Abstract
Replication stress involving collision of replisomes with camptothecin (CPT)-stabilized DNA-Topoisomerase I adducts activates an ATR-dependent pathway to promote repair by homologous recombination. To identify human genes that protect cells from such replication stress, we performed a genome-wide CPT sensitivity screen. Among numerous candidate genes are two previously unstudied proteins; the ankyrin repeat protein NFKBIL2 and C6ORF167 (MMS22L) distantly related to yeast replication stress regulator Mms22p. MMS22L and NFKBIL2 interact with each other and with FACT (facilitator of chromatin transcription) and MCM (minichromosome maintenance) complexes. Cells depleted of NFKBIL2 or MMS22L are sensitive to DNA damaging agents, load phosphorylated RPA onto chromatin in a CTIP-dependent manner, activate the ATR/ATRIP-CHK1 and double-strand break repair signaling pathways, and are defective in HR. This study identifies MMS22L-NFKBIL2 as components of the replication stress control pathway and provides a resource for discovery of addition components of this pathway.
Introduction
Genomic integrity is critical to organismal survival and is controlled by the DNA damage response (DDR) network, an elaborate signal transduction system that senses DNA damage and recruits appropriate repair factors (Harper and Elledge, 2007). Repair of DNA damage during the DNA replication stage of the cell cycle represents a particularly challenging process for the cell (Cimprich and Cortez, 2008). Such damage can take several forms but the majority of cases involve the collision of a replisome with an obstruction in the DNA, thereby blocking the replication of a particular segment of DNA. The elucidation of mechanisms used in the response to and repair of collapsed replication forks is an area of intense investigation (Cimprich and Cortez, 2008). This reflects both the extent to which endogenous and exogenous cellular stress causes replication problems and the fact that many chemotherapeutic agents activate the DNA damage pathway and trigger cell death by promoting replication fork collapse. A case in point are agents such as camptothecin (CPT), the founding member of a class of Topoisomerase I inhibitors. CPT forms a tight complex with Topoisomerase I-DNA adducts and prevents DNA re-ligation, which leads to formation of single-strand breaks (SSBs) that can be converted to double-strand breaks (DSBs) upon encounter with the replication apparatus (Pommier, 2009).
Central to the prevention and repair of collapsed replication forks is the PI3K-related protein kinase ATR, which acts in concert with its obligate partner protein ATRIP (Cortez et al., 2001) to signal DNA replication stress (Zou, 2009; Friedel et al., 2009; Zou, 2007; Paulsen and Cimprich, 2007). ATR/ATRIP is recruited to regions of single-stranded DNA (ssDNA) coated with the Replication Protein A (RPA) ssDNA binding protein complex through its ATRIP subunit (Zou and Elledge, 2003). A separate replication stress sensor is the RFC-related Rad17 complex that recognizes RPA-ssDNA complexes adjacent to a dsDNA-ssDNA junction and loads the PCNA-related 911 complex (RAD9-HUS1-RAD1) together with TopBP1 onto sites of DNA damage. The interaction of the 911 and ATR sensors activates ATR kinase activity and sets in motion the DNA stress response signal transduction pathway (Zou and Elledge, 2001; Zou et al., 2002; Paulsen and Cimprich, 2007; Zou, 2007; Cimprich and Cortez, 2008). Activated ATR phosphorylates many substrates including RPA2, and the CHK1 protein kinase (Liu et al., 2000), which then phosphorylate additional effectors involved in cell cycle checkpoint responses. ATR is structurally related to the ATM protein kinase, which plays a primary role in the response to DSBs, and directly activates the CHK2 protein kinase (Harper and Elledge, 2007). ATM and ATR share related substrate specificities and overlapping targets, and can function in concert to signal particular kinds of DNA damage (Matsuoka et al., 2007). For example, DSB formation initially sensed by ATM is followed later by resection, which allows RPA loading and activation of ATR.
The available evidence indicates a preferential role for ATR-CHK1 in response to and repair of collapsed forks generated by CPT and related Topoisomerase I inhibitors in proliferating cells (Flatten et al., 2005). Given that CPT treatment leads to 911 clamp loading in addition to CHK1 activation (Loegering et al., 2004), and that functional inactivation of 911 components RAD9 and HUS1 renders cells hypersensitive to CPT derivatives (Loegering et al., 2004), it appears likely that CPT-dependent fork collapse induces a largely canonical ATR signaling pathway. One possibility is that exposed ssDNA generated by DSB resection allows RPA binding and signaling to ATR. The ability of ATR to reactivate collapsed replication forks may explain why functional inactivation of this pathway has more severe consequences upon cell survival after replication stress as compared to that observed with ATM/CHK2 inactivation (Flatten et al., 2005). In contrast, the response of post-mitotic cells to Topoisomerase I inhibitors is exclusively ATM-dependent due to the absence of ATR expression (Sordet et al., 2010), suggesting that mitotic and post-mitotic cells employ distinct mechanisms for repair.
In order to gain further insight into the mechanisms controlling signaling and repair of collapsed replication forks, we performed a genome-wide CPT sensitivity screen in mammalian cells using recently developed barcode-based drop-out screening technology (Schlabach et al., 2008). Among the proteins displaying the strongest sensitivity to CPT upon depletion was a previously unstudied 120 kDa protein encoded by C6ORF167. Proteomic analysis of C6ORF167 complexes revealed the presence of a previously unstudied human ankyrin, TPR, and leucine-rich repeat containing protein called NFKB Inhibitor-Like 2 (NFKBIL2), which was identified independently in our CPT synthetic lethality screen, and is distantly related to Arabidopsis BRU1/TONSOKU, a protein previously implicated in the S-phase DDR and epigenetic regulation (Takeda et al., 2004). U2OS and HeLa cells depleted of C6ORF167 or NFKBIL2 were exquisitely sensitive to DNA damaging agents that block replisome progression, and more weakly to ionizing radiation (IR). Depletion of C6ORF167 or NFKBIL2 in the absence of exogenous DNA damage led to activation of ATR-dependent signaling cascades, formation of 53BP1, γH2AX, and phospho-RPA-positive foci, a delay in DDR signaling inactivation after DNA damage, and defects in homologous recombination (HR). Interestingly, the C6ORF167-NFKBIL2 complex also associated with the nucleosome regulatory FACT complex composed of the ATM/ATR substrate SSRP1 (Matsuoka et al., 2007) and SUPT16H, as well as the replicative helicase MCM (minichromosome maintenance) complex. C6ORF167 is distantly related to and shares functional parallels with yeast Mms22p/Mus7p (Duro et al., 2008; Dovey et al., 2009; Yokoyama et al., 2007), and for this reason, we will refer to C6ORF167 hereafter as MMS22-Like (MMS22L). Mms22p is required for resistance to replication stress and replication re-start after repair in yeast. Mms22p interacts with the CUL4-DDB1-like ubiquitin ligase Rtt101p-Mms1p (Zaidi et al., 2008) and has recently been shown to be degraded by the proteasome after Rtt101p-Mms1p-dependent ubiquitination, but does not appear to associate with proteins related to NFKBIL2. Thus, this work reveals the MMS22L-NFKBIL2 complex as a component of the machinery required for mammalian cells to respond to replicative stress.
Results
A Genetic Screen for Proteins that Confer Resistance to Replisome Stress
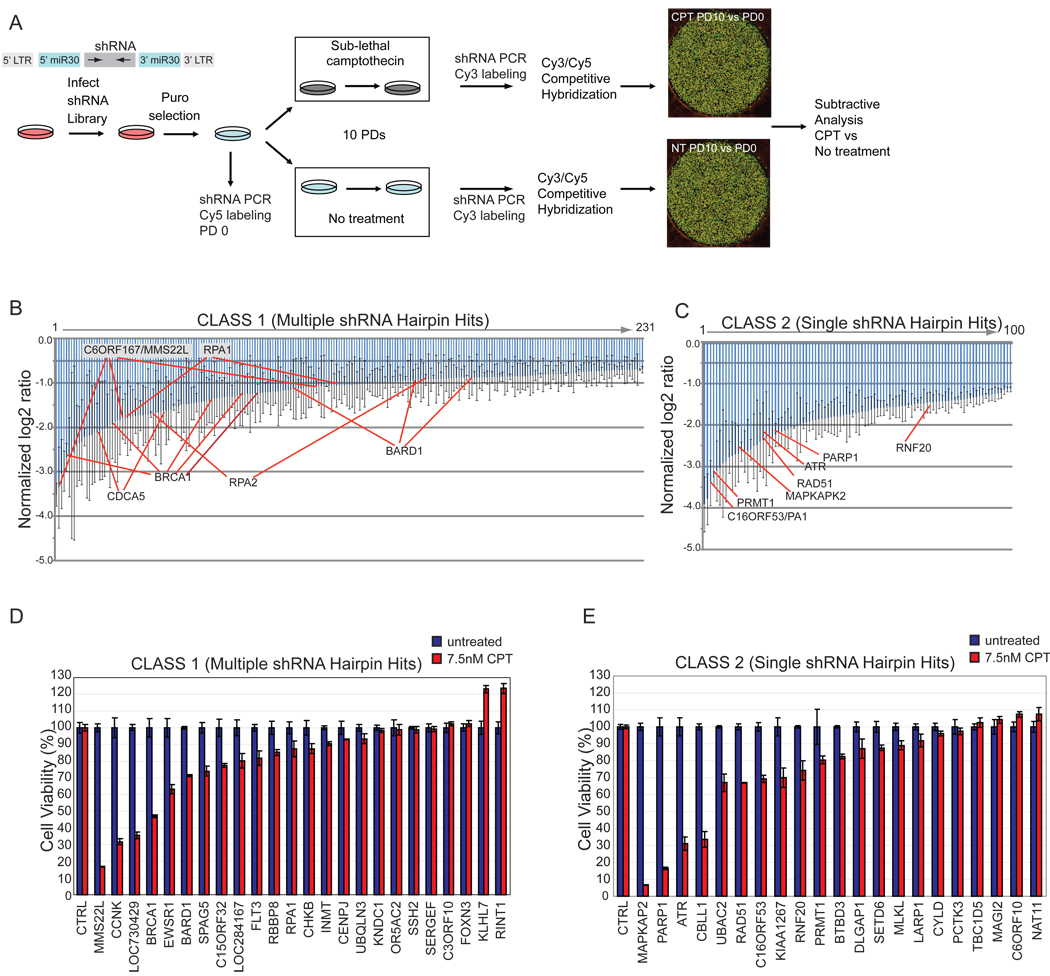
We performed a genome-wide shRNA screen to identify proteins whose depletion alters the response of human cervical carcinoma (HeLa) cells to CPT, a Topoisomerase I inhibitor that blocks replisome progression through stabilization of DNA-Topoisomerase I adducts and activates the ATR-CHK1 pathway (Figure 1A). Our screen was conducted using a library of 74,905 shRNAs targeting 32,293 unique human transcripts. The library was subdivided into 6 pools and HeLa cells were transduced in triplicate with each pool. Cell lines stably expressing shRNA pools were cultured either in the absence or presence of a sub-lethal dose of CPT (7.5 nM) for 10 population doublings (PD), and relative shRNAs abundance determined by half-hairpin barcode microarray hybridization (see Experimental Procedures). Using a strict scoring criteria (see Experimental Procedures), we identified 331 shRNA’s targeting 209 unique genes whose abundance was reduced in CPT treated populations (Table S1), as would be expected for genes whose depletion leads to reduced proliferation. We subdivided candidates into two classes based on whether multiple shRNA’s (Class1) or single shRNA’s (Class 2) for a given gene passed our criteria. (Figure 1B,C, see Experimental Procedures). Gene Ontology (GO) analysis revealed an enrichment of proteins involved in the DNA damage response, DNA replication, chromatin, and phosphorylation (p < 10−13). Proteins known to be involved in the DDR included ATR, RPA1, RPA2, RAD51, BRCA1, BARD1, RBBP8 (CTIP), MAPKAP2 and PARP1 among others (Figure 1B,C). Five independent shRNA’s targeting BRCA1 and 3 independent shRNA’s targeting its binding partner, BARD1, were identified in the screen, underscoring the importance of this repair complex in the response of cells to CPT (Table S1). While not the focus of this study, we identified several shRNA’s targeting Topoisomerase I that were enriched in CPT-treated cells (data not shown), as expected based on targeting of Topoisomerase I by CPT.
Figure 1. A genome-wide shRNA screen to identify genes necessary for resistance to CPT.
(A) Schematic of the primary screen.
(B, C) Normalized log2 ratios for candidate genes. Error bars indicate the standard error of the mean for triplicate hybridization signals for each shRNA. Individual shRNAs for selected genes are indicated.
(D, E) MCA in HeLa cells transduced with shRNAs targeting the indicated Class 1 (panel D) or Class 2 (panel E) genes with or without CPT (7.5 nM). Error bars represent standard deviation (STDEV) for triplicate assays.
Candidate Gene Validation
We performed multicolor competition assays (MCA) (Smogorzewska et al., 2007) on a subset of candidate genes from each class to retest for CPT sensitivity in an independent assay. GFP-expressing HeLa cells were transduced with shRNA’s targeting candidate genes and co-cultured with an equal number of unlabeled HeLa cells transduced with a control shRNA in the absence or presence of sub-lethal doses of CPT (7.5 nM). After 7–10 days, the ratio of GFP-positive to unlabeled cells was determined. As expected, cells transduced with shRNA’s targeting known DDR genes such as ATR were highly sensitive to CPT (Figure 1D,E and data not shown). In total, 13 of 24 Class 1 shRNA’s (Figure 1D) and 14 of 21 Class 2 shRNA’s (Figure 1E) tested displayed enhanced CPT sensitivity upon depletion (<90% viability compared with control cells), including several genes not previously linked with the DDR.
MMS22L is Required for Resistance to a Variety of DNA Damaging Agents
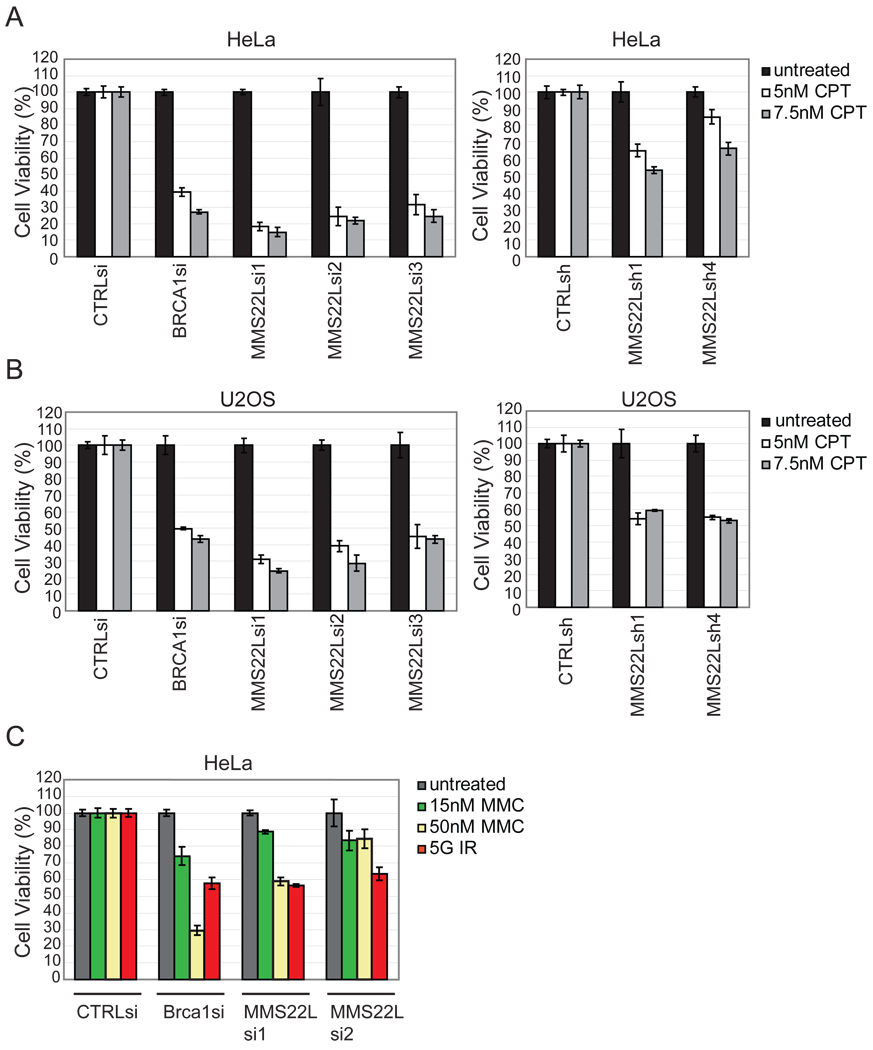
Among the Class 1 proteins with the strongest CPT sensitivity phenotype was the previously uncharacterized protein C6ORF167 (Figure 1B, D). C6ORF167 is distantly related to yeast Mms22p/Mus7p, a protein required for resistance to IR and CPT (Bennett et al., 2001), and will be referred to as MMS22L to reflect this relationship as well as shared phenotypes (see Discussion and Figure S1). Depletion of MMS22L with 3 siRNAs and 2 shRNAs led to sensitivity to CPT (5 or 7.5 nM) in both HeLa and U2OS cells (Figure 2A, B). Loss of MMS22L also resulted in an increased sensitivity of cells to MMC and to IR, indicating that MMS22L may play a broad role in the cellular response to DNA damage (Figure 2C). Based on the strength of the phenotype seen upon depletion of MMS22L, which was comparable to that seen with BRCA1 (Figure 2A, B), this gene was chosen for further study.
Figure 2. MMS22L promotes resistance to DNA damaging agents.
(A, B) MCA in HeLa or U2OS cells using the indicated RNAis with or without CPT (5 or 7.5 nM). CTRLsi is a control siRNA (see Table S1). Error bars represent STDEV for triplicate assays.
(C) MCA in HeLa cells transduced with RNAis with or without MMC (15 nM or 50 nM) or IR. Error bars represent STDEV for triplicate assays.
MMS22L Interacts with with NFKBIL2, FACT, and MCMs
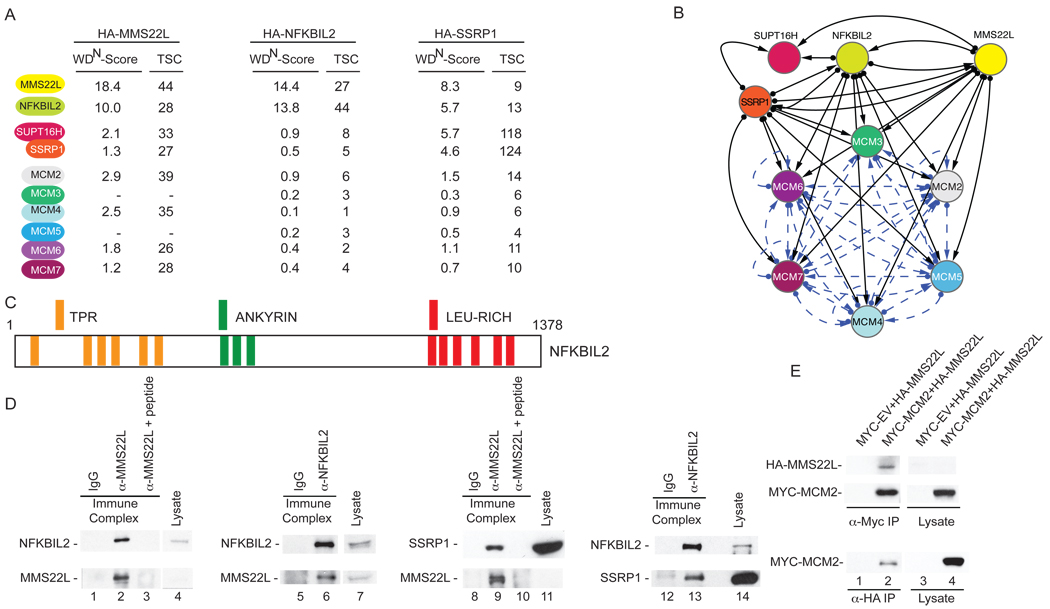
We took two parallel approaches to examine MMS22L. First, as described subsequently, we performed a phenotypic analysis of its role in DNA repair. Second, we undertook a proteomic analysis of MMS22L using our recently reported CompPASS platform (Sowa et al., 2009; Behrends et al., 2010) in order to identify proteins that might cooperate with or antagonize its function. In the later approach, the top scoring interacting proteins included NFKBIL2, both components of the FACT complex (SSRP1 and SUPT16H), as well as several members of the MCM complex, which functions as the replicative helicase (Figure 3A,B, Table S2). Interestingly, NFKBIL2, a previously unstudied human protein containing N-terminal TPR repeats, central ankyrin repeats, and C-terminal leucine rich repeats (Figure 3C) was independently identified as a Class 1 DDR protein in our CPT sensitivity screen (Table S1). Reciprocal proteomic analysis of HA-NFKBIL2 identified MMS22L, SSRP1, SUPT16H, and MCMs (Figure 3A,B and Table S2). Moreover, MMS22L, NFKBIL2, and MCMs were identified in immunoprecipitates of HA-SSRP1 (Figure 3A,B and Table S2). Indeed, previous studies have found a functional interaction between FACT and the MCM complex (Tan et al., 2006; Tan et al., 2010). NFKBIL2 and SSRP1 were found in α-MMS22L immunoprecipitates, but were absent when a competing antigenic peptide was pre-incubated with α-MMS22L (Figure 3D, first and third panel). Moreover, α-NFKBIL2 immune complexes contained MMS22L and SSRP1 (Figure 3D, second and fourth panels). We note, however, that only a very small fraction of the abundant SSRP1 protein was associated with NFKBIL2 or MMS22L (Figure 3D), consistent with roles for FACT that are independent of the MMS22L-NFKBIL2 complex such as transcription-coupled chromatin remodeling. Furthermore, MYC-MCM2 associated with HA-MMS22L in a reciprocal fashion (Figure 3E). Proteomic analysis of MMS22L in the presence and absence of DNA damage (CPT) indicated the existence of a pre-assembled complex consisting of MMS22L, NFKBIL2, FACT, and MCMs, the composition of which is not altered appreciably 1 h post CPT treatment based on spectral counting (Table S2). Taken together, this analysis indicates that both MMS22L and NFKBIL2 interact with the FACT complex and with the MCM complex. The ability of MMS22L, NFKBIL2, and SSRP1 to associate with each of the other proteins as well as the MCM complex suggests the possibility that each of these components interact simultaneously to form a large complex, however we cannot exclude the possibility that MMS22L and NFKBIL2 interact with each other but interact independently with FACT or MCM complexes. Further studies are required to fully define the molecular organization of the complex.
Figure 3. MMS22L interacts with NFKBIL2, the FACT complex, and the replicative helicase MCM complex.
(A) HA-tagged proteins were stably expressed in 293T cells, immunoprecipitated with α-HA, and analyzed by mass spectrometry. Total spectral counts (TSCs) were analyzed using CompPASS, and normalized weighted D-scores (WDN-scores) determined (Behrends et al., 2010). Proteins with WDN-scores >1.0 are considered high confidence candidate interacting proteins.
(B) Interaction network for proteins identified in association with MMS22L, NFKBIL2, and SSRP1. Solid lines: this study. Dotted lines: BIOGRID.
(C) Domain structure of NFKBIL2.
(D) Validation of interaction in 293T cell extracts using endogenous co-immunoprecipiation with the indicated antibodies with or with a competing antigenic peptide for MMS22L.
(E) Extracts from 293T cells expressing the indicated proteins were subjected to immunoprecipitation with the indicated antibodies and blots probed with α-MYC or α-HA.
NFKBIL2 is Required for Resistance to DNA Damaging Agents
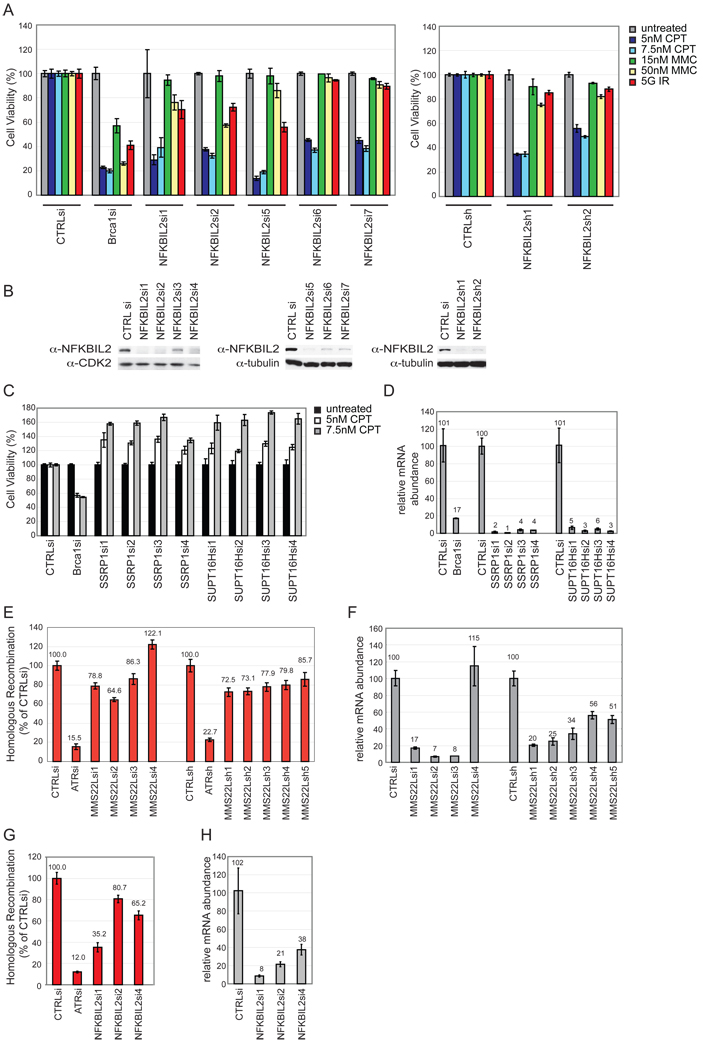
The fact that NFKBIL2 also was identified in our screen and associated with MMS22L prompted us to validate its involvement in the DDR using the MCA assay. We performed MCAs using 5 siRNAs and 2 additional shRNAs independent of those identified in the screen and several DNA damaging agents. Depletion of NFKBIL2 resulted in strong sensitivity to CPT with the most potent depletion leading to the strongest phenotypes (Figure 4A, B). Modest sensitivity was observed with higher levels of MMC and with IR in combination with the most potent siRNAs (Figure 4A,B), a result that parallels that found with MMS22L (Figure 2).
Figure 4. NFKBIL2 is required for resistance to DNA damaging agents, and both MMS22L and NFKBIL2 promote HR.
(A) MCA in HeLa cells using the indicated RNAis targeting NFKBIL2 or BRCA1 in the absence or presence of DNA damaging agents. CTRLsi or CTRLsh are a control RNAis (see Table S1). Error bars represent STDEV for triplicate assays.
(B) Immunoblots of crude cell extracts showing depletion of NFKBIL2 with various RNAis. Tubulin was used as a loading control.
(C, D) MCA in HeLa cells using the indicated RNAis targeting SSRP1, SUPT16H, or BRCA1 in the absence or presence of CPT (C). CTRLsi or CTRLsh are controls RNAis (see Table S1). Error bars represent STDEV for triplicate assays. mRNA depletion for each siRNA was determined using qPCR (D).
(E–H) MMS22L and NFKBIL2 promote HR. The extent of HR in DR-GFP U2OS cells subjected to MMS22L (E) or NFKBIL2 (G) depletion was determined by flow cytometry 36 h post infection with adenovirus-I-Sce1. Error bars are STDEV across 3 technical replicates. Extent of mRNA depletion by the indicated RNAi, as determined by qPCR (F and H).
NFKBIL2 and MMS22L also associated with the FACT complex. We performed MCAs on cells depleted of SSRP1 and SUPT16H with 4 independent siRNAs. Interestingly, depletion of either of these proteins led to resistance to CPT, up to 60% with the highest level of CPT tested (Figure 4C,D). While it is possible that FACT acts in some manner to oppose MMS22L-NFKBIL2 function, given roles for FACT in chromatin structure, we cannot rule out an indirect effect on the MCA assay.
MMS22L and NFKBIL2 promote Homologous Recombination
Given that HR is the primary means by which CPT, MMC and IR mediated DNA damage is repaired during S or G2 phase, we explored the effects of MMS22L and NFKBIL2 depletion on HR using a DR-GFP reporter construct in U2OS cells. The HR cassette is engineered with an I-Sce1 site, such that exposure to I-Sce1 triggers an HR event, generating an intact GFP protein only in cells that are HR competent (Xia et al., 2006). This assay does not distinguish between gene conversion and single-strand annealing as mechanisms for HR. With MMS22L, we tested 5 stable cell lines expressing shRNA’s as well as cells transiently transfected with 3 individual siRNA’s targeting MMS22L in the HR assay. Loss of MMS22L resulted in a consistent, albeit modest, defect in HR ranging from 65%–86% GFP positive cells relative to control RNAi (Figure 4E), with the effects on HR largely paralleling the extent of mRNA depletion as measured by qPCR (Figure 4F). By comparison, ATR depletion examined in parallel resulted in only 15% HR (Figure 4E). Similarly, we found that cells depleted of NFKBIL2 also displayed reduced HR activity, with the most potent siRNA reducing activity to 35% of controls and more weakly active siRNAs reducing activity to 65–81%, consistent with the possibility that low levels of NFKBIL2 support partial function (Figure 4G, H).
MMS22L and NFKBIL2 Suppress Spontaneous DDR Activation
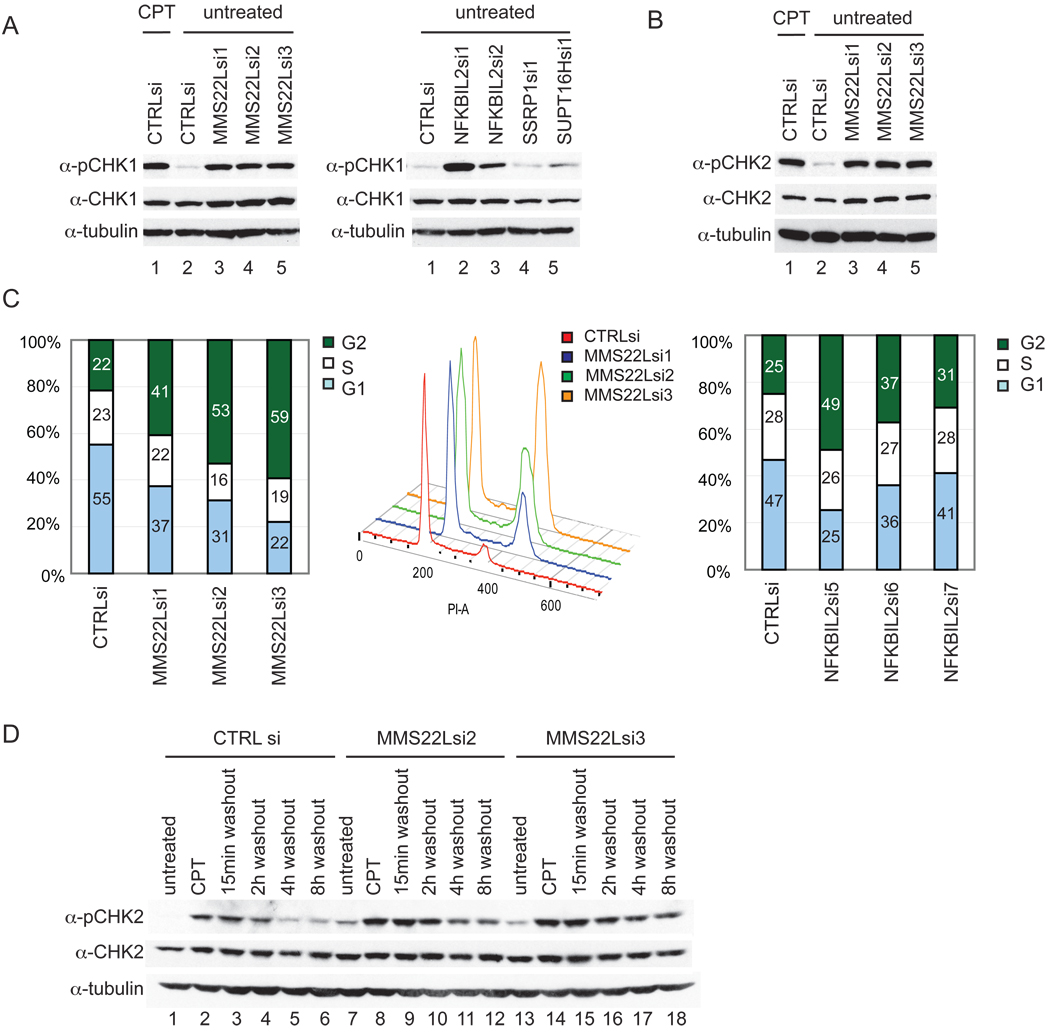
The absence of proteins that function in DNA repair often results in the accumulation of intrinsic DNA damage that can be visualized by the phosphorylation of DDR signaling components, including CHK1 (S296) and CHK2 (T68). Depletion of MMS22L with 3 independent siRNAs resulted in phosphorylation of both CHK1 and CHK2 to an extent similar to that seen upon treatment with CPT (Figure 5A,B). Similarly, depletion of NFKBIL2 also resulted in an increase in the phosphorylation of CHK1 (Figure 5A), with the most potent siRNA leading to the strongest induction of CHK1 phosphorylation (right panel, lane 2). In contrast, depletion of components of the FACT complex SSRP and SUPT16H had little effect on CHK1 phosphorylation (Figure 5A). These results are consistent with the activation of ATM/ATR in response to MMS22L or NFKBIL2 loss. DDR activation typically results in cell cycle arrest in G2. We found that cells depleted of MMS22L or NFKBIL2 accumulate in G2, as assessed using flow cytometry (Figure 5C), a phenotype also seen upon deletion of Mms22p in yeast (Baldwin et al., 2005). Depletion with the strongest MMS22L siRNAs resulted in a ~35–38% increase in G2/M cells, largely at the expense of G1 cells, and the strongest siRNA for NFKBIL2 (#1) resulted in a 24% increase in the G2/M population. Phospho-H3 staining revealed that the accumulated cells were in G2 phase as opposed to mitosis (data not shown).
Figure 5. MMS22L and NFKBIL2 suppresses intrinsic checkpoint activation and MMS22L is required for efficient recovery from the DNA damage checkpoint.
(A, B) Effect of MMS22L, NFKBIL2, or FACT depletion on CHK1 or CHK2 activation. Extracts from the indicated cells were immunoblotted with the indicated antibodies, using CPT (2 µM, 1h) as a checkpoint activation control.
(C) Effect of MMS22L or NFKBIL2 depletion on cell cycle progression. G1, S, and G2/M phases were determined by flow cytometry using propidium iodide staining, with the middle panel showing flow diagrams for MMS22L depletion.
(D) HeLa cells were transfected with the indicated siRNAs and after 72 h, cells were cultured with or without CPT (2 µM 1h). CPT was removed and extracts from cells analyzed for CHK2 and phospho-CHK2 (T68) at the indicated times with tubulin as a loading control.
MMS22L Depletion Delays Recovery from CPT Induced Damage
Budding yeast lacking Mms22p display defects in recovery from MMS induced replisome stalling as indicated by an extended phosphorylation of Rad53 as compared to wild type cells (Duro et al., 2008). In order to examine whether MMS22L promotes recovery from DNA damage, we examined CHK2 (T68) phosphorylation in HeLa cells depleted of MMS22L in both the presence of CPT and after CPT removal (Figure 5D). Compared with control siRNA treated cells, MMS22L-depleted cells displayed a delay in the decay of CHK2 T68 phosphorylation after CPT removal, indicating a likely defect in DNA repair (Figure 5D compare lanes 5,6 with lanes 11,12 and 17,18).
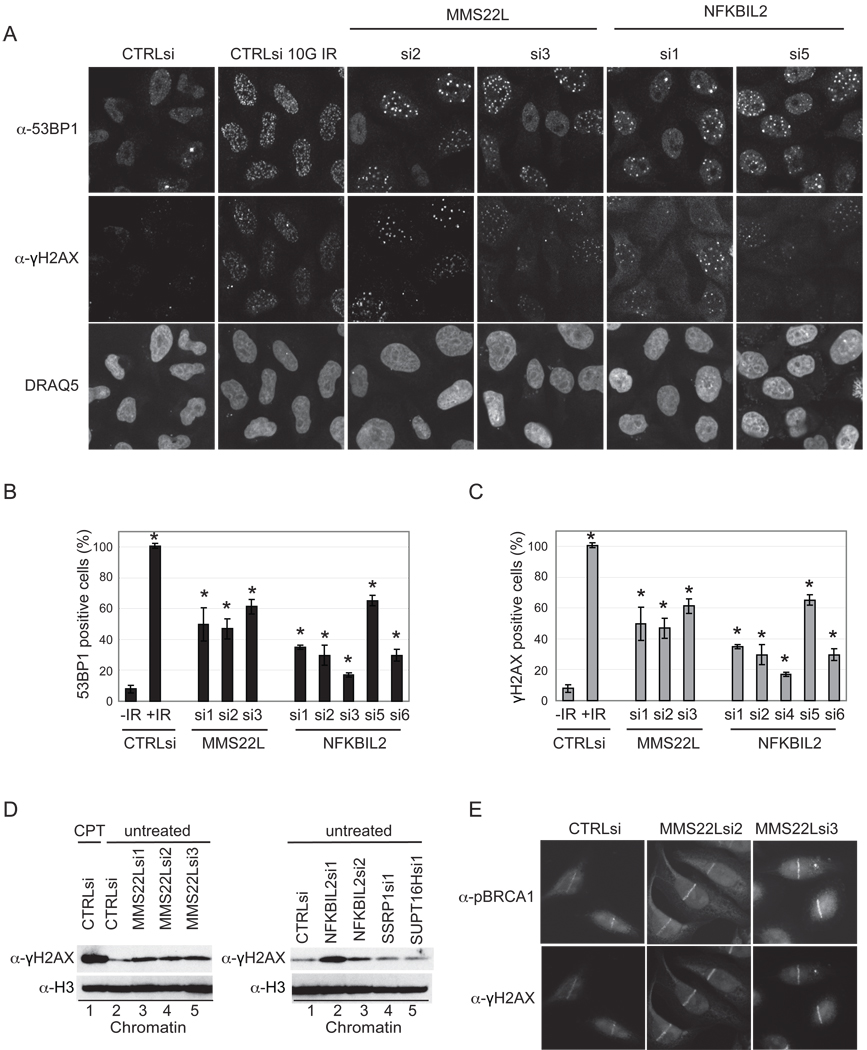
Increased 53BP1 and γH2AX Foci upon Loss of MMS22L or NFKBIL2
The mediator protein MDC1 is recruited to sites of DNA damage through interaction with γH2AX (Harper and Elledge, 2007) and becomes heavily phosphorylated by ATM and ATR (Matsuoka et al., 2007). This sets into motion a series of events leading to accumulation of several DDR proteins at discrete foci, termed IR-induced foci (IRIF). Having observed activation of checkpoint kinases (Figure 5A,B), we next sought to examine the formation of γH2AX and 53BP1 foci at sites of DNA damage after MMS22L or NKBIL2 depletion using immunofluorescence microscopy. Approximately 50–60% of MMS22L-depleted cells displayed robust 53BP1 or γH2AX foci (>5 foci per cell), relative to control cells in which <10% were IRIF positive, with multiple siRNAs giving similar results (Figure 6A–C). By comparison, IR treated cells were uniformly positive for robust IRIF formation (Figure 6A–C). Moreover, depletion of NFKBIL2 with 5 siRNAs led to 2–6 fold increase in IRIF formation with both 53BP1 and γH2AX (Figure 6A–C), in a manner that paralleled the extent of depletion (Figure 4B). Consistent with this result, we found that depletion of either MMS22L or NFKBIL2 led to increased γH2AX associated with chromatin, while depletion of either SSRP1 or SUPT16H did not lead to increased γH2AX association with chromatin (Figure 6D). In response to DNA damage, BRCA1 is heavily phosphorylated (Matsuoka et al., 2007; Stokes et al., 2007) and localizes to sites of DSBs. Given the role of BRCA1 in HR, we reasoned that the defects in HR seen upon MMS22L depletion could reflect an inability to recruit BRCA1 to sites of damage. However, using laser micro-irradiation to generate DSBs in HeLa cells, we did not observe a defect in the recruitment of phospho-BRCA1 (pS1524) to laser-induced stripes (Figure 6E). Similarly, localization of γH2AX to stripes did not require MMS22L (Figure 6E). Thus, MMS22L function appears to be after or parallel to BRCA1 recruitment to sites of DNA damage.
Figure 6. MMS22L or NFKBIL2 depletion activates DSB signaling pathways.
(A–C) Induction of 53BP1 or γH2AX foci upon MMS22L or NFKBIL2 depletion. HeLa cells transfected with the indicated siRNAs (72 h) were co-stained for 53BP1 or γH2AX, and nuclei stained with DRAQ5 (A). Cells were also subjected to IR (10 G) as a positive control for IRIF formation. (B, C) Quantification of 53BP1 (B) and γH2AX (C) foci in >200 cells (see Supplemental Methods) from quadruplicate transfections. * indicates p<.01 as determined by Students t-test.
(D) HeLa cells were depleted of the indicated proteins and γH2AX on chromatin determined by immunoblotting with histone H3 as a loading control.
(E) HeLa cells transfected with the indicated siRNAs were subjected to laser micro-irradiation and after 30 min, cells were fixed and subjected to immunoflourescence using α-γH2AX or α-phospho-BRCA1 (S1524).
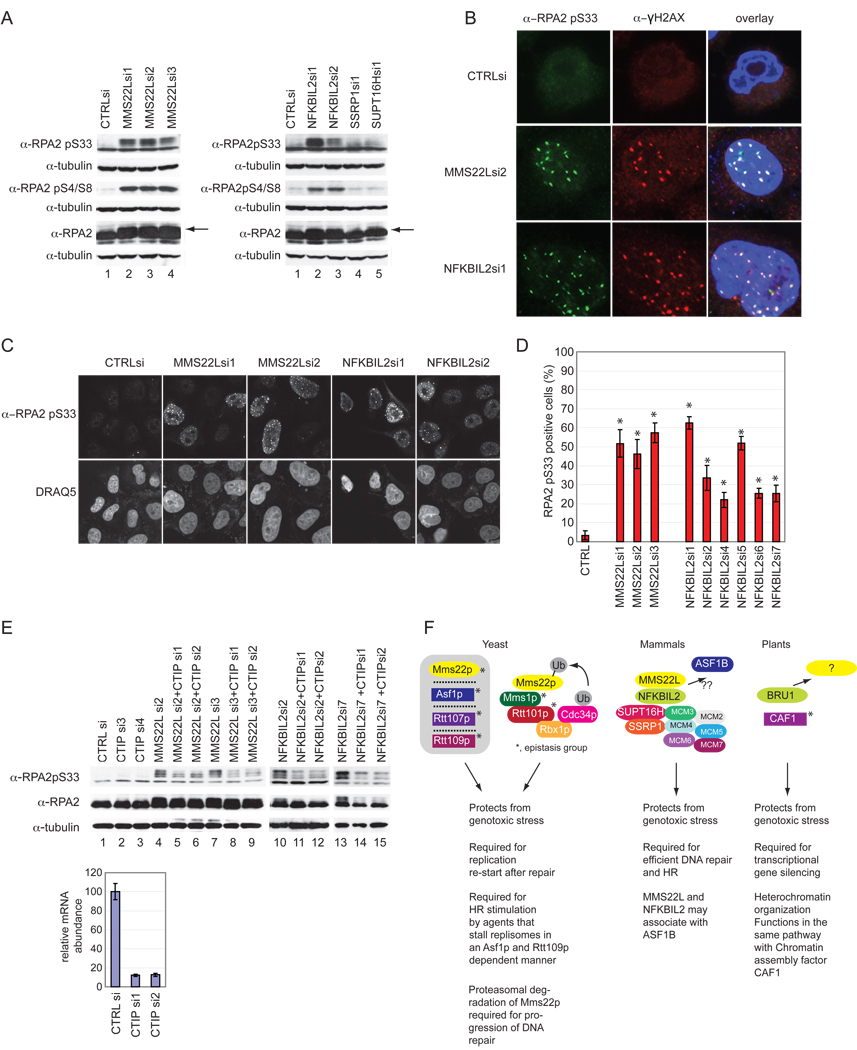
CTIP-dependent Phosphorylation of RPA2 and RPA2 Foci Formation Accompany MMS22L-NFKBIL2 Depletion
RPA exists in a heterotrimeric complex composed of RPA1, RPA2, and RPA3. RPA2 is a ssDNA binding protein that is phosphorylated upon DNA damage at several sites within the N terminal domain including S33 by ATR and S4/S8 by DNA-PK in a manner that is thought to require prior phosphorylation of S33 (Cimprich and Cortez, 2008; Anantha et al., 2007; Manthey et al., 2007; Olson et al., 2006; Vassin et al., 2009; Zernik-Kobak et al., 1997). Consistent with persistent ATR activation, we observed a substantial increase in phosphorylation of RPA2, as assessed by both a mobility shift and phosphospecific antibodies that recognize pS4/8 and pS33, upon loss of MMS22L or NFKBIL2 (Figure 7A). This, coupled with the observation that hyperphosphorylated RPA2 co-localizes with γH2AX to the exclusion of replication centers in response to replication stress (Vassin et al., 2004) led us to examine the localization of pS33 RPA2 after loss of MMS22L or NFKBIL2. Initially, we found that depletion of either MMS22L or NFKBIL2 with multiple siRNAs promotes the formation of nuclear pS33 RPA2 foci in HeLa cells (Figure 7B–D). Analysis of >200 cells revealed a 10-fold increase in the number of cells displaying pS33 RPA2 foci upon depletion of MMS22L and a 5–12-fold increase in pS33 RPA2 foci positive cells upon depletion of NFKBIL2 (Figure 7D), again consistent with the extent of depletion seen by immunoblotting (Figure 4B). These foci displayed extensive overlap with γH2AX foci that emerge upon MMS22L or NFKBIL2 depletion (Figure 7B).
Figure 7. Architecture of MMS22L-NFKBIL2 complexes in yeast, humans and plants.
(A–D) Depletion of MMS22L or NFKBIL2 promotes RPA2 phosphorylation and co-localization in γH2AX-positive IRIFs. HeLa cells were transfected with the indicated siRNAs and after 72 h, cell were either processed for immunoblotting with the indicated antibodies (A) or immunofluorescence with α-RPA2 pS33 (B,C) or α-γH2AX/α-RPA2 pS33 (B). In D, >200 cells from 4 replicate transfections were stained with α-RPA2 pS33 and cells quantified as described in the Supplemental Methods. Arrows indicate the position of phosphorylated RPA2. * indicates p<.01 as determined by Students t-test.
(E) CTIP is required for RPA2 phosphorylation in response to depletion of either MMS22L or NFKBIL2. Extracts from the indicated cells were immunoblotted with the indicated antibodies. mRNA abundance for CTIP was determined by qPCR.
(F) Known physical and genetic interactions are shown schematically and functions within each organism are summarized. See text for details.
This data suggests that MMS22L or NFKBIL2 depletion promotes DNA damage that is accompanied by ATR-dependent RPA2 phosphorylation, a modification that is known to be dependent upon RPA2 binding to ssDNA (Cimprich and Cortez, 2008). If this were the case, then resection at the DNA break would be predicted to be a pre-requisite for RPA2 phosphorylation. To test this prediction, we depleted CTIP, a protein known to cooperate with the MRN11-RAD50-NBS1 complex to promote DSB resection (Cimprich and Cortez, 2008). Co-depletion of MMS22L and CTIP or NFKBIL2 and CTIP led to a reduction of RPA2 S33 phosphorylation relative to MMS22L or NFKBIL2 depletion alone (Figure 7E), indicating that CTIP function is required for RPA2 phosphorylation and recruitment to ssDNA upon depletion of the MMS22L-NFKBIL2 complex.
Discussion
Genetic Screen for CPT Synthetic Lethality as a Resource for Discovery in the Replication Stress Pathway
While several components of the replication stress response pathway have been identified, a complete understanding of this pathway and its role in promoting HR is lacking (Cimprich and Cortez, 2008). The genetic screen we report here provides a number of candidate proteins whose analysis may facilitate a further understanding of the replication stress response pathway. In addition to known DDR proteins, including BRCA1, BARD1, ATR, PARP1, RAD51, RNF20, MAPKAP2/CHK3, RBBP8/CTIP, RPA1, and RPA2, a number of additional proteins (Table S1) with domains linking them to chromatin or signaling functions were also identified, as were proteins that contain catalytic activities. Such proteins represent candidate drug targets that may cooperate with Topoisomerase I inhibitors in anti-cancer therapy.
MMS22L-NFKBIL2 Complex Promotes Resistance to Replication Stress and Homologous Recombination
Two orthogonal approaches – RNAi screening and proteomics – led to the identification of the MMS22L-NFKBIL2 complex and the finding that both proteins are required for resistance to replication stress. The sensitivity of cells depleted of MMS22L in both the original screen and in follow-up assays was comparable to that observed with ATR, a central component of the replication stress pathway, and PARP1, a critical component of the response of cells to DSBs. MMS22L, like its yeast relatives Mms22p and Mus7p, lacks known protein interaction motifs, but its interaction with NFKBIL2, which contains TPR, ankyrin, and LRR motifs, suggests a possible scaffolding function for the complex. The phenotypes observed upon depletion of either MMS22L or NFKBIL2 from cells indicate that MMS22L and NFKBIL2 function in a common pathway to minimize endogenous replication stress. Cells depleted of MMS22L or NFKBIL2 were not only sensitive to CPT and other damaging agents, but were also defective for HR induced by a DSB (Figure 2, 4). Depletion of either protein results in phosphorylation of two ATR targets, CHK1 and RPA2, indicating the inappropriate formation of ssDNA in the absence of MMS22L-NFKBIL2 (Figure 5A, 7A). Consistent with this, the absence of MMS22L-NFKBIL2 also led to phospho-RPA2 foci that co-localized with γH2AX foci (Figure 7B). Co-depletion of MMS22L or NFKBIL2 with CTIP, a protein required for DSB resection, resulted in a loss of RPA2 phosphorylation, indicating that RPA2 phosphorylation reflects the formation of ssDNA upon depletion of MMS22L or NFKBIL2 (Figure 7E). Loss of MMS22L-NFKBIL2 also promotes the formation of DSBs, based on phosphorylation of γH2AX and recruitment of 53BP1 to foci (Figure 6A–C). Cells depleted of MMS22L also displayed a delay in the inactivation of CHK2 in response to removal of CPT, suggesting a role for the MMS22L-NFKBIL2 complex in DNA repair processes (Figure 5D).
MMS22L and NFKBIL2 Interact with each other and with FACT-MCM Implicated in DNA Unwinding
We found that MMS22L associates with NFKBIL2, and that both MMS22L and NFKBIL2 interact with both FACT and the MCM complex (Figure 3). Moreover, SSRP1, a component of FACT, reciprocally interacts with MMS22L, NFKBIL2, and the MCM complex. FACT associates functionally with MCM complexes on replication origins to promote replication initiation and DNA unwinding (Tan et al., 2006; Tan et al., 2010). The functional basis for the interaction of MMS22L and NFKBIL2 with the FACT and MCM complexes remains to be dissected but one interesting possibility is that this interaction facilitates the recruitment of the MMS22L-NFKBIL2 complex to active replication forks. In this regard, we observe only a small fraction of SSRP1 associated with MMS22L-NFKBIL2, consistent with the possibility that a specialized FACT complex, presumably in association with MCM, is selectively recognized by MMS22L-NFKBIL2. Moreover, SSRP1 is extensively phosphorylated by ATM/ATR in response to IR (Matsuoka et al., 2007), suggesting that it may respond functionally to the DDR. Additional studies are required to evaluate the dynamics of assembly or disassembly of this complex during the cell cycle or in response to DNA damage. While a role for FACT in regulating H2AX exchange has been proposed (Heo et al., 2008), we did not observe any changes in the amount of γH2AX bound to chromatin upon loss of either subunit of FACT (Figure 6D).
Divergent MMS22L-NFKBIL2 Biochemical Assemblies Across Species Despite Functional Similarities
Clear orthologs of MMS22L are found in vertebrates and Drosophila (>50% similarity) but MMS22L is only distantly related to yeast Mms22p and Mus7p (9% identity, 21% similarity), with conservation distributed across the entire length of the protein (Figure S1). This suggests the absence of individual functional domains within the MMS22/Mus7 family. Likewise, S. pombe and S. cerevisiae orthologs are rather distantly related with each other, as well, with 15% identity and 29% similarity.
Previous studies have shown that Mms22p is a component of a genetic interaction network that includes the CUL4-like protein Rtt101p, the DDB1-like protein Mms1p, the histone H3 K56 acetyl transferase Rtt109p, the histone chaperone Asf1p, and Rtt107p which is required for completion of DNA replication following fork stalling (Duro et al., 2008; Zaidi et al., 2008). Mms22p interacts physically with Mms1p and Rtt101p (Figure 7E), and has been proposed to function as a substrate adaptor of a Rtt101p-Mms1p E3 (Zaidi et al., 2008). However, recent studies suggest that Mms22p is actually a target of the Rtt101p-Mms1p E3 and is degraded by the proteasome after recruitment to chromatin (Ben-Aroya et al., 2010). Turnover of Mms22p is required for completion of DNA repair (Ben-Aroya et al., 2010). Remarkably, mammalian MMS22L is engaged in interactions distinct from that seen in yeast. We were unable to observe stable association between MMS22L and either CUL4 or DDB1 either by proteomics (Table S2) or by direct co-transfection/co-immunoprecipitation (data not shown). Thus, while we cannot fully rule out a functional association of MMS22L with a CUL4-like E3 in mammalian cells, the available data support a model wherein the most stable MMS22L containing complexes are distinct from the Mms22p containing complexes observed in yeast (Figure 7F). Interestingly, the SSRP1 component of human FACT has been shown to be ubiquitylated (Landais et al., 2006). However, we did not observe any changes in abundance of SSRP1 upon loss of MMS22L nor did we detect significant changes in the abundance of FACT or MCMs associated with MMS22L upon DNA damage in our proteomic analysis that would support a role for MMS22L in regulating turnover of either complex (Figure S3 and Table S2).
Nevertheless, like MMS22L, yeast lacking Mms22p/Mus7p are sensitive to replication stress and Mms22p is also required for HR stimulated by replisome blockage (Baldwin et al., 2005; Dovey et al., 2009; Duro et al., 2008; Raisner and Madhani, 2008; Yokoyama et al., 2007; Zaidi et al., 2008). Moreover, mms22Δ mutants display defects in recovery from replication fork stalling (Duro et al., 2008; Zaidi et al., 2008). Further studies are required to delineate how the MMS22L and Mms22p pathways are mechanistically related. Interestingly, mms22Δ mutants also interact genetically with POB3, the yeast SSRP1 ortholog (Costanzo et al., 2010), raising the possibility of a mechanistic link between Mms22p and the FACT complex in yeast. Intriguingly, Rtt101p, but not Mms1p or Mms22p, has been implicated in ubiquitination of the yeast ortholog of SUPT16H, Sup16p (Han et al., 2010). Whether or not MMS22L affects FACT modification or targeting remains to be determined. Although not confirmed by our proteomics experiments, previous high-throughput proteomic experiments in mammalian cells have suggested that transiently expressed ASF1B, one of two Asf1p orthologs in mammals, associates with both MMS22L and NFKBIL2 (Ewing et al., 2007). FACT and Asf1p are known to function together to regulate nucleosome dynamics (Krebs and Tora, 2009; Takahata et al., 2009; Kim and Haber, 2009) . Thus, it is conceivable that such an ASF1B interaction could reflect an additional parallel between MMS22L and Mms22p pathways (Figure 7F), and further suggests that these components are important for nucleosome dynamics at damaged replication forks.
While NFKBIL2 orthologs are readily identified in vertebrates and D. melanogaster (Figure S2), we were unable to identify obvious orthologs in yeast or C. elegans, as these species lacked proteins with tandem TPR-ANK-LRR domains characteristic of vertebrate NFKBIL2 (data not shown). The closest ortholog of NFKBIL2 in A. thaliana (11% identical, 22% similar) is the TPR-ANK repeat containing protein BRU1 (also called TONSOKU) (Figure S2). BRU1 has been linked to the DNA damage response, epigenetic silencing, DDR-dependent PARP2 transcription, and root and shoot apical meristem development (Suzuki et al., 2004; Takeda et al., 2004) (Figure 7E). However, we were unable to identify an obvious A. thaliana ortholog of MMS22L/Mms22p (data not shown). Consistent with a potential role in chromatin assembly, previous studies indicate that bru1 mutants share phenotypes with loss-of-function mutations in p150 and p60 subunits of the CAF1 chromatin assembly factors, and bru1 is epistatic to caf1 mutants in A. thaliana (Takeda et al., 2004). While this result provides a potential additional link with chromatin assembly, BRU1 did not associate with CAF1 in vitro (Takeda et al., 2004) and therefore, whether BRU1 functions directly in chromatin assembly remains to be established.
In summary, this work identifies a previously unrecognized complex – MMS22L-NFKBIL2 – involved in the response of cells to genotoxic stress and in suppression of spontaneous DNA damage (Figure 7F). The extreme sensitivity of cells depleted of either MMS22L or NFKBIL2 to DNA damaging agents that impede replication forks suggests an important role for this complex in control of replication-dependent DNA damage. We hypothesize that the FACT-MCM/MMS22L-NFKBIL2 interaction complexes places MMS22L-NFKBIL2 in the vicinity of replication forks. Further mechanistic studies, possibly including reconstitution of chromatin assembly at structures that mimic damaged DNA templates, will be required to understand the contribution of MMS22L-NFKBIL2 to chromatin structure and to repair of damaged replication forks.
Experimental Procedures
Detailed Experimental Procedures are provided in the Supplemental Material.
Cell Culture, plasmids, antibodies, RNAi, and flow cytometry
Cell culture and protein expression was performed as described (Svendsen et al., 2009). α-MMS22L (against residues 1243–1288) was raised in rabbits and affinity purified using immobilized antigenic peptide. Commercial antibodies employed are provided in the Supplemental Methods while qPCR primer and RNAi sequences are provided in Table S1. Cell cycle analysis was performed using propidium iodide staining and analyzed using FLOWJO (Tree Star, Inc).
RNAi screening, validation, and HR assays
HeLa cells (107) were infected with pools of retroviral shRNA libraries (12,484 shRNAs in 6 independent pools) in MSCV-miR30-puro (Schlabach et al., 2008) at a multiplicity of 1000 and selected for 3 days in puromycin (1µg/ml). Cells were cultured for 10 population doublings (PD) with or without 7.5 nM CPT. Half-hairpin barcodes were amplified from genomic DNA using the polymerase chain reaction and labeled with either Cy3 (PD10) or Cy5 (PD0). Comparative hybridizations were performed individually on triplicate samples for both treated and untreated samples using custom microarrays. Normalized log2 ratios between PD10 and PD0 were determined for each sample and then the difference in log2 ratios between treated and untreated samples were averaged. Class 1 screen hits had multiple shRNAs with average difference in log2 ratios less than −0.7 (> 1.6-fold change) and a value of the average difference in log2 ratio minus the SEM less than −0.35 (1.3-fold change). Class 2 screen hits had single shRNAs with an average difference in log2 ratio less than −1.0 (> 2-fold change) in all triplicate experiments. Additional details of the screening methodology are provided in the Supplemental Methods. MCA assays for validation were performed in U2OS or HeLa cells as described previously (Smogorzewska et al., 2007). Screening data and detailed methods are provided in Table S1 and Supplemental Methods, respectively. HR assays were performed using I-SceI-induced cleavage of a DR-GFP substrate in U2OS cells as previously described (Xia et al., 2006).
Protein purification and mass spectrometry
Proteomic analysis of HA-tagged proteins expressed in 293T cells was performed and analyzed using CompPASS as described (Behrends et al., 2010; Sowa et al., 2009; see Supplemental Methods). Chromatin extraction was performed as described in the Supplemental Methods.
Supplementary Material
Acknowledgments
We thank L. Zou (MGH Cancer Center) for helpful discussions, and D. Durocher, J. Rouse, and M. Peter for communicating results prior to publication. We thank G. Nalepa for help during initial stages of the project as well as intellectual support and J. Hernandez for technical support. This work was supported by NIH grants AG011085 and GM054137, and by a grant from the Stewart Trust to J.W.H. Additional support was provided by a grant from the NIH to S.J.E. B.C.O. was supported by a fellowship from the American Cancer Society. S.J.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- Baldwin EL, Berger AC, Corbett AH, Osheroff N. Mms22p protects Saccharomyces cerevisiae from DNA damage induced by topoisomerase II. Nucleic Acids Res. 2005;33:1021–1030. doi: 10.1093/nar/gki246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–77. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Agmon N, Yuen K, Kwok T, McManus K, Kupiec M, Hieter P. Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genet. 2010;6:e1000852. doi: 10.1371/journal.pgen.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey CL, Aslanian A, Sofueva S, Yates JR, 3rd, Russell P. Mms1-Mms22 complex protects genome integrity in Schizosaccharomyces pombe. DNA Repair (Amst) 2009;8:1390–1399. doi: 10.1016/j.dnarep.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E, Vaisica JA, Brown GW, Rouse J. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst) 2008;7:811–818. doi: 10.1016/j.dnarep.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatten K, Dai NT, Vroman BT, Loegering D, Erlichman C, Karnitz LM, Kaufmann SH. The role of checkpoint kinase 1 in sensitivity to topoisomerase I poisons. J Biol Chem. 2005;280:14349–14355. doi: 10.1074/jbc.M411890200. [DOI] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Han J, Li Q, McCullough L, Kettelkamp C, Formosa T, Zhang Z. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 2010;24:1485–1490. doi: 10.1101/gad.1887310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs A, Tora L. Keys to open chromatin for transcription activation: FACT and Asf1. Mol Cell. 2009;34:397–399. doi: 10.1016/j.molcel.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Landais I, Lee H, Lu H. Coupling caspase cleavage and ubiquitin- proteasome-dependent degradation of SSRP1 during apoptosis. Cell Death and Differentiation. 2006;13:1866–1878. doi: 10.1038/sj.cdd.4401878. [DOI] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Loegering D, Arlander SJ, Hackbarth J, Vroman BT, Roos-Mattjus P, Hopkins KM, Lieberman HB, Karnitz LM, Kaufmann SH. Rad9 protects cells from topoisomerase poison-induced cell death. J Biol Chem. 2004;279:18641–18647. doi: 10.1074/jbc.M313536200. [DOI] [PubMed] [Google Scholar]
- Manthey KC, Opiyo S, Glanzer JG, Dimitrova D, Elliott J, Oakley GG. NBS1 mediates ATR-dependent RPA hyperphosphorylation following replication-fork stall and collapse. J Cell Sci. 2007;120:4221–4229. doi: 10.1242/jcs.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD. Genomewide screen for negative regulators of sirtuin activity in Saccharomyces cerevisiae reveals 40 loci and links to metabolism. Genetics. 2008;179:1933–1944. doi: 10.1534/genetics.108.088443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Nakamura AJ, Redon CE, Pommier Y. DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle. 2010;9:274–278. doi: 10.4161/cc.9.2.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Inagaki S, Nakajima S, Akashi T, Ohto MA, Kobayashi M, Seki M, Shinozaki K, Kato T, Tabata S, et al. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. 2004;38:673–684. doi: 10.1111/j.1365-313X.2004.02074.x. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18:782–793. doi: 10.1101/gad.295404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Liu H, Lin CL, Lee SC. Functional cooperation between FACT and MCM is coordinated with cell cycle and differential complex formation. J Biomed Sci. 2010;17:11. doi: 10.1186/1423-0127-17-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci. 2009;122:4070–4080. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Yokoyama M, Inoue H, Ishii C, Murakami Y. The novel gene mus7(+) is involved in the repair of replication-associated DNA damage in fission yeast. DNA Repair (Amst) 2007;6:770–780. doi: 10.1016/j.dnarep.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J Biol Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- Zou L. Checkpoint Mec-tivation comes in many flavors. Mol Cell. 2009;36:734–735. doi: 10.1016/j.molcel.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing and signaling DNA damage: roles of Rad17 and Rad9 complexes in the cellular response to DNA damage. Harvey Lect. 2001;97:1–15. [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.