Abstract
The bZIP transcription factor C/EBPβ is important for mammary gland development and its expression is deregulated in human breast cancer. To determine whether C/EBPβ regulates mammary stem cells (MaSCs), we employed two different knockout strategies. Utilizing both a germline and a conditional knockout strategy, we demonstrate that mammosphere formation was significantly decreased in C/EBPβ-deficient mammary epithelial cells (MECs). Functional limiting dilution transplantation assays indicated that the repopulating ability of C/EBPβ-deleted MECs was severely impaired. Serial transplantation experiments demonstrated that C/EBPβ deletion resulted in decreased outgrowth potential and premature MaSC senescence. In accord, FACS analysis demonstrated that C/EBPβ-null MECs contained fewer MaSCs, the loss of luminal progenitors and an increase in differentiated luminal cells as compared to wildtype. Gene profiling of C/EBPβ-null stem cells revealed an alteration in cell fate specification, exemplified by the expression of basal markers in the luminal compartment. Thus, C/EBPβ is a critical regulator of both MaSC repopulation activity and luminal cell lineage commitment. These findings have critical implications for understanding both stem cell biology and the etiology of different breast cancer subtypes.
Keywords: C/EBP beta, mammary stem cell, lineage commitment, progenitor cell, luminal, alveolar, differentiation
Introduction
Adult mammary stem cells (MaSCs) function to maintain tissue homeostasis and provide regenerative capacity that is required during the pregnancy-lactation cycle. The mammary gland is comprised of two major types of epithelia, luminal and basal, which form a ductal network embedded within the stroma. During pregnancy, a combination of systemic hormones and local growth factors induce alveolar cell proliferation and differentiation, which are responsible for milk production and secretion during lactation. The recent identification of stem and progenitor cells in both human and mouse have provided evidence for a hierarchical model in which ductal luminal, alveolar luminal, and myoepithelial cells originate from a common progenitor [1–7]. However, the precise genetic mechanisms that regulate lineage commitment at each stage of mammary gland development are just beginning to be unraveled.
The CCAAT/enhancer binding proteins (C/EBPs) are a family of highly conserved transcription factors that regulate numerous genes involved in proliferation, differentiation, and more recently the instruction of stem cell fate in a variety of tissues (reviewed in [8]). One member of this family, C/EBPβ, is an important mediator of mammary ductal morphogenesis and lobuloalveolar development during pregnancy [9]. During puberty, C/EBPβ protein is expressed in both the cap cell layer and body cells of terminal end buds. In the virgin mammary gland, this protein resides in steroid receptor-negative luminal cells as well as myoepithelial cells [10]. The mammary glands of C/EBPβ−/− mice display delayed ductal outgrowth, enlarged ducts and decreased of C/EBP branching. Transplantation β mammary epithelial cells (MECs) resulted in severely impaired lobuloalveolar development in pregnant recipient mice, and indicated that these effects are intrinsic to the epithelial cells [11]. The misexpression of steroid hormone receptors coupled with a marked decrease in cell proliferation following estrogen and progesterone (E+P) stimulation suggested the presence of an altered alveolar cell fate program, rendering alveolar progenitor cells incapable of properly responding to hormone-induced proliferation. Additionally, C/EBPβ−/− glands exhibit increased expression of epidermal markers including a small proline-rich protein (SPRR2A) and keratin 6 (K6), suggesting an alteration in ductal progenitor cell differentiation. [12–14].
Several recent studies have begun to define a role for specific transcription factors in mammary cell lineage commitment [1, 15–17]. Here we examined the potential role of C/EBPβ in mediating MaSCs using two different knockout strategies. Utilizing various in vitro assays as well as functional in vivo transplantation assays, we show that deletion of C/EBPβ results in a lower frequency of repopulating MaSCs, suggesting that stem cell self-renewal is impaired. Further, deletion of C/EBPβ causes the ablation of luminal progenitor cells and an accumulation of committed luminal cells. In summary, our studies demonstrate that C/EBPβ is required for the expansion of normal MaSCs and is an important determinant of luminal cell lineage commitment. Elucidating the mechanisms that specify cell fate at each stage of the mammary stem cell hierarchy is a crucial prerequisite for understanding how these signals are altered during tumorigenesis.
Materials and Methods
Mouse strains and breeding
The germline C/EBPβ−/− mice were provided by Esta Sterneck (NCI, Frederick, MD) and have been described [18]. Homozygous mutant and wildtype littermates were derived by intercrossing hemizygous parents, and the resulting progeny were of mixed genetic background (C57BL/6× 129/Sv). The C/EBP βfl/fl mice were provided by Esta Sterneck [19] and were bred to ROSA26 lacZ reporter mice (R26R) and maintained in the C57BL/6 background for 12 generations. The R262R and β-actin-cyan fluorescent protein (CFP) mice were generous gifts from P. Soriano [20] and M. Lewis (Baylor College of Medicine, Houston, TX), respectively, and were maintained in C57BL/6 backgrounds. For transplantation, C57BL/6 and SCID/beige mice were obtained from Harlan Laboratories (Houston, TX) and Charles River Laboratories (Portage, WI), respectively. Mice were maintained in accordance with the NIH Guide for the Care and Use of Experimental Animals with approval from the Baylor College of Medicine Institutional Animal Care and Use Committee.
Primary MEC isolation and conditional deletion
For all experiments, MECs were derived from #3, #4 (without the lymph node) and #5 mammary glands of 10–12-week-old female mice. The glands were minced into 1mm fractions using a Vibratome Series 800-Mcllwain Tissue Chopper (Vibratome, St. Louis, MO) and digested in 2mg/ml collagenase A (Roche Applied Science, Indianapolis, IN) in F12 Nutrient Mixture containing 1X antibiotic-antimycotic (InVitrogen, Carlsbad, CA) for 1 hr at 37°C with shaking at 200rpm. Single cells were then purified as previously described [21]. For limiting dilution transplantation, mammary glands were digested in 1mg/ml collagenase A/F12 Nutrient Mixture containing 1X antibiotic-antimycotic for 14 hr at 37°C with shaking at 75rpm and isolated as described [22]. Cell preparations from germline wildtype and C/EBPβ−/− glands yielded equal numbers of MECs per gram of tissue. For all MEC isolations, viable cells were counted on a hemacytometer using trypan blue exclusion.
For conditional deletion, freshly isolated MECs were incubated with an adenovirus expressing Cre recombinase (Ad.Cre1) [23] at an MOI of 50 in 2ml of 5% FBS/DMEMF12 for 1 hr at 37°C in suspension. The cells were then centrifuged at 450×g for 5min, washed with PBS, and used immediately for mammosphere cultures or transplantation. For qPCR analysis, single cells were cultured for 3 days in primary growth medium (5% FBS, 5μg/ml insulin, 1μg/ml hydrocortisone, 10ng/ml EGF, 1X antibiotic-antimycotic, F12 Nutrient Mixture) prior to RNA isolation.
Quantitative RT-PCR
Total RNA was isolated from MECs using Trizol Reagent and DNased (InVitrogen) according to the manufacturer’s protocol. RNA (1μg) was reverse-transcribed using the iScript cDNA Synthesis Kit (Biorad, Hercules, CA) according to the manufacturer’s instructions. For qPCR, 1μl of cDNA was amplified in 1X SYBR® Green PCR Master Mix (ABI, Foster City, CA) containing specific primer pairs (Table S1) using the StepOnePlus™ Real-Time PCR System (ABI). PCR parameters were as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1min. Following amplification, melt curves were generated to confirm the specificity of each primer pair. Differences in gene expression between wildtype and C/EBPβ-null cells were determined by the Quantitative Comparative CT Method, where 18S rRNA served as the internal control.
Mammosphere and colony assays
MECs isolated from 3 sets of both wildtype or germline C/EBPβ−/−, and conditional C/EBPβ+/+;R26R or C/EBPβfl/fl;R26R (2 mice/genotype, transduced with Ad.Cre1) mice were plated in 6-well, ultra-low attachment plates at a density of 30,000 cells/well, cultured for 12 days and dissociated as previously described [24]. Single cells were re-plated at a density of 5,000 cells/well and secondary mammospheres were allowed to grow for 14 days, with feeding every 4 days. Twelve wells for each genotype were counted and the percentage of mammosphere forming cells was calculated as a measure of mammosphere efficiency.
For colony assays, feeder layers were prepared by treating NIH 3T3 cells (ATCC, Manassas, VA) with 10μg/ml of mitomycin C (Roche Applied Science) for 2 hr at 37°C. Cells were then washed 3X with PBS, plated onto 6-well plates (200,000 cells/well), and allowed to adhere overnight at 37°C. The next day, MECs isolated from 3 sets of C/EBPβ+/+;CFP+ and germline C/EBPβ−/−; CFP+ mice were plated at a clonal density (1,000 cells/well) on the feeder layers in primary growth medium. MECs were cultured for 10 days, feeding every 3 days, and CFP+ colonies were counted from 12 wells for each genotype (per experiment). Colonies were then fixed in methanol for 15min, stained with crystal violet for 30min, rinsed 3X with ddH20 and imaged on a Leica MZ16F stereoscope (Meyer Instruments, Houston, TX). Fluorescent images were captured on Leica TCS SP5 confocal microscope (Meyer Instruments).
Transplantation assays
To verify the phenotype of the conditional knockout mice, MECs isolated from 3 sets of wildtype (C/EBPβ+/+;R26R) or C/EBPβfl/fl;R26R (3 mice/genotype) were transduced with Ad.Cre1, resuspended at a concentration of 100,000 cells/10μl in a 1:1 solution of PBS and Matrigel (BD Biosciences #354230, San Jose, CA), and kept on ice during transplantation. Ten microliters of cells were injected into contralateral cleared fat pads (10 fat pads per genotype per set) of 21-day-old female C57BL/6 mice using a 26G needle and 50μl Hamilton glass syringe [25]. For limiting dilution transplantation, primary MECs derived from 5 sets of wildtype (C/EBPβ+/+;R26R) or C/EBPβfl/fl;R26R (4 mice/genotype) were transduced with Ad.Cre1, recounted on a hemacytometer and resuspended at the desired concentration in a 1:1 PBS:Matrigel solution. To verify deletion, 100,000 cells from each set were cultured for 3 days and qPCR was performed (data not shown). Cells of each genotype were injected at limiting dilutions (2500, 1000, 750, 500, or 250 cells) as described above. Animals were sacrificed 10 weeks post-transplantation and #4 glands and a #3 endogenous control gland were removed and stained with X-gal. LacZ+ glands showing at least 5% outgrowth were included in the analysis. For glands that displayed no outgrowth, the lack of epithelia was verified by Neutral Red staining, and these were included in the calculation of take rate.
For serial transplantation, 3 sets of C/EBPβ+/+;CFP+ or germline C/EBPβ−/−; CFP+ #4 contralateral glands were visualized using a Leica MZ16F fluorescent stereoscope. CFP+ epithelia located in the most distal regions from the nipple were carefully dissected with a razor blade and minced into 1 mm fractions using a tissue chopper (Vibratome). To ensure that equal amounts of epithelia were transplanted, tissue fragments were reanalyzed by fluorescence microscopy prior to transplantation. Tissue was floated in PBS and transplanted into the cleared contralateral fat pads of 21-day-old female SCID/beige mice. Eight weeks post-transplantation, #4 glands were removed and CFP+ outgrowths were analyzed by fluorescent microscopy. Glands demonstrating the best outgrowth were dissected and re-transplanted as described above.
X-gal staining and whole mount analysis
For X-gal staining, contralateral mammary glands were fixed in cold 4% paraformaldehyde (PFA) for 2 hr and stained with 1mg/ml X-gal (InVitrogen) as previously described [26]. The next day, glands were dehydrated in a series of ethanols and placed in Histoclear before imaging on a Leica MZ16F stereoscope. For fluorescent imaging, #4 glands were compressed between two glass slides and visualized using a Leica fluorescent stereoscope. For all transplantation experiments, #3 endogenous control glands were removed, fixed in 4% PFA for 2 hr on ice and stained with Neutral Red as described previously [22]. In some cases, control glands were first stained with X-gal and counterstained with Neutral Red. Glands were then embedded in paraffin, sectioned and stained with hematoxylin and eosin.
Fluorescence activated cell sorting
Four paired sets of freshly isolated single MECs derived from wildtype or germline C/EBP β−/− mice were resuspended at a concentration of 1×107 cells/ml in HBSS containing 2% FBS and 100mM Hepes (HBSS+), and incubated with primary antibodies (diluted 1:100) or isotype controls for 30min on ice. Antibodies are listed in Table S2. Cells were then washed in HBSS+ and incubated with either streptavidin-APC or streptavidin-PE-Cy7 (diluted 1:200) for 20min on ice. Cells were washed again, filtered through a 40μm cell strainer, and analyzed on a LSRII analyzer (BD Biosciences) or sorted on a FACS Aria Cell Sorting Flow Cytometer (BD Biosciences). Data were analyzed using FlowJo 8.7.3 (Tree Star, Inc, Ashland, OR).
Statistical analysis
Data from mammosphere assays, colony assays, and FACS analysis are presented as the means ± standard error of the means (SEM). C/EBPβ-null cells were compared to wildtype and significant differences were determined by an unpaired two-tailed t test (GraphPad Prism© Home, San Diego, CA). Limiting dilution transplantation results were analyzed by the single-hit Poisson model using a complementary log-log generalized linear model [27] and was validated as described previously [28]. Wald confidence intervals (95%) were calculated by the delta method for the frequency of MRUs. For both limiting dilution and serial transplantation assays, the percentages of fat pat filled were analyzed by a generalized linear model after logarithmic transformation. For serial transplantation, the percentages of fat pat filled at each generation were compared by the nonparametric Wilcoxon test between wildtype and C/EBPβ−/− outgrowths. The statistical software R was used for all transplantation analyses.
Results
C/EBPβ-null MECs exhibit decreased mammosphere formation
To investigate the role of C/EBPβ in stem cells, both a germline C/EBPβ−/− mouse as well as a conditional knockout strategy were employed. We initially sought to determine whether somatic cell deletion of C/EBPβ in the adult mammary gland resulted in a different phenotype to that observed from germline deletion. C/EBPβfl/fl mice were bred to ROSA26 reporter mice so that recombination could be monitored by lacZ expression. To facilitate the targeting of stem cells for recombination, MECs were isolated from wildtype (C/EBPβ+/+;R26R) or C/EBPβfl/fl;R26R mice, transduced in vitro with Ad.Cre1, and plated for 3 days to allow for recombination. Results from qPCR analysis showed that there was a >300-fold decrease in C/EBPβ expression in transduced MECs isolated from C/EBPβfl/fl;R26R mice as compared to wildtype (Fig. 1A). Immediately following transduction, MECs were also transplanted into the cleared fat pads of syngeneic hosts and stained with X-gal after 10 weeks of outgrowth. Glands in which C/EBPβ was deleted exhibited dilated ducts and decreased branching (Fig. 1B–1E), similar to the phenotype observed in germline knockout mice [11, 13]. Pregnant mammary glands (days 16–17) exhibited decreased alveolar development, illustrated by the lack of lipid droplet formation in C/EBPβ-deleted glands as compared to wildtype (Fig. 1F, 1G). LacZ expression was detected in both luminal and myoepithelial cells, indicating that recombination occurred in all cell types of the mammary gland. Furthermore, these results indicate that the phenotypes observed in somatic and germline cell deletion were morphologically indistinguishable.
Figure 1.
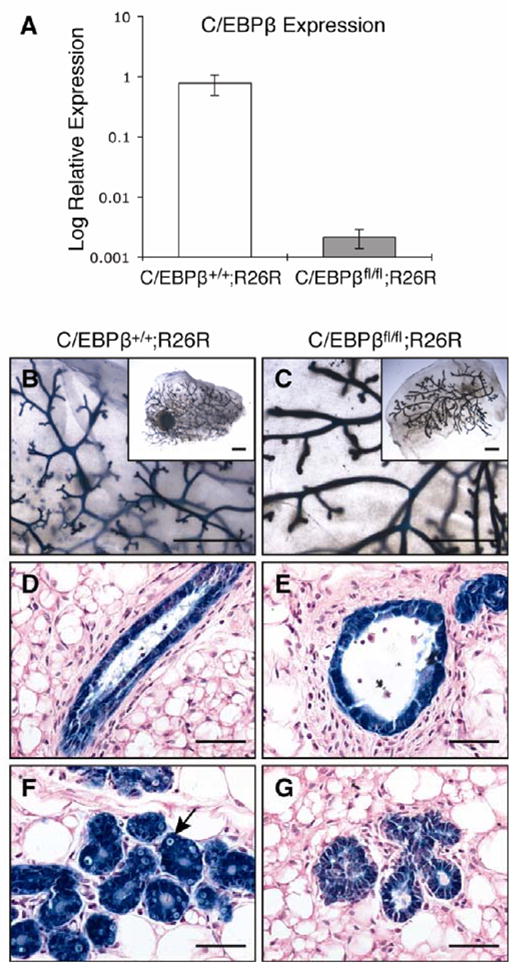
Conditional deletion of C/EBPβ results in altered ductal morphogenesis and decreased lobuloalveolar development. MECs were isolated from 10-week old wildtype (C/EBPβ+/+;R26R) and C/EBPβfl/fl;R26R mice, transduced with Ad.Cre1 in vitro and Cre-mediated recombination was examined (n=3). (A) qPCR for C/EBPβ showed that the expression of C/EBPβ was decreased >300-fold in transduced C/EBPβfl/fl;R26R MECs as compared to wildtype. (B–G) Transduced cells were transplanted into syngeneic hosts and stained with X-gal after 10 weeks of outgrowth. Large, dilated ducts were evident in virgin C/EBPβ-deleted glands as compared to wildtype by whole mount analysis (B,C) and H&E staining of paraffin-embedded sections (D,E). H&E staining of pregnant glands (P16–17) illustrated decreased alveolar development in C/EBPβ-null glands (G) as compared to wildtype (F), the latter of which contained lipid droplets. Uniform lacZ expression was observed throughout the mammary gland, indicative of a high extent of Cre-mediated recombination. Scale bars, 5 mm (B,C) or 20μm (D–G).
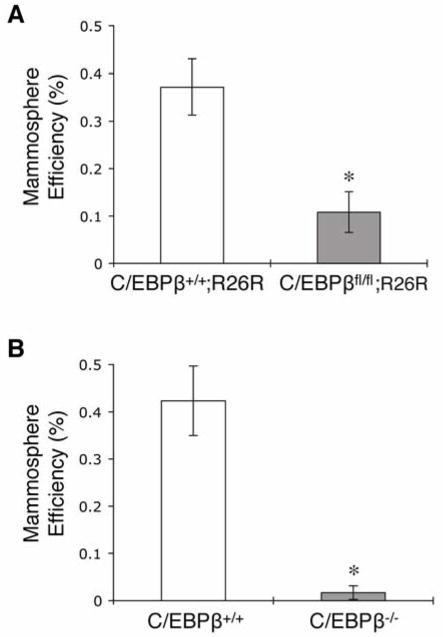
Previous studies have demonstrated that MECs cultured in serum-free media under non-adherent conditions form mammospheres that are enriched for stem/progenitor cells [24, 29]. To test whether deletion of C/EBPβ altered stem/progenitor cells, mammosphere assays were performed utilizing both germline and conditional knockout models. Conditional deletion of C/EBPβ resulted in a significant decrease in secondary mammosphere formation as compared to wildtype (Fig. 2A). There were no observed differences in mammosphere size or shape between the two groups (data not shown). Strikingly, MECs isolated from germline C/EBPβ knockout mice rarely formed secondary mammospheres, while wildtype mammospheres developed with the expected frequency (Fig. 2B). These results indicate that the proportion of mammosphere-forming cells is decreased in C/EBPβ-null MECS, suggesting a reduction of stem/progenitor cells.
Figure 2.
Decreased mammosphere formation in C/EBPβ-null MECs. Graphs illustrate the number of secondary mammospheres formed per 5000 cells expressed as mammosphere efficiency using either (A) conditional wildtype (C/EBPβ+/+;R26R) and C/EBPβfl/fl;R26R cells transduced with Ad.Cre1 or (B) germline wildtype (C/EBPβ+/+) and C/EBPβ−/− MECs. The ability of C/EBPβ-null cells to form secondary mammospheres was significantly inhibited using both these models (*p<0.0001). Data represent mean ± SEM of 1 representative experiment and the experiment was performed 3 times.
Deletion of C/EBPβ decreases the frequency of MaSCs in vivo
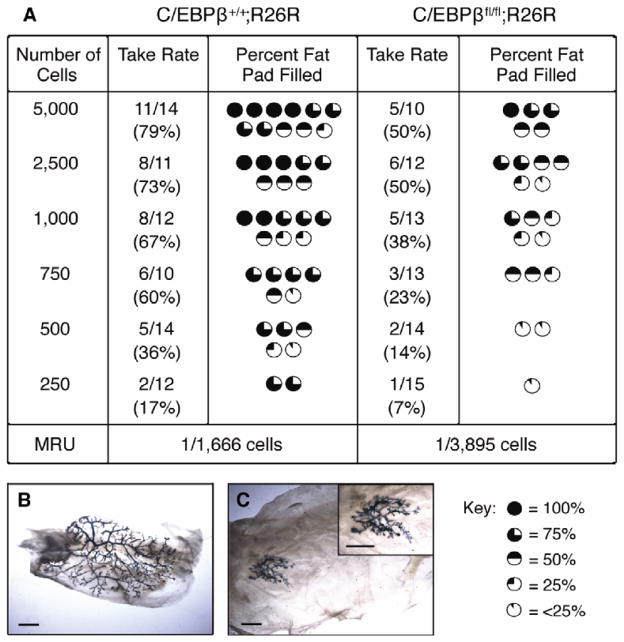
While mammosphere assays have emerged as a powerful surrogate method to evaluate stem/progenitor cells, previous studies demonstrated that as few as 15–30% of mammosphere cells contain regenerative stem cell activity [22]. Therefore, we performed functional limiting dilution transplantation experiments to determine the effects of C/EBPβ deletion on MaSC repopulating activity. MECs were isolated from wildtype (C/EBPβ+/+;R26R) or C/EBPβfl/fl;R26R mice, transduced with Ad.Cre and transplanted into the cleared fat pads of syngeneic hosts at decreasing cell numbers. At clonal cell numbers, the outgrowth potential of C/EBPβ-null MECs was significantly decreased as compared to wildtype (Fig. 3). Using a single-hit Poisson distribution, the mammary repopulating unit (MRU) was determined to be 1 stem cell per 1,666 cells in wildtype epithelium. In contrast, the frequency of MaSCs was decreased >2-fold in C/EBPβ-deleted outgrowths, with a calculated MRU of 1 in 3,895 (p=0.017). Importantly, the ability to completely reconstitute the entire mammary fat pad was severely reduced in outgrowths lacking C/EBPβ (p=0.0005). These findings suggest that ablation of C/EBPβ in mammary epithelia leads to both decreased MaSC repopulation activity as well as a reduction in MaSC frequency.
Figure 3.
Decreased MRUs in C/EBPβ-null outgrowths when transplanted at limiting dilution. MECs were isolated from wildtype (C/EBPβ+/+;R26R) and C/EBPβfl/fl;R26R glands, transduced with Ad.Cre1 and transplanted into syngeneic hosts at limiting dilutions. (A) Outgrowth potential was significantly decreased in C/EBPβ-deleted glands as compared to wildtype (p=0.017). The ability of C/EBPβ-null MECs to fill the fat pad was also significantly impaired as compared to wildtype (p=0.0005). Photomicrographs depict whole mount analysis of glands injected with 250 of wildtype (B) or C/EBPβ-null (C) cells. Scale bars, 5 mm.
C/EBPβ deletion results in premature senescence of MaSCs
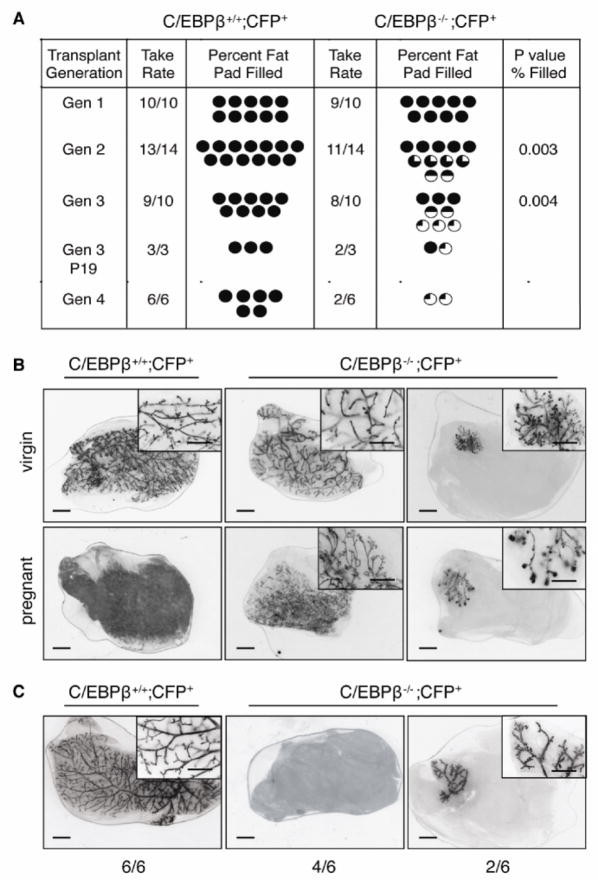
Previous studies have shown that decreased outgrowth potential and senescence occurs following 6–7 transplant generations [30], providing strong evidence for the existence of MaSCs with long-term repopulating activity that eventually become exhausted after multiple transplant generations. To further investigate MaSC repopulating activity, serial transplantation experiments were performed. For these experiments, mice harboring the germline deletion of C/EBPβ were bred to mice constitutively expressing CFP to permit visualization of the ductal epithelium. CFP+ epithelia from donor mice were carefully dissected and transplanted into 3-week old SCID/beige recipient mice. After 8 weeks of outgrowth, CFP+ glands were analyzed and cells at the leading edge of the outgrowths were re-transplanted for subsequent generations. As expected, transplanted tissue from wildtype (C/EBPβ+/+CFP+) mice completely filled the available fat pads of recipient mice when sequentially transplanted for three generations (Fig. 4). Surprisingly, the number of outgrowths remained unchanged between wildtype and C/EBPβ−/−CFP+ outgrowths. However, the ability of C/EBPβ−/− tissue to fill recipient fat pads was significantly decreased in secondary grafts (p=0.003), although ductal outgrowth was variable, reconstituting 50–100% of recipient fat pads. By the third generation, outgrowth potential was severely impaired, as half of the ductal outgrowths did not surpass 25% of the available fat pads (p=0.004). Further, using a generalized linear model of statistical analysis, the decreasing rate of the percentages of fat pad filled was also significantly reduced in C/EBPβ −/− CFP+ outgrowths (p=0.002). Third generation transplants were also examined in late pregnant recipients to determine whether pregnancy affected outgrowth potential. In glands that demonstrated complete outgrowth, lobuloalveolar development was impaired to a similar extent to that reported previously [11]. Importantly, impaired ductal outgrowth was also observed in these mice, indicating that pregnancy did not rescue the decreased ability of C/EBPβ−/− CFP+ cells to reconstitute the mammary fat pad (Fig. 4A,B).
Figure 4.
Decreased outgrowth of C/EBPβ−/− serial transplants. CFP+ epithelia were dissected from wildtype (C/EBPβ+/+;CFP+) or germline C/EBPβ−/−; CFP+ glands and transplanted sequentially for 3 consecutive generations, harvesting tissue at each generation after 8 weeks of outgrowth. (A) While take rate remained similar between wildtype and C/EBPβ−/−; CFP+ tissue, the ability to reconstitute the fat pad significantly decreased with subsequent generations (p=0.002) in C/EBPβ-null glands as compared to wildtype (Gen 2 p=0.003, Gen 3 p=0.004). (B) Fluorescent micrographs (inverted images) depict representative images of generation 3 outgrowths of wildtype and C/EBPβ−/−; CFP+ glands in mature virgin mice (top) or late pregnant (P19) glands (bottom). (C) Images depict generation 4 outgrowths from wildtype and stunted C/EBPβ−/− CFP+ donor glands. The number of outgrowths represented by each micrograph is depicted below each image. Scale bars, 5 mm.
To further address whether C/EBPβ-null MaSCs were undergoing early senescence, generation three stunted outgrowths from C/EBPβ−/− CFP+ mammary glands were transplanted for an additional generation. In four out of six recipient mammary fat pads, C/EBPβ-null epithelium failed to grow, suggesting the exhaustion of MaSCs (Fig. 4C). Notably, two transplanted mammary glands recapitulated stunted outgrowths, suggesting that C/EBPβ may regulate progenitor cell proliferation and/or differentiation. As expected, transplanted wildtype tissue resulted in complete outgrowths in all recipient mice. These findings indicate that deletion of C/EBPβ results in premature senescence of MaSCs, and are consistent with a role for C/EBPβ in both MaSC expansion and progenitor cell function.
C/EBPβ−/− mammary glands contain fewer luminal progenitor cells
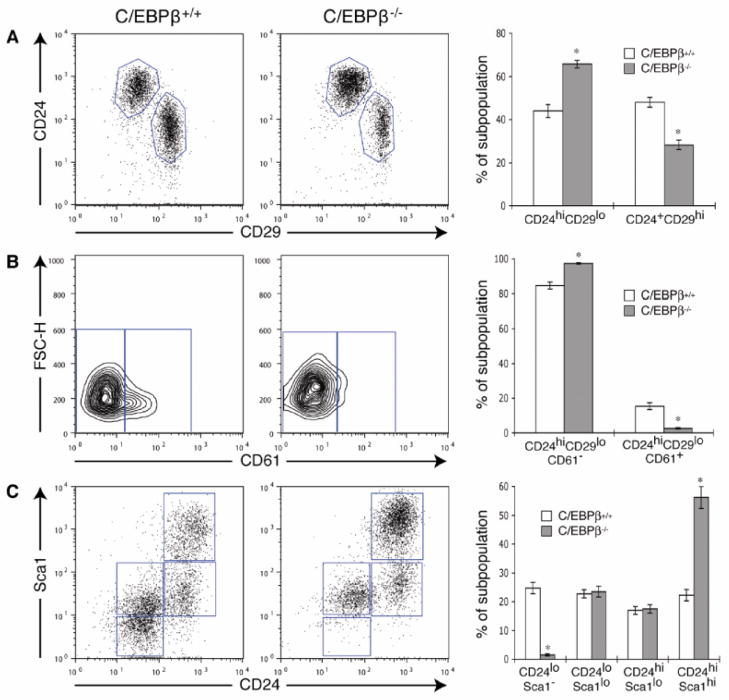
Our results suggest that either germline or conditional deletion of C/EBPβ results in a change in the frequency of stem and progenitor cells in the adult mammary gland. Therefore, we next used flow cytometry to examine various stem/progenitor cell populations in wildtype and C/EBPβ−/− MECs using previously well-defined markers. The LIN−CD24+CD29hi subpopulation, which was previously shown to contain MRUs [2], decreased nearly 2-fold in C/EBP was β−/− cells, while the LIN−CD24hiCD29lo subpopulation was increased as compared to wildtype (Fig. 5A). The latter subpopulation has been shown to contain luminal progenitor cells that are characterized by CD61 (β3 integrin) expression [1]. Surprisingly, the CD61 luminal progenitor population was markedly depleted in the C/EBPβ−/− MECs (14% to 3%), while the majority of the cells were CD61−, suggesting a switch to a more committed luminal cell (Fig. 5B). Likewise, an increase in LIN−CD24hiSca1hi cells, which has been shown to represent a more differentiated population that lacks significant outgrowth potential [3], was also observed, while the LIN−CD24loSca1− subpopulation was lost (Fig. 5C). We also examined C/EBPβ expression in the various stem/progenitor cell populations in wildtype mice. Intriguingly, C/EBPβ mRNA was enriched in LIN−CD24loSca1− cells, further demonstrating an important role for C/EBPβ in this subpopulation (Fig. S1). Collectively, these results suggest that when C/EBPβ is deleted, there is a depletion in MRUs, a decrease in luminal progenitors, and an accumulation of more differentiated luminal cells.
Figure 5.
Altered stem/progenitor cell populations in C/EBPβ−/− mammary glands. (A) Dot plots depict that the percentage of LIN−CD24+CD29hi cells, which contain MRUs, is decreased in C/EBPβ−/− MECs, while the LIN−CD24hiCD29lo subpopulation is increased. (B) The LIN−CD24hiCD29lo subpopulation was gated and CD61 expression was examined within this subpopulation. C/EBPβ−/− MECs express a lower percentage of LIN−CD24hiCD29loCD61+ cells as compared to wildtype (C/EBPβ+/+). (C) The LIN−CD24hiSca1hi subpopulation is increased in C/EBPβ−/− glands as compared to wildtype, which is accompanied by the loss of LIN−CD24loSca1− cells. For all FACS plots, lineage-positive cells were excluded using a mouse lineage panel kit plus biotin-conjugated CD31 and CD140a antibodies. Each dot/contour plot represents 1 experiment, and bar graphs depict the mean ± SEM of 4 independent experiments (*p<0.0001).
Visvader and colleagues previously demonstrated that LIN−CD24hiCD29loCD61+ cells are enriched for colony forming ability, while CD61− cells have a limited ability to form colonies on Matrigel or fibroblast feeder layers [1]. Therefore, we next tested whether the loss of CD61+ cells in C/EBPβ−/− MECs correlates with decreased colony forming ability. MECs were isolated from either wildtype or C/EBPβ−/−; CFP+ mice and plated on fibroblast feeder layers for 10 days. As expected, colony formation was significantly decreased in C/EBPβ−/− MECs as compared to wildtype (Fig. 6). Notably, colonies derived from C/EBPβ−/−; CFP+ cells appeared to be smaller than wildtype colonies. These results support the notion that CD61− MECs have decreased colony forming ability, and suggest that CD61+ luminal progenitor cells are lost in C/EBPβ−/− mammary glands.
Figure 6.
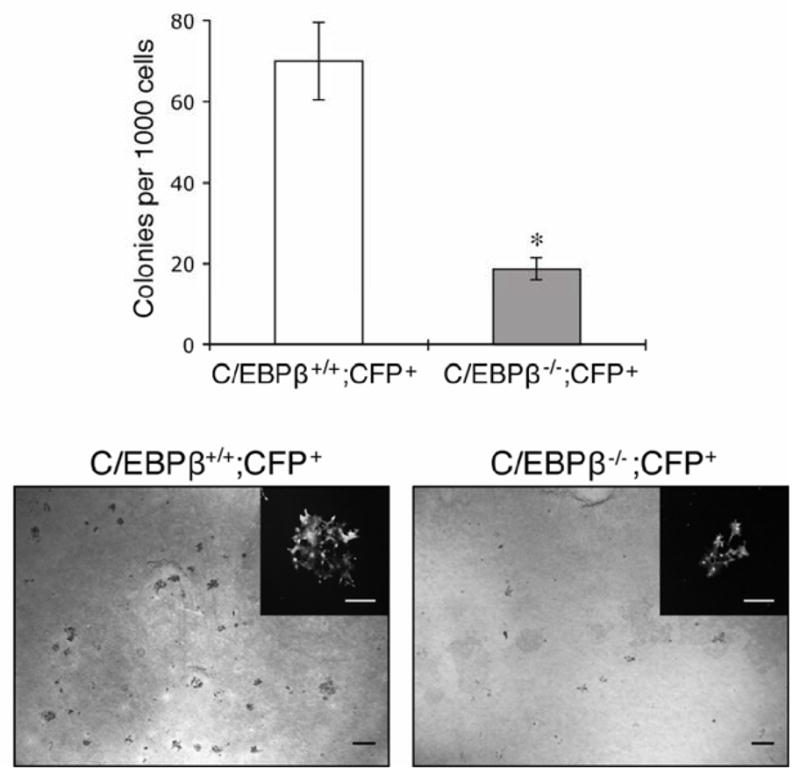
Decreased colony formation in C/EBPβ-null MECs. Colony forming ability on cultured feeder layers was significantly decreased in C/EBPβ−/−;CFP+ MECs as compared to wildtype (C/EBPβ+/+;CFP+). Data represent mean ± SEM of 3 independent experiments (*p<0.0001). Images depict crystal violet-stained colonies in wildtype and C/EBPβ−/−;CFP+ cells, and insets demonstrate a single CFP+ colony. Scale bars, 5 mm (large panels) and 250 μm (insets).
Disruption of luminal cell fate in C/EBPβ mammary glands
To identify potential signaling pathways regulated by C/EBPβ in stem/progenitor cells, microarray analysis was performed. For this analysis, subpopulations defined by LIN−CD24+CD29hi and LIN−CD24hiCD29lo were FACS-sorted wildtype and C/EBP from β −/− glands, and RNA was isolated from each group. A heat map was generated using supervised clustering of genes significant (ANOVA p<0.001, fold change >1.4) in C/EBPβ−/− cells as compared to wildtype in each subpopulation (Fig. S2). There were 181 common probe sets (ps) identified that were differentially expressed either of these subpopulations in C/EBP for β −/− MECs as compared to wildtype (Table S3). Interestingly, numerous members of both Notch and Wnt/β-catenin signaling were significantly changed in C/EBPβ−/− subpopulations. A small group of selected genes previously shown to be involved in cell fate and commitment were validated by qPCR (Table 1).
Table 1.
Real-time PCR validation of selected genes
| Gene Name | Molecular Pathway/Function | Fold Change in LIN− CD24hiCD29lo subpopulationa | Fold Change in LIN−CD24+CD29hi subpopulationa |
|---|---|---|---|
| Pre-B-cell leukemia homeobox-1 (Pbx-1) | Pluripotency, maintenance of hematopoiesis | + 14.6 ± 0.2 | |
| Keratin 15 (Krt15) | Epidermal stem cells | + 9.0 ± 0.2 | |
| Keratin 5 (Krt5) | Basal cells | + 1.9 ± 0.2 | |
| Stem cell antigen-1 (Sca-1) | Progenitor cell differentiation | − 1.5 ± 0.1 | − 2.0 ± 0.2 |
| Transgelin (Tagln) | Actin binding, invasion | − 15.9 ± 0.1 | − 2.2 ± 0.5 |
| Eyes absent-1 homolog (Eya-1) | Cell fate commitment and differentiation | + 5.2 ± 0.3 | + 51.0 ± 0.2 |
| ΔNp63 | Mammary basal cell differentiation | + 3.3 ± 0.3 | − 6.0 ± 0.2 |
| Notch 3 | Stem cell self-renewal, luminal cell commitment | − 2.5 ± 0.3 |
Mean ± sd of the mRNA fold change of the selected genes in the C/EBPβ−/− subpopulations as compared to wildtype.
The LIN−CD24+CD29hi subpopulation that contains MRUs is comprised primarily of ER− basal cells, characterized by K5 and K14 expression [31, 32]. Interestingly, these basal markers were slightly upregulated in the LIN−CD24hiCD29lo subpopulation of C/EBPβ−/− cells (Table S3). In agreement, p63, which regulates basal cell differentiation in the mammary gland, was downregulated in the LIN−CD24+CD29hi cells and increased in the LIN−CD24hiCD29lo luminal subpopulation of C/EBPβ−/− glands. K15, a known epidermal basal stem cell marker, was also increased in the luminal subpopulation of C/EBPβ−/− cells, indicating a change in cell fate (Table 1). These findings illustrate that there is aberrant expression of basal cell markers in the luminal cell population of C/EBPβ−/− mice, indicating that luminal cell fate is disrupted.
Discussion
While considerable progress has been made in defining MaSCs, the precise genetic mechanisms that regulate stem/progenitor cell self-renewal, maintenance and differentiation remain ill-defined. Recent studies have begun to define a role for several critical transcription factors, such as Gata-3, Elf5 and promyelocytic leukemia protein (PML), in the MaSC hierarchy. Here, we demonstrate an important role for C/EBPβ in both stem cell repopulation activity and luminal cell lineage commitment.
To investigate whether C/EBPβ regulates mammary stem/progenitor cell self-renewal, we utilized a combination of in vitro mammosphere assays as well as in vivo functional limiting dilution transplantation techniques. The results of these studies collectively suggest that C/EBPβ mediates MaSC expansion. These studies were extended by performing serial transplantation experiments. The ability of mammary epithelium to reconstitute the fat pad upon multiple serial transplantations provides strong evidence for the existence of a population of cells that contain high regenerative potential in the adult mammary gland [30]. Although this method provides a powerful tool for understanding regenerative MaSC ability and maintenance [33], very few studies have utilized this technique to analyze the effect of specific genes on repopulating activity. Here, we show that the outgrowth potential of C/EBPβ−/− tissue progressively and significantly decreased with each generation as compared to wildtype, resulting in nearly a complete loss of outgrowth (Fig. 4). While it is not possible to distinguish between symmetric and asymmetric division at present, our results suggest that deletion of C/EBPβ impedes MaSC repopulation activity, stem cell maintenance, and causes premature senescence. C/EBPβ was recently shown to mediate oncogene-induced senescence of Ras-transformed mouse fibrobasts, which required the loss of p19Arf [34]. As p16Ink4a/p19Arf has been shown to be involved in Bmi1 regulation of MaSCs [35], it will be of interest to determine if p19Arf also mediates MaSC senescence as observed in C/EBPβ−/− mammary glands.
Upon serial transplantation, a proportion of the cells retained the ability to completely reconstitute the mammary fat pad in second and third transplant generations. This effect may be due to compensatory mechanisms by other transcription factors, including other C/EBP family members. For example, C/EBPβ and Runx2 synergistically cooperate to promote osteogenesis in the bone. However, bone defects are not observed in C/EBPβ−/− mice, suggesting that functional redundancy of other C/EBP family members may be important [8, 36]. As other C/EBPs are expressed in the mammary gland, including C/EBPα and C/EBPδ, it will be important to investigate the collaboration of other transcription factors with C/EBPβ as well as functional redundancy of the C/EBP family in the regulation of MaSCs.
Over the past five years, numerous putative hierarchical models that delineate mammary stem cell differentiation have been proposed [37–39]. The luminal progenitor cell has been postulated to be CD24hiCD29loCD61+ and to give rise to both ductal luminal and alveolar luminal cell lineages. The differentiation of these cells is thought to be regulated by Gata-3, as deletion of Gata-3 in the virgin mammary gland results in an accumulation of CD61+ cells and impaired ductal development [1]. This population was characterized by increased colony forming ability and the expression of K18. Furthermore, the percentage of CD61+ cells was reported to progressively decrease with age, and was nearly lost in pregnant glands, consistent with an undifferentiated luminal cell. Here we show that deletion of C/EBPβ results in the ablation of the CD24hiCD24loCD61+ subpopulation and a consequent increase in CD61− cells. One caveat to this analysis, and most of the currently published studies, is the possibility that C/EBPβ regulates the expression of these cell surface markers. To address this issue, conditionally-deleted MECs were cultured for 4 days and the expression of these markers was analyzed by FACS. The expression CD24, CD29, CD61 and Sca-1 remained unchanged when C/EBPβ was deleted, suggesting that C/EBPβ alone likely does not regulate these markers (data not shown). Collectively, these findings suggest that C/EBPβ is required for the generation and/or maintenance of CD61+ cells.
Impaired lobuloalveolar development is a phenotype shared among several mouse knockout models, including Gata-3, Stat5, Id2, PrlR, Elf5 and C/EBPβ. The alveolar lineage, comprised of both luminal and myoepithelial cells, has been suggested to arise from a common bipotent luminal progenitor cell. However, there is evidence for the existence of distinct ductal-limited and lobule-limited progenitor cells [40–42]. The luminal cell lineage can be subdivided based on ER and Sca1 expression, where Sca1− cells differentiate into milk-producing alveoli. This subpopulation was postulated to be ER−, and to express CD24, CD29, CD49f (α6 integrin), CD49b (α2 integrin), CD14 and CD61 [3, 38]. Watson and colleagues recently demonstrated a role for PML in ductal morphogenesis and luminal cell lineage commitment. In this study, the percentage of ER+ cells was markedly increased PML−/− mammary glands, as well as the proportion of CD24hiSca1+ cells [16]. In the present study, we show that the CD24loSca1− subpopulation was depleted in C/EBPβ−/− MECs, while the percentage of CD24hiSca1hi cells was increased 2.5-fold. The Sca1hi subpopulation, therefore, most likely represents the luminal ER+ progenitor cell previously described [3]. Accordingly, C/EBPβ−/− mammary glands display increased expression of ER+ cells in E+P-treated mice [10]. Thus, while we can speculate that the loss of CD24loSca1− cells in C/EBPβ−/− glands results in the loss of alveolar progenitors, causing a block in lobuloalveolar development, it is not yet possible to distinguish between ductal and alveolar progenitor cells with the currently existing mammary lineage markers.
Our results support the existence of a common progenitor (multipotent or bipotent) that requires C/EBPβ for luminal, and potentially alveolar, cell differentiation. Similar to Bmi1 [35] and the Notch pathway [43], C/EBPβ presumably acts on more than one stage of mammary stem cell differentiation: first, the expansion of MaSCs, and later, the generation of luminal progenitors. Alternatively, C/EBPβ may be required to maintain the luminal cell lineage, so that in the absence of C/EBPβ, MaSCs are exhausted and lose repopulating capacity. C/EBPβ is likely only one of a number of genes that coordinately regulates these processes. The precise definition of the cell lineages regulated by C/EBPβ will require the identification of new markers to distinguish luminal ductal and alveolar progenitors.
To identify potential C/EBPβ target genes that may be involved in MaSCs, gene profiling was performed on stem/progenitor cell populations in wildtype and C/EBPβ−/− mammary glands. Intriguingly, members of the Notch pathway were altered in the CD24+CD29hi subpopulation of C/EBPβ−/− cells as compared to wildtype. Notch 3, which was decreased in CD24+CD29hi cells of C/EBPβ−/− glands, was recently shown to be critical for the commitment of human bipotent progenitors to the luminal cell lineage [44]. In another study, Notch signaling was shown to restrict MaSC expansion and to instruct the fate of MaSCs to the luminal cell lineage [43]. The Notch pathway has also been shown to maintain luminal alveolar cell fate during pregnancy [45]. Deletion of RBP-J led to the expression of basal markers K14 and p63 in the luminal compartment during pregnancy. Our microarray data suggest that luminal cells may acquire basal cell characteristics in the absence of C/EBPβ, as K5, K14 and p63 were upregulated in the CD24hiCD29lo subpopulation. Additionally, RBP-J-deleted glands contained a large percentage of K6+ cells [45], similar to the phenotype observed in the glands of E+P-treated C/EBPβ−/− mice [12]. The misexpression of K6 in C/EBPβ−/− and RBP-J-null mammary glands may represent the inability of luminal progenitor cells to properly differentiate. Increasing evidence suggests that C/EBPβ can interact with the Notch pathway [46, 47]. Although it is unclear whether C/EBPβ and Notch regulate each other in the mammary gland, these proteins have been shown to mediate common downstream targets [48, 49]. These reports suggest the potential for cross-talk between the Notch pathway and C/EBPβ, although further experiments are required to test this postulate.
CONCLUSION
C/EBPβ is a master regulator of cell differentiation in numerous tissues, and its function in stem cell biology clearly extends beyond the mammary gland. C/EBPβ was reported to induce the transdifferentiation of committed pancreatic cells into hepatocytes [50] and was recently shown to be important in the development of leukemias in mice [51]. Thus, C/EBPβ is likely a key instructor of cell fate during numerous processes, such as hematopoiesis, osteogenesis, adipogenesis and hepatogenesis.
In conclusion, we provide definitive evidence that C/EBPβ is one of several critical transcription factors that specifies MaSC fate during mammary gland development. An understanding of the normal MaSC hierarchy will be critical for our understanding of the etiology of the multiple breast cancer subtypes. Increasing evidence from numerous tissues including the mammary gland suggests that normal stem/progenitor cells are targets for tumorigenesis; furthermore, C/EBPβ has been shown to be critical for Ras-induced carcinogenesis [52]. C/EBPβ is also misregulated in human breast cancer, especially those associated with poor prognosis. Altered expression of the C/EBPβ protein isoform LIP is associated with poorly differentiated, ER− breast cancers [53], and C/EBPβ has been shown to play a critical role in the TGFβ-mediated cytostatic switch required for breast cancer metastasis [54]. Elucidating the mechanism by which C/EBPβ regulates normal MaSCs may provide key insights into how these regulatory cues are altered during tumor development and progression.
Supplementary Material
Acknowledgments
This work was supported by the American Cancer Society Tricam Industries Postdoctoral Breast Cancer Fellowship (PF-06-252-01-MGO; H.L.L.) and a National Institutes of Health Grant (CA030195-22; J.M.R.).,
The authors would like to thank Drs. Peter Johnson, Esta Sterneck, Mike Lewis and Philippe Soriano for generously supplying various mouse strains for this study. We thank John Landua for technical assistance with confocal imaging, Rachel Atkinson for assistance with statistical analysis, and the Baylor College of Medicine Microarray Core for technical assistance. We thank Drs. Dan Medina, Cindy Zahnow and Jason Hershkowitz for scientific discussion and critical review of this manuscript. This work was supported by the BCM Cytometry and Cell Sorting Core with funding from the NIH (NCRR S10RR024574 and NCI P30CA125123), by the American Cancer Society Tricam Industries Postdoctoral Breast Cancer Fellowship (PF-06-252-01-MGO; H.L.L.) and a National Institutes of Health Grant (CA16303; J.M.R.).
Footnotes
The authors declare that there are no conflicts of interest.
Author contributions: H.L.L.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; A.P.V.: collection and assembly of data; C.J.C: data analysis and interpretation; H.L.: data analysis and interpretation; Y.Z.: data analysis and interpretation; F.B.: conception and design, data analysis and interpretation; J.M.R.: conception and design, financial support, final approval of manuscript.
See www.StemCells.com for supporting information available online.
References
- 1.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 2.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 3.Sleeman KE, Kendrick H, Robertson D, et al. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stingl J, Eaves CJ, Zandieh I, et al. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67:93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- 5.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 6.Eirew P, Stingl J, Raouf A, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 7.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 8.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Gigliotti AP, DeWille JW. Lactation status influences expression of CCAAT/enhancer binding protein isoform mRNA in the mouse mammary gland. J Cell Physiol. 1998;174:232–239. doi: 10.1002/(SICI)1097-4652(199802)174:2<232::AID-JCP10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 10.Grimm SL, Rosen JM. The role of C/EBPbeta in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:191–204. doi: 10.1023/a:1025900908026. [DOI] [PubMed] [Google Scholar]
- 11.Robinson GW, Johnson PF, Hennighausen L, et al. The C/EBPbeta transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 1998;12:1907–1916. doi: 10.1101/gad.12.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm SL, Seagroves TN, Kabotyanski EB, et al. Disruption of steroid and prolactin receptor patterning in the mammary gland correlates with a block in lobuloalveolar development. Mol Endocrinol. 2002;16:2675–2691. doi: 10.1210/me.2002-0239. [DOI] [PubMed] [Google Scholar]
- 13.Seagroves TN, Krnacik S, Raught B, et al. C/EBPbeta, but not C/EBPalpha, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 1998;12:1917–1928. doi: 10.1101/gad.12.12.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seagroves TN, Lydon JP, Hovey RC, et al. C/EBPbeta (CCAAT/enhancer binding protein) controls cell fate determination during mammary gland development. Mol Endocrinol. 2000;14:359–368. doi: 10.1210/mend.14.3.0434. [DOI] [PubMed] [Google Scholar]
- 15.Kouros-Mehr H, Slorach EM, Sternlicht MD, et al. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Ferguson BJ, Khaled WT, et al. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc Natl Acad Sci U S A. 2009;106:4725–4730. doi: 10.1073/pnas.0807640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oakes SR, Naylor MJ, Asselin-Labat ML, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterneck E, Zhu S, Ramirez A, et al. Conditional ablation of C/EBP beta demonstrates its keratinocyte-specific requirement for cell survival and mouse skin tumorigenesis. Oncogene. 2006;25:1272–1276. doi: 10.1038/sj.onc.1209144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 21.Welm BE, Dijkgraaf GJ, Bledau AS, et al. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moraes RC, Zhang X, Harrington N, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 23.Rijnkels M, Rosen JM. Adenovirus-Cre-mediated recombination in mammary epithelial early progenitor cells. J Cell Sci. 2001;114:3147–3153. doi: 10.1242/jcs.114.17.3147. [DOI] [PubMed] [Google Scholar]
- 24.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deome KB, Faulkin LJ, Jr, Bern HA, et al. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 26.Wagner KU, Wall RJ, St-Onge L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonnefoix T, Bonnefoix P, Verdiel P, et al. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. J Immunol Methods. 1996;194:113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
- 28.Bonnefoix T, Bonnefoix P, Callanan M, et al. Graphical representation of a generalized linear model-based statistical test estimating the fit of the single-hit Poisson model to limiting dilution assays. J Immunol. 2001;167:5725–5730. doi: 10.4049/jimmunol.167.10.5725. [DOI] [PubMed] [Google Scholar]
- 29.Dontu G, Wicha MS. Survival of mammary stem cells in suspension culture: implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:75–86. doi: 10.1007/s10911-005-2542-5. [DOI] [PubMed] [Google Scholar]
- 30.Daniel CW, Young LJ, Medina D, et al. The influence of mammogenic hormones on serially transplanted mouse mammary gland. Exp Gerontol. 1971;6:95–101. doi: 10.1016/0531-5565(71)90053-2. [DOI] [PubMed] [Google Scholar]
- 31.Asselin-Labat ML, Shackleton M, Stingl J, et al. Steroid hormone receptor status of mouse mammary stem cells. J Natl Cancer Inst. 2006;98:1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]
- 32.Taddei I, Deugnier MA, Faraldo MM, et al. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat Cell Biol. 2008;10:716–722. doi: 10.1038/ncb1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paguirigan A, Beebe DJ, Alexander CM. Simulating mouse mammary gland development: cell ageing and its relation to stem and progenitor activity. Cell Prolif. 2007;40:106–124. doi: 10.1111/j.1365-2184.2007.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebastian T, Johnson PF. RasV12-mediated down-regulation of CCAAT/enhancer binding protein beta in immortalized fibroblasts requires loss of p19Arf and facilitates bypass of oncogene-induced senescence. Cancer Res. 2009;69:2588–2598. doi: 10.1158/0008-5472.CAN-08-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pietersen AM, Evers B, Prasad AA, et al. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18:1094–1099. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- 36.Hata K, Nishimura R, Ueda M, et al. A CCAAT/enhancer binding protein beta isoform, liver-enriched inhibitory protein, regulates commitment of osteoblasts and adipocytes. Mol Cell Biol. 2005;25:1971–1979. doi: 10.1128/MCB.25.5.1971-1979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaMarca HL, Rosen JM. Minireview: hormones and mammary cell fate--what will I become when I grow up? Endocrinology. 2008;149:4317–4321. doi: 10.1210/en.2008-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009;217:229–241. doi: 10.1002/path.2457. [DOI] [PubMed] [Google Scholar]
- 39.Visvader JE, Lindeman GJ. Mammary stem cells and mammopoiesis. Cancer Res. 2006;66:9798–9801. doi: 10.1158/0008-5472.CAN-06-2254. [DOI] [PubMed] [Google Scholar]
- 40.Alvi AJ, Clayton H, Joshi C, et al. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 2003;5:R1–8. doi: 10.1186/bcr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Booth BW, Boulanger CA, Smith GH. Alveolar progenitor cells develop in mouse mammary glands independent of pregnancy and lactation. J Cell Physiol. 2007;212:729–736. doi: 10.1002/jcp.21071. [DOI] [PubMed] [Google Scholar]
- 42.Wagner KU, Boulanger CA, Henry MD, et al. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 43.Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Raouf A, Zhao Y, To K, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 45.Buono KD, Robinson GW, Martin C, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 46.Chen H, Chong Y, Liu CL. Active intracellular domain of Notch enhances transcriptional activation of CCAAT/enhancer binding protein beta on a rat pregnancy-specific glycoprotein gene. Biochemistry. 2000;39:1675–1682. doi: 10.1021/bi991786k. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Pasolli HA, Williams T, et al. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol. 2008;183:37–48. doi: 10.1083/jcb.200804030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustafson TL, Wellberg E, Laffin B, et al. Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPbeta. Oncogene. 2009;28:1561–1568. doi: 10.1038/onc.2008.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Y, Puzhko S, Wabitsch M, et al. Transcriptional regulation of the human growth hormone receptor (hGHR) gene V2 promoter by transcriptional activators and repressor. Mol Endocrinol. 2009;23:373–387. doi: 10.1210/me.2008-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tosh D, Shen CN, Slack JM. Conversion of pancreatic cells to hepatocytes. Biochem Soc Trans. 2002;30:51–55. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 51.Costinean S, Sandhu SK, Pedersen IM, et al. Ship and C/ebp{beta} are targeted by miR-155 in B cells of E{micro}-miR-155 transgenic mice. Blood. 2009 doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu S, Yoon K, Sterneck E, et al. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahnow CA, Younes P, Laucirica R, et al. Overexpression of C/EBPbeta-LIP, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J Natl Cancer Inst. 1997;89:1887–1891. doi: 10.1093/jnci/89.24.1887. [DOI] [PubMed] [Google Scholar]
- 54.Gomis RR, Alarcon C, Nadal C, et al. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 55.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Creighton CJ, Casa A, Lazard Z, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.