of this phosphorylation.
Fig. 3.
Differential genetic interactions identify novel DNA damage–dependent pathways. (A) Full profile of CBF1 genetic interactions with the strongest positive genetic interactions in MMS highlighted. (B) Fluorescence-activated cell sorting (FACS) analysis of cell cycle progression in alpha-factor–arrested wild-type and cbf1Δ cells released into media with or without MMS. In MMS, cbf1Δ cells bypass cell cycle checkpoints and eventually accumulate in S phase (between 1N and 2N DNA content). (C) γH2AX levels for wild-type and CBF1 overexpression at times indicated after exposure to MMS. Pgk1 is used as a loading control. (D) Effect of CBF1 overexpression on cell cycle progression using FACS. (E) Correlation coefficients between the CBF1 genetic interaction profile and that of each gene in the epistasis map, in MMS versus untreated conditions. (F) Top changes in abundance of phosphorylated peptides in pph3Δ versus wild-type cells by phosphoproteomic profiling. (G) Histogram of autocorrelation coefficients. For each gene, the genetic interaction profiles before and after MMS exposure are compared by Pearson correlation. (H) Correlation plot as in (E) for HTZ1. (I) Representative genetic interactions ofHTZ1, MEC1, and SWR-C members. Clustering based on similarity of profiles is shown. (J) The acetylation of multiple Htz1 N-terminal lysine residues (K8, K10, or K14) measured in an Htz1-3HA strain after MMS exposure in wild-type (WT) and hda1Δ backgrounds. HA represents total amount of Htz1. Rpn8 is a loading control. (K) Acetylation status of Htz1-K14 in response to MMS in WT and mec1Δ backgrounds. γH2AX is a downstream marker of DNA-damage signaling by Mec1.
As another means of mapping DNA repair pathways, we identified genes with genetic interaction profiles that were conditionally disrupted by MMS, suggesting a shift in gene function. We observed that most genes had high correlation between their genetic interaction profiles measured in the presence or absence of MMS (high “genetic autocorrelation”) (Fig. 3G). However, several genetic interaction profiles were markedly disrupted in MMS (low autocorrelation), including those of RAD52, a critical factor in homologous recombination–mediated DNA repair (18), and HTZ1, encoding the histone variant H2A.Z, whose role in DNA repair is less well understood (19, 20). In untreated conditions, the HTZ1 profile correlated with members of the SWR complex (SWR1, SWC5, VPS71, and VPS72) (11), responsible for incorporating Htz1 into chromatin (21). This correlation was lost in MMS (Fig. 3H), which suggested a functional disassociation between Htz1 and the SWR-C. Conversely, HTZ1 became correlated with the DNA-damage checkpoint kinase MEC1 upon MMS treatment (Fig. 3, H and I), and a mec1Δhtz1Δ strain showed synthetic sensitivity to MMS (fig. S7), suggesting a damage-dependent functional link between the two proteins. Htz1 is acetylated on its amino terminus by histone acetyltransferase NuA4 (22) and deacetylated by histone deacetylase Hda1 (19). We found that the degree of Htz1 acetylation at multiple lysine residues was strongly reduced in response to MMS (Fig. 3J). This effect was dependent on both Hda1 (Fig. 3J) and Mec1 (Fig. 3K), which suggested that the regulation of Htz1 acetylation contributes to the DNA damage response.
We next investigated the association between differential genetic interactions and known yeast pathways and protein complexes (i.e., modules) (table S1). In contrast to static genetic interactions, which are enriched within modules, we found that differential genetic interactions are not (fig. S8A). Rather, differential genetic interactions are much more likely to occur among pairs of genes connecting two different modules than among pairs of genes within the same module (fig. S8B). These findings were corroborated by an alternative analysis in which modules were defined through hierarchical clustering of the treated and untreated genetic interaction data. Genes that clustered into the same module in both conditions were much more likely to physically interact than genes that coclustered in one condition only (fig. S8C). These results suggest that known protein complexes tend to be stable across conditions—it is the genetic interactions between these modules that are reprogrammed in response to perturbation (Fig. 4A).
Fig. 4.
Module-based interpretation of differential genetic interactions. (A) MMS induces a set of positive interactions between DNA damage pathways (top right) that are not evident in untreated conditions (bottom left). (B) Module map of protein complexes and pathways connected by differential genetic interaction bundles. Node color represents the most severe single-deletion phenotype among members of a module (table S1). (C) Detailed view of differential genetic interactions between protein complexes corresponding to selected modules in (B) (dotted node borders). For clarity, only physical interactions and differential genetic interactions with P < 0.01 are shown. Thickness is scaled with increasing significance of the P value (10).
On the basis of these findings, we constructed a global map of gene modules and their dynamic genetic interactions in response to DNA damage. Using an established method (23), we defined modules as dense clusters of physical and static genetic interactions. Module-module interactions were characterized by heavy enrichment for many differential genetic interactions across the two modules (table S4) (10). The resulting map of 56 multigene modules and 66 module-module interactions (Fig. 4, B and C) provides a global resource of pathways and complexes that are reconfigured in response to DNA damage–induced stress, many of which have not been previously linked to DNA repair.
Large-scale genetic interaction networks have proved extremely powerful for mapping the pathways that regulate essential cell functions. In this study, we have shown that differential genetic networks are comparable in size to static networks, yet access a very different set of interactions governing a dynamic cellular response. Given that most gene functions arise in response to changing conditions, the differential network revealed here offers a glimpse into a much larger universe of genetic interactions that are condition-, cell type–, or tissue-specific.
Supplementary Material
Acknowledgments
The authors thank S. Collins, H. Hombauer, A. Desai, S. Gasser, X. Shen, and S. Choi for helpful discussions and strains. This work was funded by NIH grants R01-ES14811 and R01-GM084279. Additionally, W-K.H. and M-K.S. were funded by the 21C Frontier Functional Proteomics Project (FPR08A1-060), R.A. and B.B. were supported by SystemsX.ch and the Swiss National Science Foundation, and M-C.K. was supported by the NCI (P30CA013330). N.J.K. is a Keck Young Investigator Fellow and a Searle Fellow. T.I. is a David and Lucille Packard Fellow. R.D.K. is a paid consultant to On-Q-ity.
Footnotes
References and Notes
- 1.Boone C, Bussey H, Andrews BJ. Nat Rev Genet. 2007;8:437. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 2.Costanzo M, et al. Science. 2010;327:425. [Google Scholar]
- 3.Pan X, et al. Cell. 2006;124:1069. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Nat Genet. 2006;38:896. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 5.Roguev A, et al. Science. 2008;322:405. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Game JC, Mortimer RK. Mutat Res. 1974;24:281. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 7.St Onge RP, et al. Nat Genet. 2007;39:199. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Methods. 2006;40:344. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 9.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supporting material on Science Online.
- 11.Collins SR, et al. Nature. 2007;446:806. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 12.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Nature. 2001;411:41. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 13.Jaimovich A, Rinott R, Schuldiner M, Margalit H, Friedman N. Bioinformatics. 2010;26:i228. doi: 10.1093/bioinformatics/btq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elledge SJ, Davis RW. Mol Cell Biol. 1987;7:2783. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai M, Davis RW. Cell. 1990;61:437. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 16.Heideker J, Lis ET, Romesberg FE. Cell Cycle. 2007;6:3058. doi: 10.4161/cc.6.24.5100. [DOI] [PubMed] [Google Scholar]
- 17.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proc Natl Acad Sci USA. 2007;104:10364. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pâques F, Haber JE. Microbiol Mol Biol Rev. 1999;63:349. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin YY, et al. Genes Dev. 2008;22:2062. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papamichos-Chronakis M, Krebs JE, Peterson CL. Genes Dev. 2006;20:2437. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogan NJ, et al. Mol Cell. 2003;12:1565. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 22.Keogh MC, et al. Genes Dev. 2006;20:660. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. PLOS Comput Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Mol Cancer Res. 2002;1:103. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.