Abstract
In Treponema pallidum, homopolymeric guanosine repeats of varying length are present upstream of both Subfamily I (tprC, D, F and I) and II (tprE, G and J) tpr genes, a group of potential virulence factors, immediately upstream of the +1 nucleotide. To investigate the influence of these poly-G sequences on promoter activity, tprE, G, J, F and I promoter regions containing homopolymeric tracts with different numbers of Gs, the ribosomal binding site and start codon were cloned in frame with the green fluorescent protein reporter gene (GFP), and promoter activity was measured both as fluorescence emission from Escherichia coli cultures transformed with the different plasmid constructs and using quantitative RT-PCR. For tprJ, G and E-derived clones, fluorescence was significantly higher with constructs containing eight Gs or fewer, while plasmids containing the same promoters with none or more Gs gave modest or no signal above the background. In contrast, tprF/I-derived clones induced similar levels of fluorescence regardless of the number of Gs within the promoter. GFP mRNA quantification showed that all of the promoters induced measurable transcription of the GFP gene; however, only for Subfamily II promoters was message synthesis inversely correlated to the number of Gs in the construct.
Keywords: syphilis, tpr genes, transcriptional regulation
Introduction
The genome of Treponema pallidum ssp. pallidum, Nichols strain, includes the T. pallidum repeat (tpr) family of 12 paralogs (Fraser et al., 1998). This gene family comprises ~2% of the 1.138-Mb T. pallidum genome and is divided in three subfamilies based upon predicted amino acid homology: Subfamily I (tprC, D, F and I), Subfamily II (tprE, G and J) and Subfamily III (tprA, B, H, K and L). In the rabbit model of syphilis infection, initial semiquantitative RT-PCR studies (Centurion-Lara et al., 1999) followed by densitometric analysis of the tpr messages after limiting dilution RT-PCR (Hazlett et al., 2001) suggested differential expression of the tpr genes. Specifically, mRNAs for Subfamilies I and II members were weakly and variably detected (Centurion-Lara et al., 1999; Hazlett et al., 2001) with respect to Subfamily III tprs. More recent microarray and real time RT-PCR studies in the Nichols, Chicago, Bal 73-1 and Sea 81-4 isolates also support differential expression of the tpr genes both within and among T. pallidum strains (Smajs et al., 2005; Giacani et al., 2007). Leader et al. (2003), analyzing the antibody responses against the Tpr antigens in the rabbit model, showed variability in the presence, time of appearance and patterns of antibody reactivity against each Tpr in rabbits infected with the Nichols, Chicago or Bal 73-1 isolates. For example, a weaker and delayed reactivity was reported toward most of the Tprs in Nichols-infected rabbits compared with Bal 73-1- and Chicago-infected animals. Differential reactivity patterns were shown also for Tpr antigens (e.g. TprF and TprI) whose sequences are invariant in all syphilis isolates examined so far (Sun et al., 2004); the absence of antigenic differences among these Tprs suggests that differences in antibody reactivity might be due to differential gene expression among these isolates.
Although computer predictions localize TprA, C, D, F, I, J and K to the outer membrane (http://www.psort.org/psortb/), the definitive cellular localizations of the Tpr antigens remain controversial (Hazlett et al., 2001; Giacani et al., 2005b), and their function is still unknown. Strong antibody and T-cell responses to TprF/TprI during infection (Leader et al., 2003; Sun et al., 2004) and protection experiments in the rabbit model with recombinants encompassing the TprF/TprI amino terminal region (immunization significantly alters lesion development after homologous challenge, although it does not prevent infection) (Sun et al., 2004; Giacani et al., 2005b) strongly suggest that these proteins play a significant role in the immune response to T. pallidum and are probably involved in syphilis pathogenesis.
In bacterial pathogens, homopolymeric repeats located within or in the proximity of promoter regions are known to regulate expression of a variety of genes coding for virulence determinants and surface antigens (van der Ende et al., 1995; Saunders et al., 1998; van der Woude & Baumler, 2004). Variation in the length of these homopolymeric tracts is a well-recognized mechanism for controlling amount of message and ON–OFF states of transcription. During DNA replication or repair, these repeats can expand or contract via a phenomenon known as slipped-strand mispairing (SSM). Because misalignment can occur on either the leading or the lagging strands, SSM can respectively result in an increase or decrease of the number of repeated nucleotides in the new DNA molecule (Levinson & Gutman, 1987). Classic examples of modulation of gene expression by single nucleotide repeats are the porA and opc loci of Neisseria meningitidis, in which variation in the length of the poly-G and poly-C repeats in their promoters induces high, moderate or no expression of these genes (Sarkari et al., 1994; van der Ende et al., 1995; Arhin et al., 1998), as described below in more detail.
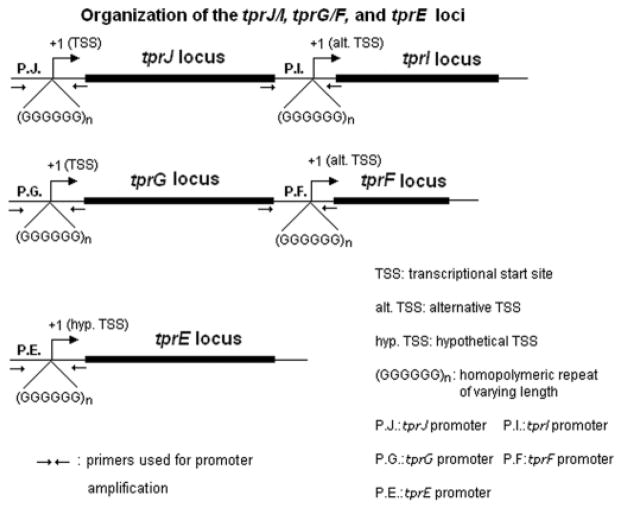
Analysis of the genome sequence of the syphilis spirochete (Nichols strain) (Fraser et al., 1998; Weinstock et al., 1998; Giacani et al., 2005a) revealed the presence of homopolymeric guanosine (G) repeats in the promoter regions of Subfamily I (tprC, D, F and I) and II (tprE, G and J) tprs, located immediately upstream of the tpr transcriptional start sites (TSSs) (Giacani et al., 2005a). These poly-G repeats vary in length within and among treponemal isolates and, by analogy, are likely to modulate transcription of the tpr genes by influencing promoter activity; homopolymeric repeats are in the range 8–12 G residues for tprE, 8–11 for tprG, 7–11 for tprJ, and 8–10 for tprF and I. In the Nichols strain, tprG and F are members of the same operon, as are tprJ and I (Fig. 1) (Giacani et al., 2005a), with tprF and tprI located immediately downstream of tprG and tprJ, respectively. Transcriptional analysis (Giacani et al., 2005a) of these loci revealed that tprG/F and tprJ/I are cotranscribed from promoters located upstream of tprG and J, respectively. Additionally, internal promoters upstream of both tprI and tprF (Fig. 1) can potentially generate suboperonic messages (Giacani et al., 2005a). The aim of this study was to investigate the activity of tprE, G, J, F and I promoters containing homopolymeric tracts with different numbers of G residues. Because of the inability to grow T. pallidum outside of a living host and the lack of genetic systems for this spirochete, we examined the role of the poly-G repeats in transcriptional regulation of tprE, G, J, tprF and I using a Cycle 3 green fluorescent protein (GFP) reporter system in Escherichia coli, and by quantitative real time RT- PCR. Our findings indicate that changes in the length of the poly-G repeats modulate the activity of promoters upstream of the tprE gene and the tprJ/I and G/F operons, resembling mechanisms of transcriptional regulation reported for other bacterial pathogens. In contrast, activity of the internal promoters (upstream of tprF and I) is not influenced by changes in the poly-G region. Overall agreement between promoter activity as determined by our GFP reporter system and transcriptional analysis of the tpr genes in T. pallidum during experimental infection suggests that poly-G regions are likely to be functional in vivo.
Fig. 1.
Schematic representation of the tprJ, tprI, tprG, tprF and tprE loci in the Nichols strain of Treponema pallidum. Location of the TSSs and poly-Gs is indicated along with the position of the primers used for promoter amplification (for promoter sequence see Fig. 2, for primer sequence see Fig. 2 or Table 1). As previously reported (Giacani et al., 2005a) transcriptional analysis of these loci showed that promoters upstream of tprJ and tprG (P.J and P.G in the figure) generate messages encompassing both tprJ/I and tprG/F sequences, while alternative promoters upstream of tprF and tprI (P.F and P.I in the figure) may generate suboperonic messages. The tprE TSS has not been identified yet using in vitro transcription assay or 5′-RACE (rapid amplification of cDNA ends) as for tprJ, I, G and F (Giacani et al., 2005a); therefore, its TSS is hypothetical. However, it has been mapped using RT-PCR analysis within the first 250 nucleotides upstream of the tprE start codon. The 3′ boundaries of these transcription units are still unknown.
Materials and methods
Treponema pallidum strain propagation and nucleic acid extraction
Treponema pallidum ssp. pallidum Nichols, Sea 81-4, Chicago and Bal 73-1 strains were propagated in New Zealand white rabbits as previously reported (Lukehart et al., 1980). The Nichols strain was provided by James N. Miller (University of California, Los Angeles) in 1979. The Sea 81-4 strain was isolated in 1981 at Harborview Medical Center (Seattle, WA) from a primary chancre, and Chicago and Bal 73-1 strains were supplied by Paul Hardy and Ellen Nell, (Johns Hopkins University, Baltimore, MD). For DNA and RNA extraction, rabbits were infected with 5 × 107 T. pallidum cells per testicle (Nichols, Chicago and Bal 73-1 strains) and 8 × 106 organisms for Sea 81-4. Treponemes were harvested at peak orchitis from infected rabbit testes at day 10 (Nichols and Chicago strains), day 20 (Bal 73-1 strain) or day 25 (Sea 81-4 strain) postinfection. Collected organisms were separated from host cellular gross debris by low-speed centrifugation (250 g for 10 min at room temperature); the supernatants were spun in a microcentrifuge for 30 min at 12 000 g at 4 °C. Pellets were resuspended in 200 μL of 1 × lysis buffer (10 mM Tris, pH 8.0; 0.1 M EDTA; 0.5% sodium dodecyl sulfate) if intended for DNA isolation or 400 μL of Ultraspec buffer (Biotecx Laboratories Inc., Houston, TX) if intended for RNA isolation. DNA extraction was performed as previously reported (Centurion-Lara et al., 1996) using the QIAamp DNA Mini Kit (Qiagen Inc., Chatsworth, CA). Detailed protocols for RNA extraction and DNaseI treatment to obtain DNA-free RNA samples were also as previously reported (Giacani et al., 2007).
Amplification and cloning
To determine the degree of variation in length of the poly-G tracts among and within strains, regions of c. 300 bp encompassing the tprG and J promoters were amplified, cloned and sequenced. Amplicons for tprF and I promoters were c. 500 bp long (Table 1). Transcriptional analysis (data not shown) to determine the presence of mRNA spanning the 5′-flanking region of tprE was performed using the same approach previously used for tprG and J (qualitative RT-PCR; Giacani et al., 2005a, b) and allowed us to identify an untranscribed putative promoter region (containing a poly-G tract) which was also cloned and sequenced (Table 1 and Fig. 2). All PCR amplifications were performed in 50 μL reactions containing 200 μM each dNTP, 20 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 50 mM KCl, 400 nM of each primer, and 2.5 U of Taq DNA Polymerase (Promega, Madison, WI) with c. 100 ng of DNA template in each reaction. Cycling conditions were denaturation for 5 min at 95 °C, followed by 1 min at 95 °C, annealing for 1 min at 60 °C and extension for 1 min at 72 °C for a total of 45 cycles. Final extension was for 10 min at 72 °C. Products were evaluated on 2% agarose gels and then cloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Twenty clones containing inserts were extracted for each promoter for each strain using the Qiagen Plasmid Mini Kit (Qiagen) and both strands sequenced with the Applied Biosystems dye terminator sequencing kit (Perkin-Elmer, Foster City, CA). Primers and amplicon sizes are listed in Table 1.
Table 1.
Primers used in this study
| Purpose | Name | Sequence (5′–3′) | Size (bp)* |
|---|---|---|---|
| Cloning into pCRII-TOPO-vector† | tprG-For | CGCGTACCCACTTCTCTCTC | 287 |
| tprJ-For | AAAGAAAAAGGATTTCCGCA | 284 | |
| tprE-For | CGGGATGAGGATCAGTTTAC | 306 | |
| tprE/G/J-Rev | GTGGCAGAGCCAGTTAGCTT | ||
| tprF-For | GTGCGTGGCATTCAGGAAAA | 519 | |
| tprI-For | CCTTCCGGCGGTTCCTC | 504 | |
| tprF/I-Rev | ACTGACCTGCGGAGTGAGTA | ||
| Cloning into pGlow-TOPO-vector† | tprG-For | CGCGTACCCACTTCTCTCTC | 247 |
| tprJ-For | AAAGAAAAAGGATTTCCGCA | 244 | |
| tprG/J-Glow-Rev | TCATTCCCCCGCTCGCTCCT | ||
| tprE-For | CGGGATGAGGATCAGTTTAC | 266 | |
| tprE-Glow-Rev | TCATTCCCCCGCTCGCTCCTA | ||
| tprF/I-For | CTGGTATTTCAGCCAGTCT | 209 | |
| tprF/I-Glow-Rev | TCATTCACTCCCCCTCCT | ||
| t47-Sense | CGTGTGGTATCAACTATGG | 313 | |
| t47-Antisense | TCAACCGTGTACTCAGTGC | ||
| lacPro-Sense | GTGAGCGCAACGCAATTAAT | 130 | |
| lacPro-Antisense | TCATAGCTGTTTCCTGTGTG | ||
| Sequencing | M13 Rev‡ | CAGGAAACAGCTATGAC | |
| M13 For‡ | GTAAAACGACGGCCAG | ||
| GFP Rev§ | GGGTAAGCTTTCCGTATGTACG | ||
| T7 promoter§ | TAATACGACTCACTATAGGG | ||
| RT-PCR/real-time PCR | GFP-Sense | CTACCTGTTCCATGGCCAAC | 262 |
| GFP-Antisense¶ | TGTGTCCGAGAATGTTTCCA | ||
| Ampr-Sense | GCTATGTGGCGCGGTATTAT | 223 | |
| Ampr-Antisense¶ | GGTTAGCTCCTTCGGTCCTC | ||
| t47-Sense | CGTGTGGTATCAACTATGG | 313 | |
| t47-Antisense | TCAACCGTGTACTCAGTGC | ||
| tprE-Sense | CGGCAAAGTCCTGTTCGGCAA | 358 | |
| tprE-Antisense | GCTCAACACGCTGTCGTATAGTA | ||
| tprG-Sense | GAAGGTGTTCATTACCGACCCT | 359 | |
| tprG-Antisense | TTGTAGCCTCAGCCGTAAGCTT | ||
| tprJ-Sense | TCTTCACACCCCGCAGGGAA | 364 | |
| tprJ-Antisense | CGTTATTTCCGTTCGCATCATC | ||
| Sea 81-4|| tprJ-Sense | CCTACAATGTCGCCTTCGAT | 277 | |
| tprJ-Antisense | CAATGTGCCCTGAGGAACC |
Amplicon sizes were calculated according to the genome sequence of the Nichols strain (Fraser et al., 1998), without taking into account variability within the poly-G repeats.
In each set of grouped primers, different forward (For) primers were used with the same reverse (Rev) primer.
Used to sequence inserts in the pCRII-TOPO vector (Invitrogen).
Used to sequence inserts in the pGlow-TOPO vector (Invitrogen).
This primer was used both as gene-specific primer for reverse transcription and as reverse primer for PCR reactions.
The Sea 81-4 strain carries a hybrid tprG/J allele in the tprJ locus (Giacani et al., 2005a, b).
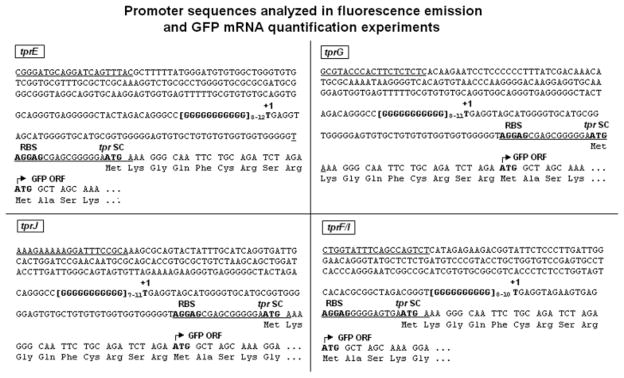
Fig. 2.
tprE, G, J and F/I promoter sequences cloned into the pGlow-TOPO vector and used for the GFP reporter assay. Regulatory elements are in bold. Underlining indicates the primers used to amplify the promoter regions. Numbers to the right of the homopolymeric repeats represent the number of G residues tested for each tpr promoter. As explained in Fig. 1, the tprE TSS (+1) is hypothetical and based on analogy with the other Subfamily II tprs. +1: transcriptional start site (experimentally determined); RBS: ribosomal binding site (putative); tpr SC: tpr gene start codon; GFP ORF: green fluorescent protein ORF. Amino acids (Lys–Gly–Gln–Phe–Cys–Arg–Ser–Arg) between the tpr putative SC and the GFP ORF start are vector-encoded.
GFP reporter assay
PCR products for the tprE, G, J and F/I promoters were obtained from the above-described clones using primers specifically designed for expression of a GFP fusion protein using the pGlow-TOPO system (Invitrogen) (Table 1). Amplicons were gel purified using the QIAquick gel extraction kit (Qiagen) and cloned into the pGlow-TOPO vector according to the manufacturer’s instructions. For each tpr promoter, the amplicon included the DNA region upstream of the poly-G tract (179, 160, 157, and 169 bp for tprE, G, J, and F/I promoters, respectively; Fig. 2), the putative tpr gene ribosomal binding site (RBS, -AGGAG-), and the GTG initiation codon in frame with the GFP coding sequence in the vector (Fig. 2). The start codons (SC) of these tpr genes have not yet been experimentally demonstrated, therefore we selected putative tpr RBSs and SCs for the pGlow-TOPO constructs separated 8–12 nt from each other in order to provide E. coli with the optimal spacing for translation. Furthermore, to facilitate recognition of the T. pallidum SC (GTG) by E. coli, the GTG triplet was mutated to an ATG in all clones (Fig. 2). Expression of GFP from these constructs resulted in the addition of nine extra amino acids to the actual GFP peptide, encoded by the tpr SC and eight additional codons (Fig. 2) already present in the vector sequence. In total, five different constructs were obtained for both tprE (with repeats 8–12 G nucleotides long) and tprJ (7–11 Gs) promoters, four constructs for tprG (with 8–11 Gs) and three (8–10 Gs) for tprF/I promoters (Fig. 2). A construct containing both the lac promoter and operator upstream of the GFP gene was used as a positive control. As a negative control, to determine possible background fluorescence due to nonspecific GFP expression, a ~300-bp fragment of the tpN47 (TP0574) coding sequence with neither promoter activity nor RBS was inserted out-of-frame with the GFP fusion coding sequence into the pGlow-TOPO vector. To evaluate background fluorescence due to cellular components, both untransformed E. coli cells (not carrying any vector plasmid) and cells transformed with a non-GFP encoding vector (pCRII-TOPO, Invitrogen) were tested. All constructs were sequenced in both strands to verify correct orientation, reading frame and absence of mutations, as well as to ensure that the length of the G repeats did not change after amplification and propagation in E. coli. Constructs were subsequently used to transform TOP-10 E. coli cells (Invitrogen); this strain was selected because it does not require isopropyl β-D-1 thiogalactopyranoside (IPTG) to induce expression from the lac promoter due to the deletion of the lacI gene (ΔlacX74).
For GFP fluorescence measurements, cells were inoculated from a Petri dish into 4 mL of LB medium at room temperature containing 100 μg mL−1 ampicillin and grown at 37 °C for 4 h. OD600 nm was then measured using a biophotometer (Eppendorf, GmbH, Germany) and cultures further diluted to similar ODs (0.5 absorbance units, AU). Fluorescence readings were performed in quadruplicate every hour until cultures reached an OD600 nm of c. 2 AU. Briefly, 400 μL of culture was spun for 4 min at 12 000 g and resuspended in an equal volume of phosphate-buffered saline (PBS); cells were then divided in four wells (100 μL well−1) of a black OptiPlate-96F (Perkin Elmer, Boston, MA) for top fluorescence reading. Excitation and emission wavelength were 405 and 505 nm, respectively, and readings were performed in a Fusion Universal Microplate Analyzer (Perkin Elmer). Before each fluorescence reading, the OD600 nm of the cultures was recorded again. Reported data represent fluorescence readings (expressed in arbitrary units, Ar.U) normalized to the OD of the culture. Background values obtained from each experiment (using E. coli cells transformed with the tpN47-pGlow TOPO vector, as explained in more detail in the ‘Results’) were subtracted from the sample values. At the end of the assay, plasmid DNA was extracted and sequenced again to ensure absence of mutations and changes in the length of the G repeats. Differences between levels of GFP expression were compared using Student’s t-test, with significance set at P < 0.05.
RNA extraction, reverse transcription, qualitative RT-PCR and quantitative real-time amplification
To confirm our observations obtained with the GFP reporter assay, 400 μL of each E. coli culture was spun immediately after fluorescence measurements and cells resuspended in an equal volume of Ultraspec buffer (Biotecx Laboratories) for RNA isolation. RNA was extracted following the manufacturer’s guidelines and followed by DNase I treatment (Turbo-DNase, Ambion, Austin, TX) according to the protocol provided. DNase-treated RNA was tested for residual plasmid DNA contamination by qualitative amplification using specific primers for the GFP gene. The same amplification conditions described above were used with primers GFP-S and GFP-As (Table 1). DNA-free RNA was stored in aliquots at −80 °C until use. Extractions were performed taking precautions to prevent cross-contamination between samples.
Reverse transcription of total RNA was performed using the Superscript II First Strand Synthesis Kit (Invitrogen) with gene-specific primers (GFP-As and Ampr-As, Table 1) according to the manufacturer’s protocol. The GFP and Ampr coding sequences are located in the plus and minus strands of the plasmid, respectively. All cDNA samples were then tested for presence of GFP-specific message using qualitative conventional RT-PCR with the GFP-S and GFP-As primers and the amplification conditions described above. For quantitative real-time amplification, cDNA samples were diluted 1 : 5 with molecular biology grade water to minimize inhibition due to components used to synthesize cDNA. Samples were then stored in 12 μL aliquots (suitable for one amplification reaction in quadruplicate) at −80 °C. A relative quantification protocol using external standards was chosen to determine GFP message levels from cell harvests. This approach normalizes the amount of target mRNA (GFP mRNA) to the message produced by a reference gene (the Ampr gene, also transcribed from the pGlow-TOPO vector but from the opposite strand). To obtain the standards, a pGlow-TOPO vector containing no inserts was purified using the QIAGEN Plasmid Mini Kit (Qiagen) and linearized by EcoRI (Promega) overnight digestion. Digested plasmid was subsequently purified with the PCR Purification Kit (Qiagen) and its concentration was measured using a ND-1000 instrument (NanoDrop Technologies, Wilmington, DE). Plasmid copy number was calculated taking into account the size of the plasmid, its concentration and the average molecular weight of a nucleotide pair. To generate standard curves, the plasmid was serially diluted over the appropriate concentration range (106–100 copies μL−1) in cDNA synthesis mixture and amplified in four replicates for each standard dilution point over the complete range, except for the 101 and 100 dilution points, which were amplified in five replicates. The threshold value for the maximum acceptable error for a standard curve was set to 0.050. Amplification reactions and data collection were carried out using the LightCycler (Roche, Basel, Switzerland) system. All reactions were performed following the manufacturer’s instructions with the Roche FastStart DNA Master plus SYBR Green Kit (Roche), which does not require MgCl2 optimization. The same primers were used for the generation of the standard curves and message quantification (Table 1); for each reaction, optimal primer concentration was found to be 0.50 μM, annealing was 8 s at 60 °C, and extension was carried out at 72 °C, for 7 and 8 s for GFP and Ampr amplifications, respectively. Acquisition temperatures were 82 and 85 °C for GFP and Ampr amplifications, respectively. Amplifications were performed using 3 μL of the final cDNA preparation; results were analyzed using LIGHTCYCLER 3.5 software (Roche). Differences between levels of GFP message were compared using Student’s t-test, with significance set at P <0.05.
Real-time quantification of tpr mRNA levels during experimental infection
To determine whether there are correlations in vivo between mRNA levels of tprE, G and J and the distribution of the G residues in their poly-Gs, total RNA was extracted from Nichols, Bal 73-1, Sea 81-4 and Chicago strain samples collected from rabbit testes in parallel to those used to assess the degree of variability of the poly-G tracts. cDNA was synthesized using the Superscript II First Strand Synthesis Kit (Invitrogen) with random hexamers according to the provided protocol. A previously reported relative quantification method based on real-time PCR external standards (Giacani et al., 2007) was then used to determine amounts of message for Subfamily II tprs. The 47-kDa lipoprotein mRNA (encoded by the TP0574 locus) was used for normalization purposes (Giacani et al., 2007). Primers (targeting the tpN47 gene and Subfamily II tprs) are listed in Table 1; the Sea 81-4 tprJ locus harbors a hybrid tprG/J allele (Giacani et al., 2005a, b), which required specific primers (Giacani et al., 2007).
Results
Length variability of the G homopolymeric repeats in the promoters of tprF, I, E, G and J
Sequence analysis of the tprE, G, J, F and I and promoters revealed a wide range of variability in the number of residues in the poly-Gs in all isolates (Nichols, Sea 81-4, Chicago, and Bal 73-1), although not all of the variants were always obtained from each strain (Fig. 3a–e). Repeats of 8–12 residues were found in the tprE promoter; 8–11 in tprG and 7–11 Gs in the tprJ promoter. For Subfamily II promoters, different lengths were not found to be equally distributed among strains, but repeats with eight Gs were consistently the least represented ones. In this set of experiments, no repeats containing eight Gs were seen for tprJ in a total of 80 clones; however, such sequence was obtained from related experiments using the Chicago C isolate and included in the GFP reporter assay.
Fig. 3.
Distribution of the number of G residues within the homopolymeric repeats located upstream of the tprE (a), tprG (b), tprJ (c), tprF (d) and tprI (e) ORFs. DNA was extracted from treponemal strains harvested from rabbits at peak orchitis and promoter regions amplified (using primers schematically represented in Fig. 1 and reported in Table 1). Twenty clones were sequenced for each tpr for each strain (Nichols, Chicago, Bal 73-1 and Sea 81-4) to analyze the range of variability of the poly-G tracts. For clarity, some symbols were slightly offset to prevent overlap.
Within tprF and I promoters only repeats containing 8-10 nucleotides were identified (Fig. 3d and e). Differently from Subfamily II promoters, some strains exhibited no variability in the poly-G length associated with Subfamily I members (tprF in Nichols and tprI in both Chicago and Bal 73-1 strains) (Fig. 3d and e). Besides variation in length of the homopolymeric repeats, no other sequence differences were found in the cloned promoters within and among strains. In additional experiments (data not shown), in which we spiked rabbit tissue with a linearized plasmid containing a tpr promoter with a known number of Gs, we found no changes in the length of the G repeats after DNA re-extraction, PCR amplification, recloning and sequencing. This further supports our previous statement that the variability in length of the poly-G repeats is not artefactual.
Promoter activity as determined by GFP reporter assays
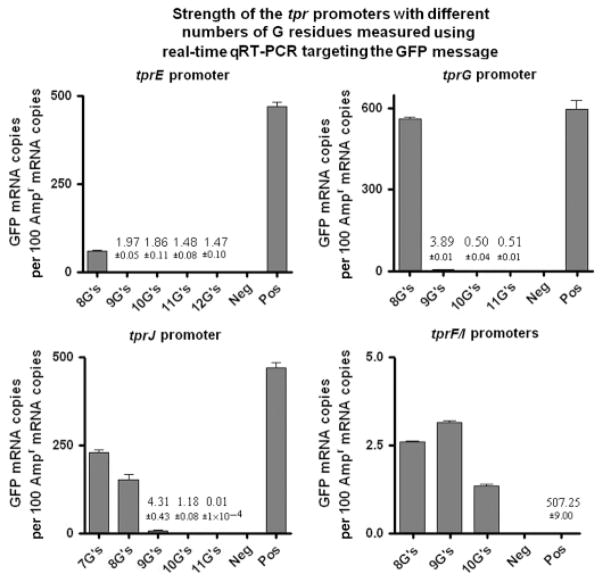
GFP fluorescence was used to determine the relative promoter strength for each of the tpr promoter-pGlow constructs. Five constructs each were tested for tprE (8–12 Gs) and tprJ (7–11 Gs) promoters, four for tprG (8–11 Gs) and three for tprI/F (8–10 Gs). Equal background fluorescence emission was seen in cultures carrying negative control plasmids (pGlow-TOPO vector with the tpN47 fragment, hence without promoter and RBS; pCRII-TOPO vector) and in untransformed E. coli cells (data not shown) indicating absence of signal due to GFP expression. For Subfamily II-derived clones (tprE, G and J promoters), GFP fluorescence was found to be significantly higher than the background with clones containing eight Gs, while clones with nine or more G residues gave only weak or no signal after background subtraction (Fig. 4a–c). Furthermore, the tprJ promoter with seven Gs was found to elicit significantly higher fluorescence than with eight G residues (Fig. 4c). This pattern was true for cultures obtained at different times of growth (Fig. 4a–c).
Fig. 4.
Strength of the tpr promoters containing different numbers of G residues measured with the GFP reporter assay. Data represent fluorescence readings (expressed in arbitrary units, Ar.U) normalized to the OD of the culture after subtraction of background fluorescence. Subfamily II tprE and G promoters containing eight Gs show the highest activity (a, b). No promoters with seven Gs have been identified yet. tprJ promoter with seven Gs induces higher fluorescence than the same promoter with eight Gs (c). Subfamily I tprF/I promoter activity is not influenced by the number of Gs (d). tprF and tprI promoters are identical. Fluorescence signal induced by the lac promoter, always >27 000 Ar.U/OD600 nm, is not shown to allow visualization of sample bars. Means ± SE of quadruplicate values are represented.
Interestingly, tprE promoters with 9–11 Gs were found to induce a signal above background only during the first two timepoints of the experiment (Fig. 4a). Analysis of the OD600 nm values at five different timepoints at which fluorescence measurements were taken (0.58 ± 0.01, 0.80 ± 0.03, 1.10 ± 0.07, 1.80 ± 0.10 and 2.40 ± 0.12, for the five time-points, respectively) showed that the cultures were still within the logarithmic phase at the last timepoint, ruling out that changes in transcription were induced by transition to the stationary phase. In contrast to Subfamily II-derived promoters, the tprF/I alternative promoters induced similar low fluorescence readings, although clearly above background, independently of the number of Gs within the homopolymeric tract (Fig. 4d).
Promoter activity as determined by qualitative standard and quantitative real time RT-PCR
GFP-specific message was analyzed using both qualitative conventional RT-PCR amplification and quantitative real-time PCR. Qualitative RT-PCR detected in every E. coli culture (except for cultures carrying the promoterless GFP plasmid) GFP mRNA, regardless of the number of Gs within the promoter region. Nonetheless, as expected for Subfamily II (tprE, G and J) but not for Subfamily I (tprF and I) internal promoters, there was a progressive reduction in the amplification signal as the number of Gs increased within their homopolymeric repeats (data not shown). Quantitative analysis of the same samples using real-time RT-PCR confirmed that GFP transcription induced by Subfamily II promoters decreases as the number of Gs within the homopolymeric repeats increases (Fig. 5); in fact, tprJ, G and E promoters containing nine or more G repeats were shown to induce extremely low levels of GFP message compared with promoters containing eight Gs (Fig. 5), and tprJ promoter containing eight Gs was shown to induce lower GFP transcription than the same promoter containing seven Gs (Fig. 5). By contrast, all tprF/I promoters, regardless of the number of Gs, yielded similar but very low numbers of mRNA copies (note y-axis values, Fig. 5).
Fig. 5.
Strength of the tpr promoters containing different numbers of G residues measured by GFP quantitative real-time RT-PCR. This analysis shows for Subfamily II promoters (tprE, tprG, and tprJ) a decrease in the amount of GFP mRNA as the number of G residues increases. Similar low levels of GFP mRNA (note values on y-axis) are induced by Subfamily I promoters (tprF and tprI) regardless of the number of Gs. Values represent the means ± SE of quadruplicate measurements. Where the bars are not visible, the mean values are shown as numbers. Positive control is not shown (value reported) so that tprF/I values could be visualized. Negative and positive controls were RNA extracted from Escherichia coli cultures transformed with a promoterless pGlow-TOPO vector and lac promoter/pGlow-TOPO vector, respectively.
These observations show that qualitative and quantitative RT-PCR are more sensitive methods than fluorescence emission assays in detecting GFP expression in E. coli cultures carrying promoters with nine or more Gs, where readings are not different from the background fluorescence.
Subfamily II tpr message quantification during experimental infection
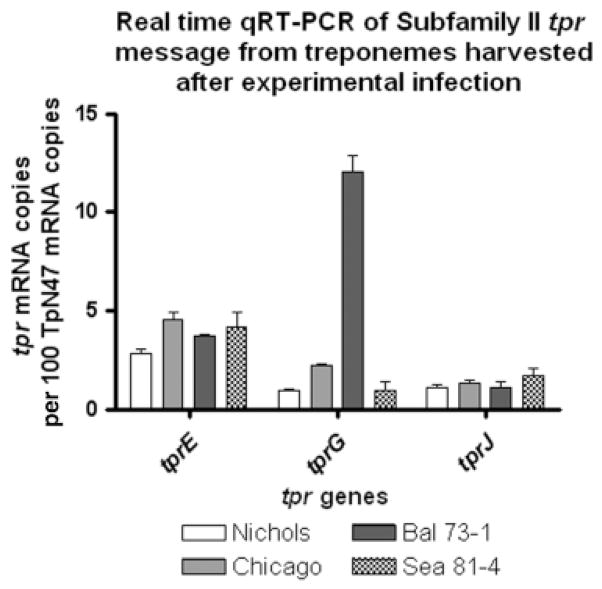
To determine whether there are correlations in vivo between mRNA levels of tprE, G and J and the length of the poly-G repeats in their promoters, we performed message quantification on the same treponemal harvests used to obtain the data on poly-G length distribution (Fig. 3), with the assumption (inferred by both fluorescence and GFP message quantification results) that strains carrying a higher percentage of poly-Gs with eight or fewer G residues would have higher levels of message. The results are shown in Fig. 6. From the promoter sequencing data (Fig. 3), for example, we expected to see the highest values for tprE in Chicago, intermediate levels in Bal 73-1 and Sea 81-4, and the lowest in Nichols. tprE transcription patterns were consistent with our hypothesis in both Chicago and Nichols (P <0.05); tprE mRNA quantification in both Bal 73-1 and Sea 81-4 yielded intermediate values between Nichols and Chicago, although not significantly different (P > 0.05) from either strain (Fig. 6). Good correlations were seen (P <0.05) for tprG in both Bal 73-1 and Nichols strains, which were expected to yield the highest and the lowest values, respectively. Chicago tprG message was expected and shown to be higher (P <0.05) than in Nichols. According to the poly-G length distribution, however, it was expected that Bal 73-1 tprG message levels would be similar to Chicago. In contrast, Bal 73-1 contained very high levels of tprG message. Finally, tprJ levels were not statistically different (as predicted) in Nichols, Bal 73-1 and Chicago. Although tprJ was expected to be higher in Sea 81-4 than in the three other strains (Fig. 6), differences among these values were not found to be statistically significant (P > 0.05). In summary, a correlation between poly-G tract length distribution and levels of mRNA for Subfamily II genes was seen in vivo in T. pallidum to a good extent, although this correlation was not as strong as would be expected if poly-G length alone affected transcription, suggesting other factors involved in transcriptional regulation.
Fig. 6.
Real time quantitative RT-PCR analysis of Subfamily II tpr message levels in treponemes harvested at peak orchitis from rabbits experimentally infected with the Nichols, Sea 81-4, Chicago and Bal 73-1 strains of Treponema pallidum. Message levels were correlated with the distribution of G residues within tprE, G and J promoters reported in Fig. 3. Reported values represent the mean ± SE of triplicate assays for each cDNA sample; number of copies of tpr transcript are expressed per 100 copies of the 47 kDa lipoprotein message. The Sea 81-4 tprJ locus harbors a tprG/J hybrid molecule, which required specific primers (Table 1).
Discussion
Although our understanding of regulation of gene expression in T. pallidum is still very limited, this work provides a first insight on a mechanism most likely to modulate activity of promoters associated with transcription of five potential T. pallidum virulence factors. Using the pGlow-TOPO GFP reporter system in E. coli, we showed that variation in length of the poly-G tracts within Subfamily II tpr (tprE, G and J) promoters modulates their transcriptional activity. Preliminary in silico analysis of the T. pallidum genome has revealed a wide variety of ORFs preceded by poly-G repeats which may potentially influence their expression (Table 2), suggesting that this mechanism could regulate several processes in this spirochete. Furthermore, it is well recognized that homopolymeric tracts are often found within bacterial promoters associated with virulence factors, and variation in the length of these homopolymeric sequences is a well-recognized mechanism for phase variation, the ON–OFF switching of gene expression (van der Woude & Baumler, 2004). Although extremely low mRNA levels were found for the tprE, G and J constructs with nine or more Gs, no absolute OFF state was induced by any of the tpr constructs as determined by real-time GFP message quantification (Fig. 5), and thus the behavior of the tpr promoters does not meet a strict definition of phase variation. Figure 5, however, clearly shows that tprE, G and J promoters with nine G residues induced GFP mRNA levels which were, respectively, 99.3%, 96.7% and 96.3% lower with respect to the same promoters with eight G residues, which intuitively does not seem very distant from an OFF state. The ability to detect a very small amount of message could be due to our choice to use real-time RT-PCR to quantitate GFP message instead of less sensitive techniques such as Northern blotting. Furthermore, a comparison between these similar mechanisms helped to formulate new hypotheses on how the changes in the poly-G tracts affect promoter strength of Subfamily II tprs in T. pallidum in vivo. In N. meningitidis, for instance, the promoters upstream of the porA and opc loci contain poly-G and poly-C repeats, respectively, which besides inducing ON–OFF states, also control the volume of transcription of these genes (Sarkari et al., 1994; van der Ende et al., 1995) as do the poly-G repeats in the tpr promoters. In the porA promoter, the poly-G tract is located between the −10 and −35 signatures, while the opc poly-C region encompasses the − 35 region. Changes in the poly-G length in the porA promoter regulate gene transcription by varying the optimal spacing of 17 nucleotides between the consensus sequences to 15 residues (van der Ende et al., 1995); in contrast, the homopolymeric repeats located in the − 35 consensus most likely modulate transcription of the opc loci by influencing the binding of a regulatory protein (Sarkari et al., 1994; van der Woude & Baumler, 2004).
Table 2.
Treponema pallidum ORFs associated with poly-G repeats*
| ORF | Gene product | Poly-G tract location |
|---|---|---|
| TP0026 | Flagellar motor switch protein (fliG-1) (Treponema denticola) | 5′-flanking region |
| TP0040 | Methyl-accepting chemotaxis protein (mcp1) | Within ORF |
| TP0067 | Conserved hypothetical protein (Borrelia burgdorferi) | Within ORF |
| TP0084 | Hypothetical protein | 5′-flanking region |
| TP0107 | licC protein (Haemophilus influenzae) | 5′-flanking region |
| TP0126 | Hypothetical protein | Within ORF |
| TP0127 | Hypothetical protein | Within ORF |
| TP0257 | Glycerophosphoryldiester phosphodiesterase (glpQ) | Within ORF |
| TP0279 | Ribosomal protein S1 (rpsA) (Escherichia coli) | Within ORF |
| TP0347 | Hypothetical protein | Within ORF |
| TP0379 | Preprotein translocase subunit (secA) (Borrelia burgdorferi) | 5′-flanking region |
| TP0381 | Hypothetical protein | 5′-flanking region |
| TP0479 | Hypothetical protein | Within ORF |
| TP0697 | Hypothetical protein | Within ORF |
| TP0798 | Methionyl-tRNA synthetase (metG) (Borrelia burgdorferi) | Within ORF |
| TP0859 | Hypothetical protein | Within ORF |
| TP0860 | Hypothetical protein | Within ORF |
| TP0969 | Hypothetical protein | Within ORF |
Only ORFs associated with homopolymeric tracts of nine or more G residues are reported. The number of genes is higher if a lower number of Gs is used as threshold.
In the tpr promoters, which contain no typical −10 and − 35 signatures, the poly-G repeats are located immediately upstream of their experimentally determined TSSs (Giacani et al., 2005a). Consequently, variation in the length of the repeats would not alter the spacing between these signatures, ruling out a porA-like mechanism. Also, to date, no experimental data are available to support any interaction between this hypervariable region of tprE, G and J promoters with regulatory proteins. Because the poly-G regions associated with Subfamily II tprs are not transcribed (they are located upstream of the TSS), one could hypothesize that variation in the number of Gs might affect conversion of double-stranded DNA (closed complex) into single-stranded DNA (open complex) driven by the RNA polymerase (RNAP) to initiate transcription. However, studies on DNA base pair opening rates attribute an overall higher instability of the double helix to DNA tracts rich in G nucleotide repeats (Dornberger et al., 1999; Bayliss et al., 2004). Consequently, a sequence with a high content in G repeats would alter the double helix conformation and decrease the RNAP ability to recognize and bind the DNA region in which the poly-G is contained. According to this hypothesis, it seems plausible that a lower number of G residues within the tpr promoter region would decrease the DNA double helix instability, perhaps facilitating the binding of the RNAP to the closed complex and the subsequent formation of the open complex, which leads to transcription initiation.
The function and location of the Tpr antigens studied here is still unknown. Cellular location analysis using Psortb (http://www.psort.org/psortb/) predicts that TprJ, TprF and TprI are outer membrane proteins. Furthermore, comparative protein modeling analysis using known bacterial crystal structures predicts homology of TprF and TprI to bacterial outer membrane porins involved in the transport of ions and sugar. Modulation of expression of these antigens could perhaps provide the pathogen with a strategy to adapt to different environments. During the establishment of the disease, T. pallidum quickly disseminates from the site of primary infection into a wide variety of organs (Turner & Hollander, 1957); in this context, a mechanism able generate tpr promoters with different numbers of Gs would also generate a mixed population of bacteria containing subgroups of organisms expressing the phenotype necessary for a successful adaptation to the new conditions. To address these hypotheses, ongoing research in our laboratories is investigating association between poly-G length and variation of tpr mRNA levels both in different tissues and as syphilitic lesions evolve over time in the rabbit model.
The partial correlation seen in vivo between poly-G tract length distribution (Fig. 3) and levels of mRNA for Subfamily II genes (Fig. 6) suggests that tpr transcription is not exclusively regulated by variation in the poly-G tracts. In fact, using a weight matrix based upon known binding motifs (available at http://www.tractor.Incc.br) we identified putative cAMP receptor protein (CRP) binding sites in the promoter regions of tprE (centered at nucleotide position −236 upstream of the TSS), G (− 388), J (− 233), F (−329 and −32) and I (−328 and −31). Because the goal of this study was to investigate the promoter activity only in relation to changes in the poly-G tracts, the upstream CRP binding sites associated with Subfamily I (−329 and −328 for tprF and tprI, respectively) and Subfamily II tprs were not included in our GFP constructs. Using electrophoretic mobility shift assays (EMSAs, not reported), we have recently demonstrated that a T. pallidum recombinant CRP binds Subfamily II tpr promoters. Furthermore, using a heterologous expression system similar to that described here, in which E. coli is expressing T. pallidum CRP, we have shown that expression of T. pallidum CRP in vivo significantly increases tprJ promoter strength, but in a fashion that is still dependent on the poly-G length. These findings, which are currently being extended to the other Subfamily II tpr members and to tprF and I, strongly support our hypothesis that tpr transcription is influenced by changes in the length of the poly-G tracts as well as by transcription factors.
Variation in the number of G residues occurs naturally in T. pallidum, and is not the consequence of PCR amplification, sequencing reactions or plasmid replication into E. coli. All of the promoters studied here were initially cloned into a generic TA vector (as described in the ‘Materials and methods’) and once selected according to their poly-G length were re-amplified from the plasmid template to be cloned into the GFP reporter vector. These amplifications never induced mutations of any sort into the poly-Gs. Finally, sequencing of both strands of the reporter vectors harboring the tpr promoters was performed after synthesis of the constructs and repeated at the end of the fluorescence and GFP mRNA quantifications, always showing the expected number of G residues.
We acknowledge that the heterologous E. coli GFP reporter system described here might not always accurately reflect transcriptional regulation in T. pallidum in vivo during infection, as has been shown in Borrelia burgdorferi (Sohaskey et al., 1997). Nonetheless, the usefulness of our GFP reporter system in the study of transcriptional regulation of the tpr promoters is supported by our previous observation that tprI, F, G and J promoters are recognized in vitro by the E. coli RNA polymerase-sigma70 complex, and that the TSSs for these genes are identical when they are identified using in vitro transcription assays with E. coli RNA polymerase holoenzyme or in RNA extracted from T. pallidum cells (Giacani et al., 2005a).
In summary, these studies report for the first time the development of tools for studying promoter strength of T. pallidum genes, and our results strongly support the involvement of the poly-G tracts in modulating expression of Subfamily II tprs, probably in concert with other regulatory transcription factors.
Acknowledgments
We are grateful to Heidi Pecoraro for manuscript preparation, Dr Martin Tompa for the in silico analysis of the DNA regions upstream of the tpr ORFs, Maritza Puray-Chavez for helping with the analysis of the Chicago tprG promoter, and Charmie Godornes and Cristina Guerra Giraldez for the preliminary EMSAs. This work was supported by NIH grants AI42143 and AI63940.
References
- Arhin FF, Moreau F, Coulton JW, Mills EL. Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can J Microbiol. 1998;44:56–63. [PubMed] [Google Scholar]
- Bayliss CD, Dixon KM, Moxon ER. Simple sequence repeats (microsatellites): mutational mechanisms and contributions to bacterial pathogenesis. A meeting review. FEMS Immunol Med Microbiol. 2004;40:11–19. doi: 10.1016/S0928-8244(03)00325-0. [DOI] [PubMed] [Google Scholar]
- Centurion-Lara A, Castro C, Van Voorhis WC, Lukehart SA. Two 16S–23S ribosomal DNA intergenic regions in different Treponema pallidum subspecies contain tRNA genes. FEMS Microbiol Lett. 1996;143:235–240. doi: 10.1111/j.1574-6968.1996.tb08486.x. [DOI] [PubMed] [Google Scholar]
- Centurion-Lara A, Castro C, Barrett L, Cameron C, Mostowfi M, Van Voorhis WC, Lukehart SA. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J Exp Med. 1999;189:647–656. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornberger U, Leijon M, Fritzsche H. High base pair opening rates in tracts of GC base pairs. J Biol Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Giacani L, Hevner K, Centurion-Lara A. Gene organization and transcriptional analysis of the tprJ, tprI, tprG and tprF loci in the Nichols and Sea 81-4 Treponema pallidum isolates. J Bacteriol. 2005a;187:6084–6093. doi: 10.1128/JB.187.17.6084-6093.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacani L, Sambri V, Marangoni A, et al. Immunological evaluation and cellular location analysis of the TprI antigen of Treponema pallidum subsp. pallidum. Infect Immun. 2005b;73:3817–3822. doi: 10.1128/IAI.73.6.3817-3822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacani L, Molini B, Godornes C, Barrett L, Van Voorhis WC, Centurion-Lara A, Lukehart SA. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect Immun. 2007;75:104–112. doi: 10.1128/IAI.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KRO, Sellati TJ, Nguyen TT, Cox DL, Clawson ML, Caimano MJ, Radolf JD. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J Exp Med. 2001;193:1015–1026. doi: 10.1084/jem.193.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader BT, Hevner K, Molini BJ, Barrett LK, Van Voorhis WC, Lukehart SA. Antibody responses elicited against the Treponema pallidum repeat proteins differ during infection with different isolates of Treponema pallidum subsp. pallidum. Infect Immun. 2003;71:6054–6057. doi: 10.1128/IAI.71.10.6054-6057.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lukehart SA, Baker-Zander SA, Lloyd RM, Sell S. Characterization of lymphocyte responsiveness in early experimental syphilis. II. Nature of cellular infiltration and Treponema pallidum distribution in testicular lesions. J Immunol. 1980;124:461–467. [PubMed] [Google Scholar]
- Sarkari J, Pandit N, Moxon ER, Achtman M. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing polycytidine. Mol Microbiol. 1994;13:207–217. doi: 10.1111/j.1365-2958.1994.tb00416.x. [DOI] [PubMed] [Google Scholar]
- Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- Smajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, Weinstock GM. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol. 2005;187:1866–1874. doi: 10.1128/JB.187.5.1866-1874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Arnold C, Barbour AG. Analysis of promoters in Borrelia burgdorferi by use of a transiently expressed reporter gene. J Bacteriol. 1997;179:6837–6842. doi: 10.1128/jb.179.21.6837-6842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ES, Molini BJ, Barrett LK, Centurion-Lara A, Lukehart SA, Van Voorhis WC. Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect. 2004;6:725–737. doi: 10.1016/j.micinf.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Turner TB, Hollander DH. Biology of the Treponematoses. World Health Organization; Geneva: 1957. [PubMed] [Google Scholar]
- van der Ende A, Hopman CT, Zaat S, Essink BB, Berkhout B, Dankert J. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the − 10 and − 35 regions of the promoter. J Bacteriol. 1995;177:2475–2480. doi: 10.1128/jb.177.9.2475-2480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM, Hardham JM, McLeod MP, Sodergren EJ, Norris SJ. The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microb Rev. 1998;22:323–332. doi: 10.1111/j.1574-6976.1998.tb00373.x. [DOI] [PubMed] [Google Scholar]