Abstract
As modulators of gene expression, microRNAs (miRNAs) are essential for normal development. Not surprisingly, aberrant expression of miRNAs is associated with many diseases, including cancer. Studies of various breast cancer subtypes have demonstrated that, like gene expression profiles and pathological differences, miRNA profiles can distinguish various tumor subtypes. Over the last few years, roles for miRNAs during many stages of breast cancer progression have been established. This includes potential breast cancer associated polymorphisms in miRNA target sites or miRNAs themselves, miRNAs that can act as tumor suppressors or oncogenes, and miRNAs that can modulate metastatic spread. Recent studies have also suggested key roles for miRNAs in regulating cancer stem cells. Thus, miRNAs have now become important therapeutic targets. This can be achieved by replacing miRNA expression where it has been lost or decreased, or conversely by inhibiting miRNA expression where it has been amplified or overexpressed in cancers. Ultimately, miRNAs should provide both important prognostic biomarkers as well as new targetable molecules for the treatment of breast cancer.
Keywords: Breast cancer, miRNA, novel therapeutics, stem cells
MicroRNAs: THE BASICS
microRNAs (miRNAs) are a specific class of endogenous non-coding RNAs that are typically conserved across species. They are single-stranded RNAs of 21–25 nucleotides in length, and function in post-transcriptional gene silencing [1]. An estimated 30%–60% of the genome is regulated by miRNA-mediated silencing [2]. Regulation of expression of these miRNAs is highly complex, as they often exist in clusters, can be encoded in intronic regions of transcribed genes, reside within intergenic regions, or can be located within large non-coding RNAs [3]. The biogenesis of miRNAs involves both nuclear and cytoplasmic components [4]. Mirtrons, a new class of miRNAs derived from short introns of pre-mRNAs, have recently been discovered in flies, nematodes and mammals. These mirtrons are spliced and debranched during pre-mRNA processing, forming pre-miRNA hairpin structures that bypass the need for Drosha cleavage, but then enter the canonical miRNA pathway [5].
In the cytoplasm, mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) complex. The miRNA-RISC complex then mediates gene silencing by targeting the 3′ untranslated region (UTR) of mRNAs and induces translational silencing, either by cleaving the mRNA or blocking ribosomal translation of the targeted mRNA. Target mRNAs are recognized by partial sequence similarity in the seed sequence of the mature miRNA [6] and often these 3′UTR target sites are evolutionarily conserved within the miRNA seed region [2, 7]. Once the target mRNA is recognized by the miRNA-RISC complex, it is unclear what mechanisms mediate mRNA repression: mRNA degradation in the P-bodies or inhibition of translation by blocking ribosomes [8–10].
Although miRNA silencing of mRNAs has traditionally been thought to occur at the 3′UTR, emerging evidence suggests that miRNAs can regulate translation via seed sequencing within the protein-coding sequences and the 5′UTR as well [2], however this 5′UTR-mediated mechanism of translational repression may differ. Additionally, reports of rare and unconventional miRNA-mediated upregulation of mRNA translation have recently been described [11, 12]. This complexity in miRNA-mediated mRNA translational regulation highlights the multiple mechanisms by which miRNAs can modulate translation of the genome.
miRNAs IN NORMAL MAMMARY GLAND DEVELOPMENT AND STEM CELLS
The requirement for normal miRNA functioning for proper stem cell maintenance has been demonstrated in mouse Dicer-deficient ES cells and tissue-specific Dicer knockout mice. Loss of mature miRNAs resulted in a failure of the cells to differentiate [13–16], providing evidence that miRNAs may also be involved in maintaining stem cell populations by keeping them in an undifferentiated state, controlling stem cell self-renewal and regulating the transition of undifferentiated stem cells to more differentiated states. It has also been demonstrated that cells originating from within the same lineage have miRNA profiles that are more similar to each other than cells of different lineages [17]. In addition, a specific set of miRNAs has been identified in ES cells that are not detectable in adult tissues [18]. These data suggest that some miRNAs act as key modulators in normal development, regulating stem cell maintenance and self-renewal by preventing differentiation. Furthermore, once lineage-specific miRNAs become expressed, they may modulate a hierarchy of transcription factors initiating differentiation along a specific lineage [19]. Finally, induction of a lineage-specific miRNA expression in an uncommitted progenitor cell may result in differentiation along specific lineages [19–24].
Many tissues, including the skin, blood and gut, contain a small population of lineage-restricted stem cells to maintain the short lived mature cells (reviewed in [25]). While mostly quiescent, these multipotent adult stem cells have the ability to self-renew and are essential for tissue homeostasis because of their ability to differentiate into the cells that make up a tissue. The existence of mammary gland stem cells was first suggested by studies performed over half a century ago in which small fragments of donor mammary tissue were transplanted into the cleared fat pad of recipient mice [26, 27]. These seminal transplantation studies demonstrated that not only could small populations of cells repopulate a full mammary gland, consisting of all the differentiated cell types of the mammary ductal tree, but also that these cells could be serially transplanted and still retain this ability [26, 28].
Identifying markers of normal mammary gland stem cells is essential for their isolation and further functional studies. Studies from two laboratories recently identified such markers within the mouse mammary gland [29, 30]. Stingl and colleagues isolated two subpopulations from the adult mammary gland that exhibited different progenitor-like behavior based on CD49f (α6-integrin)/CD24 (heat stable antigen) profiling. Single cells from the CD49fhi/CD24+ subpopulation from an adult mammary gland were able to regenerate a functional mammary gland in vivo, while the CD49flow/CD24hi subpopulation displayed clonogenic potential in vitro [30]. In related studies, the Lineage(Lin)−/CD29(β1-integrin)hi/CD24+ subpopulation from the adult mammary gland also was shown to be capable of repopulating the mammary gland [29]. It has been estimated that the stem cell activity within the CD49fhi/CD24+ or Lin−/CD29hi/CD24+ subpopulations represents only about 1 in 2000 cells, although this estimate is approximately ten-fold higher than the stem cell frequency by limiting dilution transplantation of unsorted mammary cells [31]. In humans, studies have shown that cells with high aldehyde dehydrogenase (ALDH1) activity [32] or a CD49f+/EpCAM− immunnotype have stem cell properties [33].
While miRNAs important to somatic stem cells, including hematopoietic stem cells [21, 34], cardiac and skeletal muscle stem cells [20], neural stem cells [24], and skin stem cells [22], have been identified, the role that miRNAs play in normal mammary gland stem cells has yet to be fully investigated. A recent study used a mouse mammary cell line model to identify miRNAs in the Aldefluorhi/Sca-1hi progenitor cell population found that the different cell populations have distinct miRNA profiles [35, 36], suggesting miRNAs may be involved in the regulation of mammary gland progenitor cell populations. Additionally, the miR-200 family (consisting of two clusters, miR-200c-141 and miR-200b-200a-429) and the miR-183-96-182 cluster are expressed at lower levels in both normal mammary stem cells and breast cancer stem cells. Furthermore, the investigators demonstrated that these miRNAs target the self-renewal factor BMI1, a polycomb ring finger oncogene, providing a possible mechanism for blocking self-renewal in normal and cancer stem cells [37].
miRNAs IN BREAST CANCER
MiRNAs have been shown to be associated with many of the classic hallmarks of cancer, including defects in proliferation, differentiation and apoptosis. The early studies on miRNA profiling of tumors demonstrated that aberrant expression occurs as a function of either deletions associated with frequent fragile sites or genomic amplification. MiRNAs may also be involved in carcinogenesis [38–42], suggesting that they may be critical biomarkers of cancer. Additionally, miRNA profiling can be used to cluster cancer types with the cell of origin [43]. Thus, miRNA profiling may provide useful information for classifying and diagnosing metastases of unknown origin. Furthermore, breast cancers can be classified by their miRNA profile into a specific tumor pathological phenotype (i.e., Estrogen Receptor (ER) and Progesterone Receptor (PR) status, proliferation, tumor stage, metastatic state, HER2 status) [44, 45] as well as the tumor subtype (Luminal A, Luminal B, Basal-like, HER2+ and Normal-like) [42]. Thus, miRNAs may provide additional information for the prognosis and treatment of cancer when combined with standard gene profiling. Because miRNAs play a role in directing stem cell fate, it is plausible that altered regulation of miRNA expression elicited in a stem cell by chromosomal changes, such as amplification or deletion, or epigenetic changes, may induce the transformation of a lineage-restricted stem cell to a cancer stem cell. Thus, tumor-specific miRNA differences may prove to be useful as both prognostic and predictive factors.
One of the most important prognostic markers in breast cancer is ER status. By miRNA expression profiling, miR-206 was found to be highly expressed in ER-negative breast cancers, but not in ER-positive [44]. It was subsequently shown that miR-206 regulates ERα via seed sites within the 3′UTR [46]. Additionally, transfection of miR-206 into an estrogen-dependent breast cancer cell line was shown to inhibit cell growth in a dose-dependent and time-dependent manner [47]. miR-221 and miR-222 are also direct regulators of ERα and their expression reduced sensitivity to tamoxifen when expressed in ER-positive cell lines [48]. miR-221/222 were highly expressed in tamoxifen-resistant MCF7 cell lines and HER2+ primary tumors associated with resistance to endocrine therapy [49]. While miR-206 and miR-221/222 are all capable of negatively regulating ERα, a recent study showed that their overexpression resulted in different effects and global changes in gene expression in ER-positive cell lines. miR-206 inhibited cell proliferation, while miR-221 and miR-222 increased proliferation. They also discovered a negative regulatory loop where ERα could in turn negatively modulate miR-221/222 through the recruitment of transcriptional corepressors to the miR-221/222 locus [50]. Similarly, miR-22 negatively regulates ERα via evolutionarily conserved seed sites in the 3′UTR and treatment of ERα-positive breast cancer cells with miR-22 results in repression of growth in vitro [51]. It has been proposed that some of these miRNAs may promote the transition from ER-positive to ER-negative breast tumors.
BREAST CANCER STEM CELLS
Since normal stem cells and cancer stem cells have many similar properties, including self-renewal and unlimited replicative potential, understanding the regulation of normal stem cells may be important in understanding cancer stem cells. Unlike normal stem cells, cancer stem cells may have gained mutations to make them more tumorigenic or metastatic, and may have intrinsic mechanisms for survival to evade traditional chemotherapeutic agents that target more differentiated tumor cells. Thus, traditional cancer therapies may be eliminating the non-tumorigenic differentiated cells, but leave the cancer stem cells, potentially leading to cancer relapse and/or metastasis [52]. Clinical studies have shown that these cancer stem cells, unlike the bulk of a tumor, have increased DNA damage repair activity and are thus resistant to conventional cancer therapies [52, 53].
Like normal mammary gland stem cells, putative breast cancer stem cells have been isolated based on cell surface markers. Using the normal breast stem cell profile of Lin−/CD29hi/CD24+, breast cancer stem cells were identified in a p53null genetically engineered mouse model of breast cancer [54]. Serial and limiting dilution transplantation of the tumor cells isolated by Lin−/CD29hi/CD24+ profiling indicated that they were enriched 65-fold in tumor-initiating frequency as compared with the majority of the tumor cells isolated using the other three surface marker profiles. In addition, they had characteristic stem cell properties defined by in vitro assays. Furthermore, the four CD29/CD24 subtypes could be separated based on a 710 mRNA gene signature. Since tumor types can be classified based on miRNA expression patterns [55, 56], it would be expected that cancer stem cells within breast cancers also should have unique miRNA expression profiles.
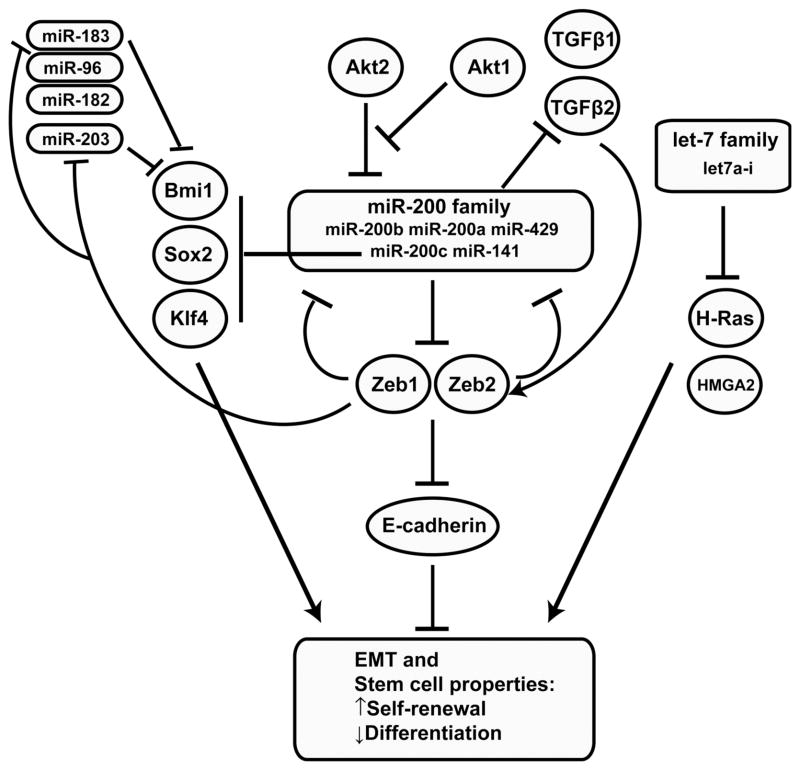
Originally isolated in 2003, human breast cancer stem cells were identified by FACS sorting for expression of a different set of cell surface markers, identifying the cancer stem cell population as CD44+/CD24−/low [57]. Studies on the CD44+/CD24−/low population in human breast cancer have shown decreased expression of the let-7 family (let-7a to let-7i) compared to differentiated cells. Additionally, induction of these cells to express let-7 was shown to inhibit tumor formation and reduce the metastatic capability of the CD44+/CD24−/low cancer stem cells. The tumor-suppressor activity of let-7 was shown to act via regulation of the oncogenic H-Ras and HMGA2 (High mobility group AT-hook 2) genes (Fig. 1). Thus, tumors expressing low let-7 had higher levels of H-Ras and HMGA2, which correlated with poor prognosis. Conversely, tumors expressing high levels of let-7 were less tumorigenic and had lower levels of metastasis [58].
Fig. 1. miRNA network regulating breast cancer stem cells.
Several miRNAs have been identified, including the miR-200 and let-7 families, that target key genes involved in regulating stem cell properties in both normal and cancer stem cells.
The miRNA expression profiles of human CD44+ CD24−/low breast cancer stem cells compared to non-tumorigenic cancer cells identified 37 differentially expressed miRNAs [37]. Included in these 37 miRNAs were three genomic miRNA clusters, miR-200c-141, miR-200b-200a-429, and miR-183-96-182 that were down-regulated in human breast cancer stem cells as well as mouse mammary stem cells and embryonal carcinoma cells, suggesting they may play key roles in the regulation of self-renewal. The miR-200 family, which is contained in two of these clusters, negatively regulates the process of epithelial to mesenchymal transition (EMT) through down-regulation of Zeb1 and Zeb2, which are transcriptional repressors of E-cadherin, and also regulated by TGFβ signaling (Fig. 1) [59–62]. Interestingly, induction of differentiated human mammary epithelial cells to undergo an EMT confers a stem cell-like phenotype [63]. In addition to Zeb1 and Zeb2, miR-200c directly targets BMI1, a known regulator of stem cell self-renewal, and miR-200c suppresses both normal mammary outgrowth and tumorigenicity of human breast cancer stem cells [37]. Furthermore, expression of the miR-200 family of miRNAs appears to depend on the balance between Akt1 and Akt2 rather than overall Akt activity [64]. In addition to a negative feedback loop whereby Zeb1 inhibits the expression of the miR-200 family, Zeb1 also represses miR-203, another stemness-inhibiting miRNA. Sox2 and Klf4, two of the pleuripotency genes used to generate iPS cells have been shown to be potential miR-200 family targets [65]. Two additional miRNAs, miR-203 and miR-183 cooperate to suppress the expression of BMI1. This may be a general mechanism of suppression of EMT and stem cell factors by a coordinated group of miRNAs, and accordingly this network represents a promising new treatment area for targeting cancer stem cells (CSCs).
ONCOGENIC miRNAs
There have been several miRNAs associated with cancers due to genomic changes, implying that miRNAs can act either as tumor suppressors or oncogenes (“oncomiRs”). Oncogenic miRNAs are up-regulated in cancer and have the potential to target many tumor suppressor genes, cell cycle regulation genes, and other genes contributing to the pathology of disease.
The long non-coding RNA B-cell Integration Cluster (BIC) was originally identified as a proto-oncogene in B-cell lymphomas because it was a common integration site for the avian leukosis virus [66]. It was later found that BIC encodes for a miRNA, miR-155, that is required for proper B and T cell immunity and for the function of antigen-presenting cells [67]. By miRNA expression profiling, miR-155 was shown to be highly expressed in breast cancers [56], as well as other solid tumors and lymphomas [68] and is one of the miRNAs that defines a 29-miRNA breast cancer-specific signature (including down-regulation of miR-10b, miR-125b and miR-145 and upregulation of miR-21 and miR-155) [44]. In breast cancer cell lines, over-expression of miR-155 led to increased growth and colony formation, increased growth in a cell transplantation assay, and miR-155 was shown to act as an oncomiR by targeting the tumor suppressor gene suppressor of cytokine signaling 1 (SOCS1), a negative regulator of the JAK/STAT pathway[69].
miR-21 is frequently found to be up-regulated in solid tumors [55, 56] and cancer cell lines [70] and is classified as an oncomiR. Targets of miR-21 include the tumor suppressor genes PTEN [71], Programmed Cell Death 4 (PDCD4) [72], maspin [73], and Tropomyosin 1 (TPM1) [74] as well as several tumor suppressor genes in glioblastomas, including p53 and TGFβ [75], which have been associated with tumor invasion and metastasis. Inhibition of miR-21 with antisense oligos was found to slow cell growth in vitro as well as inhibit tumor growth in a xenograft mouse model [74], suggesting that miR-21 may have potential as a therapeutic target.
TUMOR SUPPRESSOR miRNAs
Tumor suppressor miRNAs are frequently lost or down-regulated in cancer, and have targets that include oncogenes. Calin, et al., identified a fragile site in human chromosome 13 that is frequently deleted in B-cell chronic lymphocytic leukemia cases, in addition to several other cancers, and it was shown that miR-15-a and miR-16-1 were located at this fragile site [76]. As a result of the genomic deletion of the chromosome 13 fragile site, expression of miR-15-a and miR-16-1 was eliminated. Normal B cells have high expression of these two miRNAs and the frequent association of the loss of heterozygosity at this locus implicated these two miRNAs as essential for normal B cell function. Further investigation into miR-15-a and miR-16-1 identified that these two miRNAs target the BCL2 oncogene, in addition to other targets frequently enriched in B-cell lymphomas [77]. Genomic and miRNA profiling in human breast cancers demonstrated these miRNAs were among nine differentially expressed between luminal A and luminal B breast cancer subtypes that could be associated with genomic alterations [42].
The ErbB family of receptor tyrosine kinases is important for cell proliferation and survival. Amplification of HER2/neu and HER3 is implicated in many primary human breast tumors, and is significantly correlated with poor prognosis. Recently, it was found that HER2 and HER3 are suppressed by miR-125a or miR-125b via seed regions in their 3′UTRs. Further, over-expression of either miR-125a or miR-125b resulted in suppression of HER2 and HER3 at both the transcript and protein level in a breast cancer cell culture model selected for HER2 and HER3 dependence. This also resulted in decreased proliferation as well as impaired anchorage-dependent growth and reduced cell migration and invasion capacities, thus reducing the malignant phenotype [78]. Additionally, a germline SNP mutation in miR-125a, resulting in a block of the maturation of miR-125a, is correlated with breast cancer tumorigenesis [79]. These data suggest that the tumor suppressor activity miR-125a and miR-125b may become important therapeutic targets in HER-amplified breast cancers. It was also recently shown that miR-205, which is highly expressed in normal mammary epithelial stem cells [35], also targets HER3 in several breast cancer cell lines, thus conferring tumor-suppressor activity in these metastatic cell lines [80, 81].
CONTEXT-DEPENDENT miRNAs
Like some protein coding genes, there are miRNAs that can act either as a tumor suppressor or an oncogene, depending on cell context. For example, the polycistronic miRNA cluster miR-17–92 has been implicated as a potential oncomiR. miR-17–92 encodes a cluster of 7 miRNAs, and is within an amplified region associated with Small Cell Lung Cancer [82], as well as in B-cell lymphomas and other tumor types when over-expressed [40]. Human breast cancers, in addition to tumors in the colon, lung, pancreas, and prostate, were also observed to have amplified miR-17–92 expression [56]. In contrast, loss of this locus was observed in human breast cancer cell lines [83]. Targets of the miRNAs encoded by the miR-17–92 locus include the E2F transcription factors, which are regulators of the cell cycle and apoptosis [84]. The E2F factors, in turn, promote transcription of the miR-17–92 locus, creating a negative feedback loop [85]. Other targets of miR-17–92 include the proapoptotic genes phosphatase and tensin homolog (PTEN) and Bcl2-interacting mediator of cell death (Bim) [86, 87]. In breast cancer cell line models, miR-17-5p, a member of the miR-17–92 locus, is low, and was found to regulate the expression of the oncogene amplified in breast cancer-1 (AIB1), a member of the p160/SRC family of co-activators, implicating it as a tumor suppressor [88]. Other studies in breast cancer cell lines also identified Cyclin D1 as a miR-17–92 target, and accordingly over-expression of this cluster resulted in decreased proliferation and cell cycle arrest. Interestingly, Cyclin D1 binds to the promoter of the miR-17–92 locus, inducing its expression, indicating another negative feedback loop to modulating the oncogenic potential of Cyclin D1 expression [83]. This cluster may also be regulated in a context-dependent manner as it is directly transactivated by c-Myc, which is frequently up-regulated in cancers [89].
Conflicting reports of miR-205 expression within breast cancer subtypes illustrates the complexity of understanding miRNA function. Studies indicate that miR-205 is highly expressed in normal mammary epithelial cells, and enriched in mouse mammary stem cell populations [35], and it is also developmentally regulated [90]. In prostate and breast cancer cell lines over-expression of miR-205 lead to a reduction of growth, possibly via regulation of the oncogenes HER3, E2F1, E2F5, ZEB1/ZEB2 and PKCe [35, 60, 80, 81]. High expression of miR-205 has been observed in ER+/PR+ HER2+ breast cancers, compared to other subtypes [45] and genomic analysis indicated that miR-205 is within a genomically amplified region in some human breast cancers [42]. Other reports indicate low expression of miR-205 in human breast cancers compared to normal tissue [56], however these studies did not indicate the specific breast cancer subtype(s) analyzed, so loss of miR-205 expression in these samples should not be considered a universal breast cancer feature. Loss of miR-205 expression in breast cancer may be a late event during breast cancer progression. In fact, invasive breast cancer cell lines have lower expression of miR-205 than non-invasive breast cancer cell lines [60], and human metastatic breast cancer biopsies had low expression of miR-205 compared to non-metastatic lesions [44]. This is also supported by recent studies demonstrating that miR-205 is a negative regulator of EMT, a process required for vascular invasion and metastasis, by means of silencing ZEB2/SIP1 and ZEB1, repressors of E-cadherin and regulators of EMT [60]. Thus, over-expression of miR-205 may lead to different phenotypes depending on cell type.
miRNAs AND p53
The p53 tumor suppressor is the most commonly altered gene in human breast cancer. It is mutated in about 30–40% of all human breast tumors with much higher frequency associated with poor outcome. Aberrant cell growth can continue unchecked when p53 function is abrogated and the cells become genomically unstable. Not surprisingly, p53 has been connected recently to miRNA expression and processing (reviewed in [91]).
p53 can directly bind to promoters of the miR-34 family members, miR-34a and miR-34b/c to activate their transcription [92]. These miRNAs target a set of genes promoting cell cycle progression, thus over-expression of miR-34a and miR-34b/c results in cell cycle arrest, apoptosis and senescence. As part of a regulatory feedback loop, p53 also negatively regulates the expression of the c-Myc proto-oncogene. It was recently shown that p53 directly activates transcription of the tumor suppressor miRNA, miR-145, which in turn directly silences c-Myc via target sites in the 3′UTR. Thus, induction of miR-145 expression both in vitro and in vivo lead to suppression of tumor cell growth [93]. Other miRNAs have been found to up-regulate p53 activity. Over-expression of miR-29 is able to up-regulate p53 as a result of the reduction of two miR-29 targets, p85α and CDC42, both of which regulate p53. Thus, over-expression of miR-29 can result in p53-mediated apoptosis via the PI3K pathway [94]. Other p53-responsive miRNAs include miR-192, miR-194 and miR-215. These miRNAs target CDKN1A/p21 levels, resulting in cell cycle arrest when induced [95].
In addition to transcriptional regulation, p53 is involved in post-transcriptional maturation of miRNAs by interacting with the Drosha microprocessing complex in response to DNA damage. In this mechanism, p53 associates with Drosha via the p68 helicase (DDX5) to process pri-miRs into pre-miRs, and mutant p53 results in decreased mature miRNA expression [96]. Thus, in tumors where p53 is likely to be altered, the availability of mature miRNAs may be depleted.
BREAST CANCER METASTASIS
Complications from metastatic disease are the major reason for cancer mortality. Metastasis is multi-step process involving local invasion of cells from the primary tumor, entry into the circulation, survival in the circulation, invasion into other tissues, the establishment of micrometastases, and growth of secondary tumors. Analogous to oncomiRs and tumor suppressor miRNAs, miRNAs can also promote or suppress these steps in migration and metastasis without influencing primary tumor development. Welch and colleagues recently coined the term “metastamir” to refer to these metastasis regulatory miRNAs [97]. These metastamirs regulate key steps in the metastatic program and processes such as EMT, apoptosis, and angiogenesis. Furthermore, as recently discussed and reviewed by Calin and colleagues, there is an emerging theme that many of the same miRNAs are involved both in CSC regulation and in steering metastasis [98].
The process of EMT has been associated with tumor cell invasion and metastasis. As mentioned above, the miR-200 family, as well as miR-205, regulate EMT through targeting Zeb1 and Zeb2. While miR-205 was shown to suppress cell growth in MCF7 cells, it was shown to inhibit invasion and metastasis in MDA-MB-231 cells [81]. Most of these studies have used in vitro assays to show that miR-200 family members can suppress the invasive properties of cells in culture [60–62]. Unexpectedly, over-expression of miR-200 in the non-metastatic murine mammary tumor cell line 4TO7 enables these cells to metastasize to the lung and liver [99]. This may be due to the fact that some tumors may have the requirement of a mesenchymal to epithelial transition (MET) occurring for efficient tumor colonization at the metastatic site. Further studies using in vivo metastasis models are surely warranted to elucidate the role of the miR-200 family in metastasis. Furthermore, miR-200c targets class III β-tubulin (TUBB3), and restoration of miR-200c expression in cells with low miR-200 family levels results in increased chemosensitivity to microtubule-directed agents [100]. As a pro-metastatic miRNA, miR-9 targets E-cadherin, loss of which results in loss of epithelial characteristics, cell migration and invasion, ultimately leading to an EMT [101]. Further studies on breast cancer cells demonstrated miR-9 is directly regulated by MYC and MYCN, and over-expression of miR-9 resulted in an EMT, increased proliferation, enhanced metastasis, and induced angiogenesis when transplanted [101].
Breast cancer metastasis suppressor 1 (BRMS1) is a protein that suppresses metastasis in multiple tumor types. By analyzing the miRNA expression profiles of metastatic MDA-MB-231 and MDA-MB-435 cells compared to BRMS1-transduced non-metastatic counterparts, Welch and colleagues found that BRMS1 coordinately regulates a number of these metastamirs [102]. BRMS1 decreased metastasis-promoting miRNAs: miR-10b, miR-373, and miR-520c. Metastasis suppressing miRNAs: miR-146a, miR-146b, and miR-335 were up-regulated following BRMS1 overexpression. This shows that BRMS1-containing SIN3/HDAC complexes may be recruited to and regulate miRNA promoters in addition to coding genes involved in regulating cancer metastasis.
miR-10b, which is highly expressed in metastatic breast cancer, is also involved in breast cancer metastasis [103]. This control is due to a feedback loop where the transcription factor Twist regulates transcription of miR-10b via direct E-box binding, which in turn represses translation of HOXD10 via a highly conserved miR-10b binding site in the 3′UTR. Decreased levels of HOXD10 then resulted in increased expression the pro-metastatic gene, RHOC, although a direct connection here has not been established. miR-10b has also been shown to directly repress Tiam1 expression in breast carcinoma cells [104]. This inhibits Tiam1-mediated Rac activation suppressing migration and invasion. Furthermore, over-expression of miR-10b in non-metastatic breast cancer cells promoted metastasis, while inhibition of miR-10b in metastatic breast cancers decreased metastatic phenotypes of cells in vitro. However, systemic administration of miR-10b antagomirs to mice bearing highly metastatic 4T1 cells was not able to reduce growth of the primary lesion, but was shown to markedly suppress formation of lung metastases, representing a promising approach for an anti-metastasis agent [105].
By comparing miRNA expression profiles between MDA-MB-231 parental cells and derivatives with higher metastatic ability, Tavazoie, et al., identified miR-335 and miR-126 as low in more metastatic lines. Both miR-126 and miR-335 were significantly down-regulated in the metastatic breast cancers, and this decreased expression was correlated with poor prognosis. Induction of miR-335 expression is sufficient to suppress lung and bone metastasis in a metastatic breast cancer cell line. SOX4, which is critical in cell migration, was shown to be a functional target of miR-335, as well as the extracellular matrix component tenascin C [106]. A miR-335 six-gene target signature (SOX4, PTPRN2, MERTK, PLCB1, COL1A1 and TNC) is associated with poor metastasis-free survival in a large patient dataset. Additionally, miR-126 expression reduced overall tumor growth and proliferation. This inhibition of cell cycle progression was shown to be possibly through inhibition of Insulin Receptor Substrate-1 (IRS-1) [107]. Other studies of the metastatic MDA-MB-231 cell line indicated that miR-146 inhibits invasion and metastasis of cells by downregulating NFkB, in addition to targeting IRAK1 and TRAF6 [108].
Valastyan et al. demonstrated that miR-31, which is expressed in normal mammary cells, is specifically lost in metastatic breast cancer cell lines. Overexpression of miR-31 expression in metastatic cells can inhibit multiple steps in metastasis, and in non-metastatic cell lines, deletion of miR-31 is sufficient to confer metastatic properties. [109]. In a subsequent study, the authors demonstrate that while miR-31 is predicted to modulate the expression of >200 mRNAs, its regulation of metastasis can be accounted for primarily by three targets, integrin α5 (ITGA5), radixin (RDX), and RhoA [110]. Interestingly, each of these targets affects distinct steps in the metastatic process and re-expression of these three genes concurrently abrogates metastasis suppression by miR-31.
miR-373 and miR-520c were identified as promoters of tumor invasion and metastasis through a genetic screen using MCF7 cells subjected to a trans-well migration assay. These miRNAs can stimulate the normally non-metastatic MCF7 cell line to display metastatic behavior of cell invasion and migration, interestingly by targeting CD44 [111].
In MDA-MB-231 cells, unlike MCF7 cells, miR-21 does not affect growth of the primary tumor. It was however shown that miR-21 promotes cell invasion and lung metastasis in this model [73]. This is likely through the down-regulation of multiple targets including several other tumor suppressor genes.
miR-145 was recently shown to suppress invasion and experimental metastasis assays using metastatic breast cancer cell lines MDA-MB-231 and the LM2-4142 lung metastatic subline [112]. This was in part due to miR-145 directly targeting MUC1, which in turn down-regulates β-catenin, Cyclin D, and Cadherin 11.
Liu et al. found that the miR-17–92 cluster was expressed at higher levels in metastatic breast cancer cell lines. The expression of the cluster was diminished by treatment with a ROCK inhibitor (Y27632). They further showed that blockade of miR-17 using anti-miR-17 molecules decreased breast cancer cell invasion in vitro and lung metastasis from the orthotopic site [113]. In contrast, another group found that miR-17/20 (miR-17-5p and miR-20a) were reduced in highly invasive breast cancer cell lines and node-positive breast tumors, and in vitro miR-17/20 could inhibit migration and invasion of neighboring cells via a unique mechanism of heterotypic secreted signaling [114].
Recently, Baffa et al. performed a miRNA microarray analysis of 43 matched primary tumors (13 breast, 10 lung, 10 bladder, and 10 colon cancers) and corresponding lymph node metastasis to identify deregulated miRNAs in metastasis. They identified 32 differentially expressed miRNA [115]. These included a number of the above mentioned metastasis-regulating miRNAs including the upregulation of miR-10b and miR-21 and the down-regulation of miR-141, miR-200b, miR-200c, and miR-205 in metastatic samples.
miRNAs that regulate the process of metastasis may become useful prognostic markers and/or targets for anti-metastatic therapy. Inhibition of metastasis promoting miRNAs and/or expression of metastasis suppressing miRNAs using tools we review later in the article may, therefore, provide therapeutic strategies for metastatic breast cancer.
HYPOXIA
Tumor hypoxia results from inadequate access to vasculature and the blood supply and may be associated with poor prognosis and recurrence. This state stimulates tumor growth and may confer radiation resistance. When cancer cell lines, including MCF7 and MDA-MB-231 breast cancer cells, were kept in hypoxic conditions, the expression of miR-210 was induced [116–118], and this up-regulation was shown to be via hypoxia-inducible factor-1α (HIF-1α) at the transcriptional level [116, 118]. Not surprisingly, during miRNA profiling studies, miR-210 was highly expressed in breast cancers [44], and high expression of miR-210 in triple negative cancers was significantly associated with early relapse and poor prognosis [119]. The targets of miR-210 include the DNA repair genes RAD52 and RAD23B [117], possibly providing a mechanism for increased genomic instability by inhibiting DNA repair. Induction of miR-210 may also prevent cells from undergoing apoptosis [118]. Another consequence of hypoxia-induced miR-210 induction is increased angiogenesis, as a result of down-regulation of the miR-210 target Ephrin-A3 (EFNA3) [120]. Further analysis found that miR-210 targets the iron-sulfur cluster assembly proteins ISCU1/2, and this may affect downstream metabolic functions, such as reduced ATP generation, elevated glycolysis, and mitochondrial metabolism [121]. Furthermore, acting upstream of HIF-1α, miR-519c can act as a hypoxia-independent regulator of HIF-1α by directly binding to the 3′ UTR, leading to reduced tumor angiogenic activity [122].
GENETIC ASSOCIATION
Genetic association studies have recently uncovered a number of single nucleotide polymorphisms associated with susceptibility to common diseases including breast cancer. While most of these studies have initially focused on coding genes many of these polymorphisms may be found in regulatory regions and regions containing non-coding RNAs. A SNP (or mutation) in a miRNA could have a significant effect on its hybridization to the target sites or, alternatively, could effect the transcription or the processing of the miRNA. In addition, polymorphisms in the miRNA target sites in mRNAs can also be associated with cancer risk. For example, a polymorphism within the KRas 3′UTR encoding a let-7 binding site results in increased KRAS expression in lung cancer as a result of reduced let-7 inhibition [123]. Similarly, a SNP within the 3′UTR of SET8, a methyltrans-ferase that represses p53 activity, generates a new miR-502 binding site and is associated with early breast cancer onset in premenopausal women [124].
Using a large familial study population of BRCA1/2 mutation negative cases, Tchatchou et al. analyzed the impact on breast cancer risk of 11 miRNA target site SNPs located in the 3′UTR of cancer associated genes [125]. There was a significant association with familial breast cancer risk for a variant affecting a putative miR-453 binding site in the 3′UTR of ER. The protective effect was stronger in pre-menopausal women and in high-risk familial cases. The binding of the miRNA was predicted to be stronger when the protective allele is present, potentially leading to lower levels of ER and making biological sense with the role of estrogen and ER in breast cancer progression. Nicoloso and colleagues analyzed SNPs associated with breast cancer risk for their ability to modify miRNA binding sites [126]. They identified SNPs in TGFB1 and XRCC1 that could modulate their expression by differential interaction with miR-187 and miR-138, respectively. A genome-wide bioinformatics analysis of human HapMap data predicted approximately 64% of transcribed SNPs can increase or decrease the binding energy of putative miRNA::mRNA duplexes. To assess whether SNPs affecting miRNA target sites are implicated in breast cancer susceptibility, the authors conducted a case-control population study. They observed that occurrence of miRNA target SNPs found in BRCA1 and TGFR1 significantly varied among populations with different risks of developing breast cancer. These predictions and biological effects were validated using in vitro luciferase reporter assays as well as testing the effects of overexpression of the two interacting miRNAs (miR-638 and miR-628-5p) on protein levels in cell lines with different genotypes.
Two recent studies each found a SNP in a miRNA to have protective effect against breast cancer. Hoffman et al. performed a genetic association study by screening genetic variants in 15 miRNAs [127]. They detected a common sequence variant in miR-196a-2 that was significantly associated with decreased breast cancer risk. Interestingly, when a CpG island upstream of miR-196a-2 is hypermethylated it is associated with reduced breast cancer risk. In cell line experiments, the mutant miR-196a-2 precursor was less efficiently processed to its mature form and had a diminished ability to regulate its target genes compared to the wildtype miRNA precursor. Additionally, evaluation of 11 SNPs found in miRNAs known to be involved in breast cancer revealed a SNP located in the terminal loop of pre-miRNA-27a, possibly also affecting processing to the mature form, with a protective effect in a large familial breast cancer study [128]. The authors also showed that this effect was stronger in the subgroup of <50 years of age while not being observed in the >50 years of age group, although the reason for this difference was not determined. Most of these studies await further validation in larger study populations in multicenter collaborations. Further functional and mechanistic studies also are needed to explain some of these associations. Genetic variations in miRNAs or their binding sites may represent useful targets for prevention in high-risk individuals. In addition, the impact of miRNA and target site variants on therapeutic and clinical outcome has yet to be explored.
CLINICAL SIGNIFICANCE AND PROGNOSTIC IMPLICATIONS
As discussed previously, miRNA expression profiling can be used to classify human cancers and differentiate tumor tissue from normal tissue [42, 44, 70]. For example, expression of the let-7 family members is generally decreased in breast cancer tissue compared to normal tissue. Additionally, the lower expression of specific let-7 family members can be further used to identify the pathological features of breast cancer tumors: PR negative (let-7c), positive lymph nodes/increased metastasis (let-7f-1, let-7a-2 and let-7a-3), and increased proliferation of cells within a tumor (let-7c and let-7d) [44]. Thus, lower expression of the let-7 family members is correlated with poor prognosis. In addition to the let-7 family members, this miRNA expression signature in human breast cancers was correlated with other pathological features such as estrogen receptor positive and progesterone receptor positive, HER2 negative and tumor stage, proliferation index and metastatic state [44, 45] (Table 1).
Table 1.
Expression of miRNAs in Breast Cancer and their Targets
| miRNA | Expression in Breast Cancer Subtypes | Activity | Targets |
|---|---|---|---|
| let-7 family | ↑L, ↑Her2−, ↓NL | TS | Ras[142], HMGA2[144] |
| miR-9 | ↑ER+, ↓Her2+ | Pro-metastatic | CDH1[101] |
| miR-10a/b | ↑L, ↓M, ↑Her2− | OG/Pro-metastatic | HOXD10[103], Tiam1[104] |
| miR-17-5p | ↑B, ↑NL | TS | AIB1[88] |
| miR-15/16 | ↓NL | BCL2[153], CCND1[154] | |
| miR-21 | ↑ER+, ↓Her2+ | OG/Pro-metastatic | PTEN[71], TPM1[74], PDCD4[72], Maspin[73], BCL2[140], p63[75], HNRPK[75] |
| miR-22 | ↓ER+ | ERα[51] | |
| miR-26a/b | ↑PR+, ↑ER+ | ||
| miR-29b | ↓L, ↑Her2+ | TS | MCL1[155], TCL1[156], DNMT3[157], p85α[94], CDC42[94] |
| miR-30a-5p, -30b, -30c, -30d | ↑PR+, ↑ER+ | ||
| miR-31 | Anti-metastatic | Fzd3, ITGA5, MMP16, M-RIP, RDX and RhoA[110] | |
| miR-107 | ↑Her2− | CDK6[158] | |
| miR-125a/b | ↓Her2+ | Anti-metastatic/TS | HER2 and HER3[78] |
| miR-126 | ↓L, ↑Her2− | TS | IRS-1[107], p85β[159] |
| miR-130a | ↓NL, ↓Her2+ | GAX and HOXA5[160] | |
| miR-143 | ↑Her2− | TS | Raf1[161], ERK5[162] |
| miR-145 | ↓B, ↓L, ↑Her2− | Anti-metastatic/TS | Raf1[161], ERK5[162] |
| miR-146a/b | ↑NL | Anti-metastatic | EGFR[163], CXCR4[164], IRAK1[165], KIT[166], TRAF5[165], IL8 and IL6[108], MMP-9[108] |
| miR-150 | ↑ER+, ↓Her2+ | c-Myb[167] | |
| miR-154 | ↑Her2− | ||
| miR-182 | Pro-metastatic | FOXO3 and MITF[168] | |
| miR-185 | ↓ER+ | AKT1, CDK6 and HMGA2[158] | |
| miR-191 | ↑ER+ | ||
| miR-195 | ↑Her2− | ||
| miR-200 family | ↓M, ↑ER+ | Anti-metastatic/TS | TCF8/ZEB1 and ZEB2[60–62], BMI1[37], TUBB3[100], SOX2 and Klf4[65] |
| miR-205 | ↓M, ↑ER+, ↓Her2+ | HER3[80], E2F and PKCε[169], PTEN[35], ZEB1 and ZEB2[60] | |
| miR-206 | ↓ER+, ↑NL | Anti-metastatic/TS | ERα[46] |
| miR-210 | ↑ER+, | Pro-metastatic | EFNA3, E2F3, NPTX1, RAD52, and ACRVB1[170] RAD23B[117], EFNA3[120], ISCU1/2[121] |
| miR-212 | ↓ER+ | ZO-1[171] | |
| miR-214 | ↓L | PTEN[172] | |
| miR-221/222 | ↑Her2+ | FOXO3 and ERα[49] | |
| miR-335 | Anti-metastatic/TS | SOX4, PTPRN2, MERTK, and TNC[106] | |
| miR-373 | ↑M | Pro-metastatic/OG | CD44[111], RAD23B[117], LATS2[173] |
| miR-520c | Pro-metastatic | CD44[111] |
NL=Normal like, L=luminal, B=Basal, M=Metastatic disease, TS=Tumor Suppressor, OG=Oncogenic
miRNA THERAPEUTICS
Until recently, most cancer therapeutics have been either small molecule inhibitors designed to disrupt functional domains, such as the ATP pocket in protein kinases, or humanized monoclonal antibodies, which may inhibit cell surface receptor signaling. Since miRNAs are capable of interacting with hundreds of protein-coding genes, they are becoming attractive potential therapeutic targets. Several methods for inhibiting and mimicking miRNA function recently have been described as summarized below.
Antisense oligos that bind to the mature miRNA sequence have been exploited to inhibit specific miRNAs. Antisense oligos are 17–22 nucleotides long and are designed to have the exact nucleic acid complement of the mature miRNA sequence such that the targeted miRNA will bind to the miRNA inhibitor by means of Watson-Crick base pairing, and not to its endogenous target mRNA. One of the first methods used for inhibiting specific miRNAs was the use of 2′-O-Methyl antisense oligos. The unique design of the 2′-ribose modification made the oligo resistant to cellular nucleases, such that the oligo would have an increased stability and half-life as compared to a conventional oligo, and the inhibitor-miRNA interaction was thus irreversible [129,130]. Originally used for detection of miRNA expression by in situ hybridization and Northern Blot analysis, locked nucleic acids (LNAs) were also used as miRNA inhibitors in vitro [131, 132]. Like the 2′-O-Methyl oligos, LNAs are resistant to cellular nucleases, but the conformational change design has given the LNA/RNA interaction increased stability [133]. The use of these 2′-O-Methyl and LNA antisense oligos were effective in vitro, but the need to deliver antisense oligos in vivo has lead to the design of more advanced oligos. The addition of a cholesterol modification to the 3′ end of a 2′-O-Methoxyethyl antisense oligo, now called “antagomirs”, helped facilitate the uptake of this miRNA inhibitor into cells in vivo [134]. This method allowed injection of antagomirs intravenously, and was used to target several miRNAs in many tissues, with the exception of the brain, likely due to the blood-brain barrier [134]. Although the mechanism is not fully understood, an anta-gomir-miRNA duplex will result in the degradation of the target miRNA and subsequent recycling of the antagomir, allowing it to act on more target miRNAs [135]. Therapeutically, antagomirs have been used in mouse models of human diseases and have been shown to revert phenotypes associated with over-expression of miRNAs [134, 136–139].
As mentioned previously, the oncogenic miR-21 was found to be up-regulated in human breast cancers compared to normal tissue. The use of an antisense oligo to miR-21 in a breast cancer cell culture model was successful at inhibiting tumor growth when treated cells were transplanted into recipient mice. As expected, when miR-21 was inhibited, resulting tumors had a longer latency, were proliferating slower, had increased apoptosis and had increased susceptibility to anti-cancer drugs [140].
A second more recent approach is the use of miRNA mimics to restore a miRNA in diseases where a miRNA is lost. Mimics are generally chemically synthesized artificial miRNAs, designed to be stable and less susceptible to endogenous RNases, that can be delivered systemically. In mouse models of heart disease, several miRNAs were deregulated following a myocardial infarction. In particular, miR-29 expression was significantly lower in both mouse models and human samples of myocardial infarction. Cell culture models were employed to study the effects of replacing miR-29 with a miRNA. These resulted in the reversal of cardiac fibrosis, a result of myocardial infarction; however, these studies have not yet been validated in vivo [141]. Additionally, a miRNA mimic for the well-studied tumor suppressor miRNA let-7 was used therapeutically in mouse models of lung cancer. Expression of let-7 is frequently lower in many cancers, and is thought to act as a tumor suppressor by modulating expression of multiple oncogenes, including Ras, Myc and HMGA2 [142–144], as well as cell cycle genes Cyclin D2, Cdk6 and Cdc25 [145]. Addition of a let-7miRNA mimic to lung cancer cell culture models was shown to decrease proliferation by induction of cell cycle arrest and cell death in vitro, as well as in mouse lung cancer xenograft models. Additionally, genetically engineered mouse models of lung cancer were also inhibited by the let-7 mimic [146].
Chemotherapeutic resistance in primary and metastatic breast cancer presents a major clinical challenge, as many patients will become resistant to multiple cytotoxic agents, termed mutlidrug resistance. The use of miRNA-based therapy to enhance anticancer drug activity was recently investigated in the NCI-60 human cancer cell line panel. These investigators used the NCI-60 cell line panel and miRNA mimics and inhibitors of let7i, miR-16, and miR-21, and found that altering miRNA levels could affect the efficacy of a number of the anticancer agents tested [147]. These results provide a potential for using miRNAs in a clinical setting to improve current chemotherapy strategies. Additionally, in a cisplatin-resistant MCF-7 cell culture model of human breast adenocarcinoma, several miRNAs were found to be disregulated, including miR-345 and miR-7, which both target the human multidrug resistance-associated protein 1 (MRP1) [148]. This disregulation may, in part, account for some of the acquired drug resistance in these cells.
Although miRNAs appear to be promising therapeutic targets, it should be noted that since they have the potential to regulate hundreds of genes simultaneously, there is a greater potential for unexpected effects. Since the target repertoire of each miRNA will likely be context-, cell type- and even disease stage-dependent, more investigation into targets specificity must be done before they can be used clinically. Another potential caveat to the use of miRNA-based therapeutics is that it has been shown that increased proliferation in normal T cells is associated with an increase in expression of mRNAs terminating at upstream polyadenylation sites [149]. Likewise, compared to normal cells, cancer cells may display shorter 3′UTRs as a result of their use of alternative cleavage and polyadenylation sites [150]. These events result in shorter 3′UTRs and fewer miRNA target sites. Therefore, highly proliferative cancer cells may exhibit a resistance to miRNA-based therapeutics if the target mRNAs contain truncated 3′UTRs.
PERSPECTIVES
Breast cancer is the second most common type of cancer worldwide and the fifth most common cause of cancer deaths. Historically, breast cancers were classified by histological appearance (ductal, lobular or mixed), tumor pathology (well-differentiated low grade, poorly differentiated high grade, moderately differentiated intermediate grade), and tumor stage, including metastasis and lymph node lesions. With immunohistochemistry profiling, breast cancers were further classified by protein and gene expression status (Estrogen Receptor, Progesterone Receptor and HER2/neu) for prognostic and predictive purposes. Micro-array studies have been able to demonstrate that these tumor subtypes can be classified by gene expression profiles. Based on these classifications, at least four major tumor subtypes have been identified: estrogen receptor negative/HER2 positive; basal-like estrogen receptor negative; and estrogen receptor positive and Keratin 8/18 positive (designated as “luminal”), which has been further subdivided into “luminal A” and “luminal B” tumors [151]. These tumor subtypes have distinct mRNA gene expression signatures, prognoses, and phenotypes and are thus treated in a clinical setting differently. More recently, miRNA microarray profiling has been used to demonstrate that these breast cancer subtypes can be identified by their unique miRNA expression profiles [42, 44, 70, 152].
Since there are targeted therapies against estrogen receptor positive breast cancers (such as tamoxifen and aromatase inhibitors) and Her2/neu positive breast cancers (such as trastuzumab), these breast cancer subtypes generally have better prognoses and lower mortality. Estrogen receptor negative and Her2/neu negative breast cancers, however, are resistant to these targeted therapies, and therefore have poor prognoses with increased mortality. This heterogeneity highlights the need to understand breast cancer biology and the underlying changes within stem and progenitor cell populations that may be the cancer cell of origin to design new therapeutic targets. It is for these reasons that miRNAs represent a new and exciting set of therapeutic targets with enormous potential.
Acknowledgments
This work was supported by a Department of Defense Breast Cancer Program Predoctoral Fellowship DAMD W81XWH-06-1-0716 (S.B.G.), a Komen Post-doctoral fellowship PDF0707744 (J.I.H.), and an NIH grant CA-16303 (J.M.R.).
ABBREVIATIONS
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- UTR
Untranslated region
- Lin
Lineage
- ALDH1
Aldehyde dehydrogenase
- Sca-1
Stem cell antigen-1
- BMI1
B lymphoma Mo-MLV insertion region 1/polycomb ring finger oncogene
- HER2/neu, ErbB2
Human Epidermal growth factor Receptor 2
- ER
Estrogen Receptor
- PR
Progesterone Receptor
- FACS
Fluorescent activated cell sorting
- HMGA2
High mobility group AT-hook 2
- H-Ras
Harvey rat sarcoma viral oncogene homolog
- EMT
Epithelial to mesenchymal transition
- TGFβ
Transforming growth factor β
- Zeb1/2
Zinc finger E-box-binding homeobox 1/2
- Sox
Sex determining region Y(SRY)-box
- Klf4
Krueppel-like factor 4
- iPS
Induced Pluripotent Stem Cells
- CSC
Cancer stem cell
- BIC
B-cell Integration Cluster
- PTEN
Phosphatase and tensin homolog
- PDCD4
Programmed Cell Death 4
- TPM1
Tropomyosin 1
- BCL2
B-cell CLL/lymphoma 2
- SNP
Single nucleotide polymorphism
- Bim
Bcl2-interacting mediator of cell death
- AIB1
Amplified in breast cancer-1
- PKCε
Protein Kinase Cε
- CDC42
Cell division control protein 42
- MET
Mesenchymal to epithelial transition
- BRMS1
Breast cancer metastasis suppressor 1
- HOXD10
Homeobox D10
- RHOC
Ras homolog gene family, member C
- PTPRN2
Receptor-type tyrosine-protein phosphatase N2
- MERTK
c-Mer proto-oncogene tyrosine kinase
- PLCB1
1-Phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-1
- COL1A1
Collagen, type I, alpha 1
- TNC
Tenascin C
- IRS-1
Insulin Receptor Substrate-1
- NFkB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- IRAK1
Interleukin-1 receptor-associated kinase 1
- TRAF6
TNF receptor associated factor 6
- ITGA5
Integrin α5
- RDX
Radixin
- RhoA
Ras homolog gene family, member A
- MUC1
Mucin 1
- ROCK
Rho-associated kinase
- LNA
Locked nucleic acid
- MRP1
Multidrug resistance-associated protein 1
- HNPRK
Heterogeneous nuclear ribonucleoprotein K
- MCL1
Nduced myeloid leukemia cell differentiation protein
- TCL1
T-cell leukemia/lymphoma protein 1A
- DNMT3
DNA methyltransferase 3
- Fzl3
Frizzled 3
- MMP
Matrix metalloproteinase
- M-RIP
Myosin phosphatase Rho-interacting protein
- GAX
Growth arrest-specific homeobox
- Raf1
Murine leukemia viral oncogene homolog 1
- ERK5
Mitogen-activated protein kinase 7
- EGFR
Epidermal growth factor receptor
- CXCR4
CXC chemokine receptor
- KIT
- IL8
Interluekin 8
- FOXO3
Forkhead box O3
- MITF
Microphthalmia-associated transcription factor
- ZO-1
Tight junction protein 1, zona occludens 1
- LATS2
Large tumor suppressor, homolog 2
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22(3):165–73. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165(1):31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7(6):633–6. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B. MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}-actin gene. Nucleic Acids Res. 2008;36(19):6318–32. doi: 10.1093/nar/gkn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 13.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andl T, Murchison EP, Liu F, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16(10):1041–9. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 16.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202(2):261–9. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103(8):2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Develop Cell. 2003;5(2):351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 19.Ivey KN, Muth A, Arnold J, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–29. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452(7184):225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells (Dayton, Ohio) 2006;24(4):857–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21(6):1469–77. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 25.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 26.Daniel CW, De Ome KB, Young JT, Blair PB, Faulkin LJ., Jr The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc Natl Acad Sci USA. 1968;61(1):53–60. doi: 10.1073/pnas.61.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19(5):515–20. [PubMed] [Google Scholar]
- 28.Young LJ, Medina D, DeOme KB, Daniel CW. The influence of host and tissue age on life span and growth rate of serially transplanted mouse mammary gland. Exp Gerontol. 1971;6(1):49–56. doi: 10.1016/0531-5565(71)90048-9. [DOI] [PubMed] [Google Scholar]
- 29.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 30.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 31.Moraes RC, Zhang X, Harrington N, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134(6):1231–42. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 32.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 34.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104(8):2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010;123(Pt 4):606–18. doi: 10.1242/jcs.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21(24):3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Rev Genet. 2004;5:396. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 39.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kluiver J, Haralambieva E, de Jong D, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45(2):147–53. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 42.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33(17):5394–403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 45.Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21(5):1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 47.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68(13):5004–8. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 48.Zhao JJ, Lin J, Yang H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Leva G, Gasparini P, Piovan C, et al. MicroRNA Cluster 221–222 and Estrogen Receptor {alpha} Interactions in Breast Cancer. J Natl Cancer Inst. 2010;102(10):706–21. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong J, Yu D, Wei N, et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277(7):1684–94. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 53.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–85. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 54.Zhang M, Behbod F, Atkinson RL, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68(12):4674–82. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 56.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 59.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 61.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283(22):14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iliopoulos D, Polytarchou C, Hatziapostolou M, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2(92):ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 66.Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol. 1989;9(6):2657–64. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39(2):167–9. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 69.Jiang S, Zhang HW, Lu MH, et al. MicroRNA-155 Functions as an OncomiR in Breast Cancer by Targeting the Suppressor of Cytokine Signaling 1 Gene. Cancer Res. 2010;70(8):3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 70.Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 71.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 73.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18(3):350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 74.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282(19):14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 75.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 76.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105(13):5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282(2):1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 79.Li W, Duan R, Kooy F, Sherman SL, Zhou W, Jin P. Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet. 2009;46(5):358–60. doi: 10.1136/jmg.2008.063123. [DOI] [PubMed] [Google Scholar]
- 80.Iorio MV, Casalini P, Piovan C, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69(6):2195–200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 81.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19(4):439–48. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 83.Yu Z, Wang C, Wang M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182(3):509–17. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282(4):2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 85.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 86.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Novotny GW, Sonne SB, Nielsen JE, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17–92 cluster. Cell Death Differ. 2007;14(4):879–82. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 88.Hossain A, Kuo MT, Saunders GF. Mir-17–5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26(21):8191–201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 90.Avril-Sassen S, Goldstein LD, Stingl J, et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics. 2009;10:548. doi: 10.1186/1471-2164-10-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takwi A, Li Y. The p53 Pathway Encounters the MicroRNA World. Curr Genomics. 2009;10(3):194–7. doi: 10.2174/138920209788185270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106(9):3207–12. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16(1):23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 95.Braun CJ, Zhang X, Savelyeva I, et al. p53-Responsive MicroRNAs 192 and 215 Are Capable of Inducing Cell Cycle Arrest. Cancer Res. 2009;68(24):10094–104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 97.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69(19):7495–8. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9(4):293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 99.Dykxhoorn DM, Wu Y, Xie H, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE. 2009;4(9):e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-1046. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Edmonds MD, Hurst DR, Vaidya KS, Stafford LJ, Chen D, Welch DR. Breast cancer metastasis suppressor 1 coordinately regulates metastasis-associated microRNA expression. Int J Cancer. 2009;125(8):1778–85. doi: 10.1002/ijc.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 104.Moriarty CH, Pursell B, Mercurio AM. miR-10b targets Tiam1: Implications for Rac activation and carcinoma migration. J Biol Chem. 2010 doi: 10.1074/jbc.M110.121012. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J, Du YY, Lin YF, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377(1):136–40. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 108.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–7. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23(22):2592–7. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biol. 2008;10(2):202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 112.Sachdeva M, Mo YY. MicroRNA-145 Suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70(1):378–87. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu S, Goldstein RH, Scepansky EM, Rosenblatt M. Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res. 2009;69(22):8742–51. doi: 10.1158/0008-5472.CAN-09-1541. [DOI] [PubMed] [Google Scholar]
- 114.Yu Z, Willmarth NE, Zhou J, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA. 2010;107(18):8231–6. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219(2):214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 116.Camps C, Buffa FM, Colella S, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14(5):1340–8. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 117.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69(3):1221–9. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Foekens JA, Sieuwerts AM, Smid M, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105(35):13021–6. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fasanaro P, D’Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283(23):15878–83. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10(4):273–84. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cha ST, Chen PS, Johansson G, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70(7):2675–85. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 123.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68(20):8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Song F, Zheng H, Liu B, et al. An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;15(19):6292–300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 125.Tchatchou S, Jung A, Hemminki K, et al. A variant affecting a putative miRNA target site in estrogen receptor (ESR) 1 is associated with breast cancer risk in premenopausal women. Carcinogenesis. 2009;30(1):59–64. doi: 10.1093/carcin/bgn253. [DOI] [PubMed] [Google Scholar]
- 126.Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70(7):2789–98. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69(14):5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang R, Schlehe B, Hemminki K, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121(3):693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 129.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2(4):E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10(3):544–50. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 132.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–41. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 133.Wahlestedt C, Salmi P, Good L, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97(10):5633–8. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 135.Krutzfeldt J, Kuwajima S, Braich R, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35(9):2885–92. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fontana L, Fiori ME, Albini S, et al. Antagomir-17–5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3(5):e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 138.Ren XP, Wu J, Wang X, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119(17):2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007 May;13(5):613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 140.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 141.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105(35):13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 143.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 144.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 146.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 147.Blower PE, Chung JH, Verducci JS, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7(1):1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 148.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010 doi: 10.1002/ijc.25191. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 149.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67(24):11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 153.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 155.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 157.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104(40):15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS ONE. 2009;4(8):e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Developmental Cell. 2008;15(2):272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111(3):1217–26. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Michael MZ, SMOC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–91. [PubMed] [Google Scholar]
- 162.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 163.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69(4):1279–83. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Labbaye C, Spinello I, Quaranta MT, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10(7):788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 165.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102(52):19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lin YC, Kuo MW, Yu J, et al. c-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol Biol Evol. 2008;25(10):2189–98. doi: 10.1093/molbev/msn165. [DOI] [PubMed] [Google Scholar]
- 168.Segura MF, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106(6):1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gandellini P, Folini M, Longoni N, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69(6):2287–95. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 170.Fasanaro P, Greco S, Lorenzi M, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284(50):35134–43. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yuen T, Ruf F, Chu T, Sealfon SC. Microtranscriptome regulation by gonadotropin-releasing hormone. Mol Cell Endocrinol. 2009;302(1):12–7. doi: 10.1016/j.mce.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 173.Lee KH, Goan YG, Hsiao M, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in hu/man esophageal cancer. Exp Cell Res. 2009;315(15):2529–38. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]