SUMMARY
Three sets of in silico experiments have been conducted to elucidate the binding mechanics of two drugs, (+)-methamphetamine (METH) and amphetamine (AMP) to the single-chain variable fragment (scFv) recently engineered from anti-METH monoclonal antibody mAb6H4 (IgG, κ light chain, Kd = 11nM). The first set of in silico experiments are long time equilibration runs of scFv:drug complexes and of drug-free scFv both in solution. They demonstrate how the solution structures of scFv deviate from its crystallographic form with or without drug molecules bound to it. And they lead to the prediction that the Arrhenius activation barrier is nearly zero for transitions from the dissociated state to the bound state. The second set of in silico experiments are nonequilibrium dynamics of pulling the drug molecules out of the binding pocket of scFv and the equilibration runs for drugs to fall back into binding pocket. They demonstrate that extra water molecules (in addition to the two crystallographic waters) exist inside the binding pocket, underneath the drug molecules. These extra waters must have been evaporated from the binding pockets during the crystallization process of the in vitro experiments of structural determination. The third set of in silico experiments are nonequilibrium steered molecular dynamics simulations to determine the absolute binding free energies of METH and AMP to scFv. The center-of-mass of a drug molecule (METH or AMP) is steered (pulled) towards (forward) and away from (reverse) the binding site, sampling forward and reverse pulling paths. Mechanic work is measured along the pulling paths. The work measurements are averaged through the Brownian dynamics fluctuation dissipation theorem to produce the free-energy profiles of the scFv:drug complexes as a function of the drug-scFv separation. These experiments lead to the theoretical prediction of absolute binding energies of METH and AMP that are in agreement with the in vitro experimental results.
INTRODUCTION
Chronic or excessive (+)-methamphetamine (METH) use has been a serious problem in today's society1. Possible medical treatments currently pursued make use of monoclonal antibodies (mAbs) in various forms2–7. Of particular interest is a recently engineered single-chain variable fragment (scFv) against METH from anti-METH monoclonal antibody mAb6H4 (IgG, κ light chain, Kd = 11nM)2, 3. The affinities of scFv:METH and scFv:AMP complexes have been measured to be 10nM and 20.7μM respectively3. The crystal structure of scFv:METH has been determined to a precision of 1.9 Å2. However, questions remains: Do the solution structures of scFv deviate from its crystallographic form with or without a drug molecule bound to it? And, if yes, how and how much? What effects do the deviations have on the binding mechanics? Is there a good agreement between the in vitro experiments and the theoretical predictions of the absolute binding energy based on the standard force fields such as CHARMM8, 9? This paper attempts to answer these questions by performing three sets of in silico experiments.
The first set of in silico experiments are long time equilibration runs of scFv:drug complexes and of drug-free scFv, all in solution. They demonstrate how the solution structures of scFv deviate from its crystallographic form2 with or without a drug molecule bound to it. In particular, the conformation of the binding pocket is examined by computing the root-mean-square-distance (RMSD) of the residues that constitute the binding pocket. The bottom part of the binding pocket is found to remain close to its crystallographic form whether a drug molecule (METH or AMP) is bound in it or not. The orifice part of the binding pocket, however, deviates substantially from the crystallographic form when no drug molecule is bound in the pocket. It is enlarged so much that there is no steric hindrance to a drug molecule when it enters into the binding pocket. This theoretical prediction implies that the relaxation time for the system to reach equilibrium is very short when scFv is injected into a solution of drugs. It also indicates that the Arrhenius barrier is nearly zero along the binding pathway from the dissociated state to the bound state.
The second set of in silico experiments are nonequilibrium dynamics of pulling the drug molecules out of the binding pocket of scFv and the equilibration runs for drugs to fall back into the binding pocket. They demonstrate that extra water molecules (in addition to the two crystallographic waters) exist inside the binding pocket, underneath the drug molecules. This theoretical finding does not contradict but complement the in vitro experimental study because the extra water molecules could and must have been evaporated from the binding pockets during the crystallization process of the in vitro experiments of structural determination.2
The third set of in silico experiments are nonequilibrium steered molecular dynamics (SMD)10–16 simulations to determine the absolute binding free energies of METH and AMP to scFv. The center-of-mass z-coordinate of a drug molecule (METH or AMP) is pulled towards (forward) and away from (reverse) the binding site, sampling forward and reverse pulling paths. Mechanic work is measured along the pulling paths. The work measurements are averaged through the Brownian dynamics fluctuation dissipation theorem (DB-FDT)17, 18 to produce the free-energy profiles of the scFv:drug complexes as functions of the drug-scFv separation. These experiments lead to the theoretical prediction of absolute binding energies of METH and AMP in agreement with the in vitro experimental results. Additionally, this set of in silico experiments produce a new prediction that the Arrhenius activation barrier, namely, the free-energy barrier separating the dissociated state from the bound state, is nearly zero. This theoretical predication can be verified by in vitro experiments measuring the binding rate or the dissociation rate vs temperature.
RESULTS AND DISCUSSION
Solution structures of scFv
In order to see if and how the solution structure of scFv deviates from its crystallographic form, we performed 20 ns equilibrium molecular dynamics (MD) simulations of the drug-free scFv and 5 ns of the scFv:drug complexes each in a box of water. Significant changes in structure all happened in the first 5 ns of the in silico experiments.
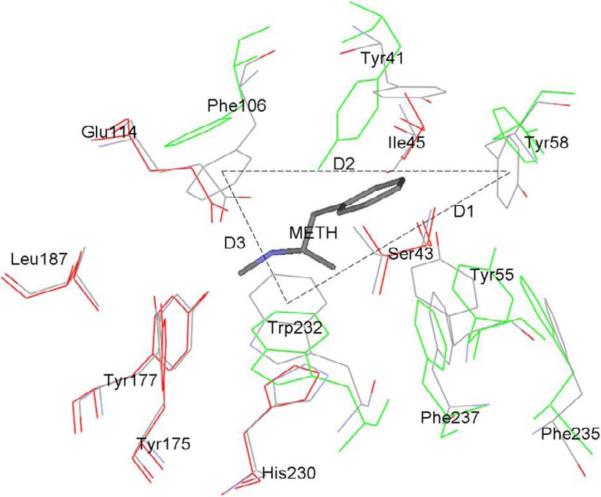
Fig. 1 illustrates the differences between the solution structures of the drug-free scFv and of the METH-bound scFv, showing the key residues that form the binding pocket. The coordinates of the two structures are the mean coordinates averaged over the 5th ns. The key residues are divided into two groups: One group (G1) consisting of seven residues (in green color) forms the orifice of the binding pocket. The other group (G2) also consisting of seven residues (in red color) form the floor of the binding pocket. The floor forming residues of the drug-free scFv nearly overlap with their counterparts in the scFv:METH complex. And they do not deviate substantially from the crystallographic structure of scFv:METH. The orifice forming residues, however, diverge significantly from their crystallographic form. In absence of a drug molecule, the orifice enlarges itself and, consequently, makes it possible for a drug molecule to fall into the binding pocket without an activation barrier to overcome along the binding pathway.
Fig. 1.
Solution structure of the drug-free scFv (in red and green colors) vs that of the scFv:METH complex (gray). Shown here are the key residues of the scFv that constitute the binding pocket. The coordinates are from averaged structure from the 5th ns MD trajectories of scFv and scFv:METH in a box of TIP3 waters. METH is shown in stick representation. The key residues are shown in line representation. The residues of scFv forming the orifice (G1) of the binding pocket are shown in green color. The residues of scFv constituting the floor of the binding pocket (G2) are shown in red color. D1, D2 and D3 are the distances between the side-chain centers-of-mass of labeled residues.
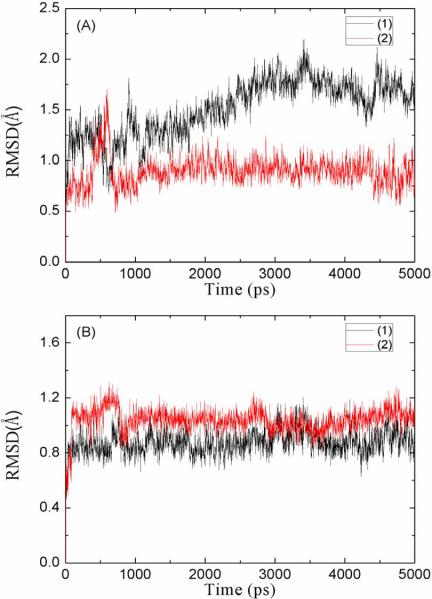
Fig. 2 shows the root-mean-square deviations (RMSD) of the G1 and G2 atoms during the first 5 ns of in silico experiments. The equilibration process of the METH-bound scFv is typical for a stable protein-ligand complex being solvated. The RMSD of G1 and that of G2 both fluctuate largely in the first 2 ns. Then they flatten out, indicating the scFv:METH structure is stable in solution. Interestingly, in presence of a drug molecule, both the orifice part and the floor part of the binding pocket do not deviate much from their crystallographic forms. However, the equilibration process of the drug-free scFv is distinctively different. While G2 (the floor part of the binding pocket) basically retains its crystallographic structure in a way similar to the drug-bound scFv, G1 (the orifice of the binding pocket) deviates from its crystallographic form continually in first 3 ns and stabilizes after 5 ns. These structural changes are measured quantitatively with three distances (D1, D2, D3 on Fig. 1) between the centers of mass of three residues side chain on the orifice of scFv. D1, D2, and D3 form a triangle that represents the size of the orifice of the binding pocket (Fig. 1).
Fig. 2.
Root-mean-square deviations (RMSD) of the selected atoms on scFv:METH complex in solution (1) and the drug-free scFv in solution (2) observed in the first 5ns of MD simulations. (A) is for the atoms on orifice of the binding pocket as shown in green color in Fig. 1. (B) is for the atoms in the floor of the binding pocket as shown in red color in Fig. 1.
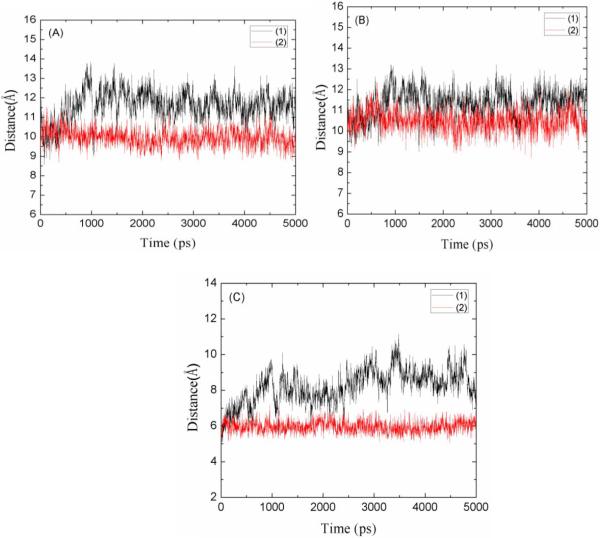
Fig. 3 shows the time-dependence of the three distances, D1, D2 and D3, for both the drug-free scFv and the scFv:METH complex. Two of the three exhibit significant increases for the drug-free scFv while all three remain close to their crystallographic values for the drug-bound scFv. The mean values of D1, D2 and D3 (averaged over the 5th ns) are, respectively, 11.6, 11.6 and 8.6 Å in the drug-free scFv and 9.9, 10.5 and 6.0 Å in the drug-bound scFv. The solution structure of the drug-free scFv differs from the solution structure of the drug-bound scFv mainly in the size of the orifice of the binding pocket. This supports the following hypothesis of the drug-binding process: When scFv's are introduced into a drug-containing solution, their binding pocket orifices open up so that drug molecules (METH or AMP) can easily move into the binding sites without encountering much steric hindrance. This, we note once again, implies the possibility that there is no activation barrier along the binding pathway of a drug molecule from the dissociated state to the bound state. This prediction of barrier-less behavior can be verified with in vitro experiments at varying temperatures.
Fig. 3.
The distances between the centers of mass of the orifice residues labeled in Fig. 1. (1) for drug-free scFv and (2) for scFv:METH complex. (A) D1 is the distance between the center of mass of Tyr58 side chain and the center of mass of Trp232 side chain. (B) D2 is the distance between the center of mass of Tyr58 side chain and the center of mass of Phe106 side chain. (C) D3 is the distance between the center of mass of Phe106 side chain and the center of mass of Trp232 side chain.
Examining the hydrophobicity/hydrophilicity of the residues that constitute the binding pocket of an scFv, we understand the mechanics of its structure changes in solution as follows: The lower half, the floor of the binding pocket (including GLU 114 and HIS 230) are strongly hydrophilic. This part of the pocket strongly attracts waters and the cationic end of a drug molecule. It is stabilized by waters in the pocket or by the cationic amine group of a drug molecule along with several waters beneath it. The upper half, the orifice of the binding pocket consists of hydrophobic aromatic residues that can interact with one another favorable via the van der Waals interactions. But, a residue on the orifice is outside the range of van der Waals interaction with the residue on the opposite side of the orifice. Here, water molecules serve to destabilize the orifice structure of the scFv crystal. Once a drug (METH or AMP) is bound in the pocket, however, the hydrophobic aromatic ring of the drug molecule is within the van der Waal range of the aromatic rings in the orifice of the binding pocket. They interact favorably to stabilize the structure of the pocket in solution.
To complete this subsection, we add that the solution structure of scFv in the scFv:AMP complex closely resemble that of the scFv:METH complex with one exception to be discussed in the next subsection.
Waters at the binding site in-between drug and scFv
In the crystallographic structure of the scFv:METH complex, there are two waters in the binding pocket between the drug's cationic amine group and GLU 114 residue of scFv. These two waters form strong hydrogen bonds with the residues surrounding them. This naturally leads to the hypothesis that the scFv:METH complex with two waters (scFv:METH:2wt) is at the global minimum of the free energy if its solution structure is stable as we have seen in the preceding subsection. However, scFv:METH:2wt could be at a local minimum of the free energy instead of the global minimum. The global minimum of the free energy could be a very different state and there could be one or more local minima in addition to the global minimum of the free energy. In fact, our in silico experiments starting from the dissociated state of drugs reveal that is indeed the case. The three in silico experiments we have performed are: (1) scFv in a box of waters without drugs that were already discussed in part in the preceding subsection, (2) scFv and METH in a box of waters, (3) scFv and AMP in a box of waters.
In experiment (1), we aim to determine the hydrophobicity/hydrophilicity of the binding pocket by running equilibration MD of scFv in drug-free water. Starting from the crystallographic state that has only two waters in the binding pocket, we observe that multiple waters move in and out of the binding pocket as the orifice of the binding pocket enlarges to its equilibrium state. Waters continue to move in and out of the binding pocket in the equilibrium state. On the average, there are 10 to 12 waters occupying the pocket. The average dwell times of the waters inside the pocket are tabulated in Table I. Note that even the two crystallographic waters are mobile in absence of drug molecules. They exchange positions with other waters once per 1.2 ns on the average. We also observe that near the floor of the binding pocket, waters are closer to one another than those near the orifice. We conclude that the bottom half of the binding pocket is strongly hydrophilic and the upper half of the pocket is hydrophobic.
Table I.
Dwell time of a water molecule inside the binding pocket of scFv
| CWT1 | CWT2 | WT3 | WT4 | WT5 | |
|---|---|---|---|---|---|
| Dwell Time | 1.2ns ± 0.4ns | 1.2ns ± 0.4ns | 0.3ns ± 0.2ns | 0.3ns ± 0.2ns | 0.3ns ± 0.2ns |
CWT1 to CWT2------Crystallographic water.
WT3 to WT5------Water inside the binding pocket but not at the crystallographic water positions.
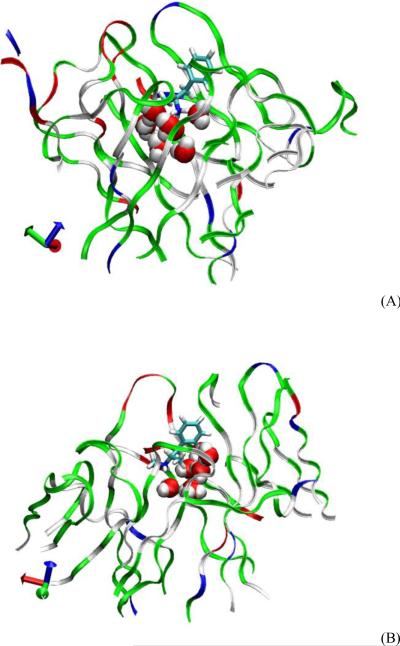
In experiments (2) and (3), we aim to determine the number of waters in the bound state inside the binding pocket between the protein and the ligand. We run and follow the equilibrium MD from the dissociated state to the bound state. In experiment (2), we observe that there are a total of seven water molecules (two of which are the crystallographic waters reported in Ref. 2) caught in the binding pocket beneath METH (Fig. 4) when the scFv:METH complex transitions from the dissociated state to the bound state (scFv:METH:7wt). These extra waters weaken the attraction between the cationic amine group of METH and the charged residues of scFv (GLU 114 in particular) but the total free energy of the whole system (scFv:METH plus waters) is actually lower in the scFv:METH:7wt state than in the scFv:METH:2wt state that has only the two crystallographic waters in the binding pocket. scFv:METH:7wt is more stable because the strong hydrogen bonds formed by the waters with each other, with the charged residues of scFv, and with the cationic amine of METH, are more than enough to compensate the weakened Coulombic attraction due to increased separation between the charged atoms caused by the presence of the five extra waters.
Fig. 4.
Waters inside the binding pocket of scFv beneath the drug molecule ((A) AMP and (B) METH). Protein in Ribbons, drug in Licorice, and water in VDW representation, respectively. Rendered with VMD21.
In experiment (3), we observe that there are a total of eight water molecules (two of which are the crystallographic waters reported in Ref. 2) caught in the binding pocket beneath AMP when the scFv:AMP complex transitions from the dissociated state to the bound state (scFv:AMP:8wt). The scFv:AMP:8wt state is more stable than the “crystallographic” scFv:AMP:3wt hypothesized in Ref. 2, thanks to the strong hydrogen bonds formed by the extra waters with each other and with their surroundings. The mechanics of scFv:AMP:8wt is similar to that of the scFv:METH:7wt. The only difference is that the eighth water in scFv:AMP:8wt takes the place of the terminal methyl group of METH in scFv:METH:7wt. This eighth water even further weakens the Coulombic attraction between the cationic amine group of AMP and the charged residues of scFv and, consequently, leads to a weaker binding energy of AMP than METH.
Ending this subsection, we emphasize that our in silico experiments are consistent with the in vitro experiments and the crystallographic structure of scFv:METH because the extra waters in the binding pocket can evaporate during the crystallization (drying-up) processes.
Absolute binding energies of METH and AMP
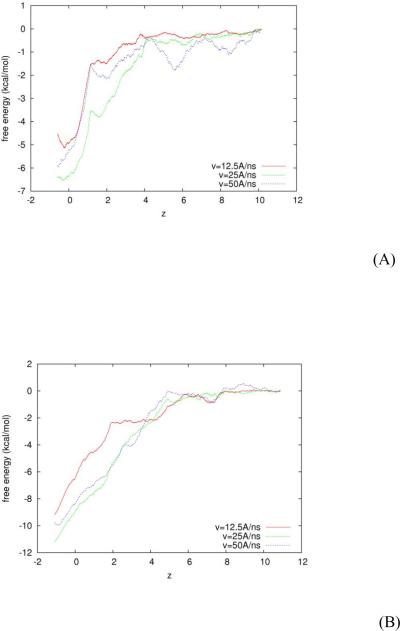
In addition to the crystallographic structure, the in vitro experiments have also determined the binding affinities of scFv with METH and AMP. In terms of free energy, the absolute binding energies of METH and AMP with scFv are, respectively, −11kcal/mol and −6.5 kcal/mol. In the third set of our in silico experiments, we aim to theoretically predict the absolute binding energies of METH and AMP to compare with in vitro experiments. Conducting SMD simulations, we pull the drug molecule (METH or AMP) towards the binding site and measure the mechanic work done to the system during this steered transition from the dissociated state to the bound state. Then we pull the drug molecule away from the binding site and measure the mechanic work done to system during the transition from the bound state to the dissociated state. We assume the scFv:drug in water system is governed by the Brownian dynamics and employ the Brownian dynamics fluctuation dissipation theorem (BD-FDT)18 to compute the free-energy differences from the work measurements. In terms of the potential of mean force (PMF), we find the free-energy landscape of scFv:METH and that of scFv:AMP along the entire binding pathway from the dissociated state to the bound state. In order to establish reliability of the predicted free-energy profiles, we perform the pulling experiments at three speeds: 12.5Å / ns, 25Å / ns, and 50Å / ns. The three PMF curves for scFv:AMP are shown in Fig. 5(A) and the same for scFv:METH in Fig. 5(B). We take the average of the three curves as the free-energy landscape and the deviations among them as the error bars.
Fig. 5.
Free energy landscapes of scFv:AMP (A) and of scFv:METH (B). z is the drug's center-of-mass z-coordinate in Å. Three curves for each system are obtained from in silico experiments with pulling speeds at 12.5 Å / ns, 25 Å / ns, and 50 Å / ns, respectively.
The absolute binding of scFv:METH is obtained as the free-energy difference between the bound state ( zb = −1.0 Å ) and the dissociated state ( zd = 13Å ) in the amount of Gb −Gd = −10.2±1.0 kcal/mol. For scFv:AMP, we have the bound state at zb =−0.3Å and the dissociated state zd = 10.2Å and the absolute binding enegy in the amount of Gb −Gd = −5.8±1.0 kcal/mol. These results are in agreement with the in vitro experiments. Moreover, the free-energy landscapes for AMP and METH are nearly monotonous descending pathways from the dissociated state to the bound state. This confirms our earlier speculation on the basis of the first set of experiments that there is no activation barrier along the binding pathway of a drug molecule.
The three sets of in silico experiments we have conducted do not contradict but complement the in vitro experiments in the current literature. Together, they lead to the following conclusions: In terms of statistics, the solution structure of scFv deviates significantly from its crystallographic form in the orifice of the binding pocket. The orifice structure of an scFv:drug complex is stabilized by the van der Waals interactions between the hydrophobic ring of a drug molecule and the aromatic residues of the orifice of the binding pocket. There are five extra water molecules in the binding pocket beneath the drug molecule in addition to the crystallographic waters. These additional waters form strong hydrogen bonds with the charged residue GLU 114, with the cationic amine group of a drug molecule, and with one another. These hydrogen bonds significantly lower the total free energy of entire system. Meanwhile, these extra waters substantially weaken the electrostatic attraction between the drug molecule and GLU 114. Without their presence, the strong electrostatic attraction would make the binding free energy far above the experimental values. In terms of dynamics, when introduced into a solution, an scFv opens up the orifice of its binding pocket so that a drug molecule falls into the pocket without encountering much steric hindrance. Quantitatively, along the pathway from the dissociated state to the bound state, there is no barrier in the free-energy landscape. Therefore the equilibration process is very short when scFv's are injected into a solution of drugs. The equilibration time does not have a strong Arrehnius type of dependence upon the temperature. This point can be verified in future in vitro experiments at varying temperatures.
EXPERIMENTAL PROCEDURES
System setup
We use the crystallographic structure of Celikel et al2 (PDB code: 3GKZ). We keep all the crystallographic waters and solvate the protein-drug complex with additional 13,526 TIP3 waters. We use CHARMM 27 force field9 for intra- and inter-molecular interactions. We use periodic boundary conditions and set the system's pressure and temperature at 1 bar and 300 K respectively. We use NAMD 2.619 as the molecular dynamics engine with a time step of 1 or 2 fs with all hydrogen's covalent bonds set rigid. The damping constant γ = 5.0 / ps. The long range forces are cut off at 12Å with a switching distance of 10Å. We run Langevin dynamics at constant temperature and constant pressure. After the system reaches its equilibrium state, the dimensions of the system are 72Å×70Å×86Å.
Solution structures of scFv
In order to determine how the solution structure of scFv deviates from its crystallographic form, we perform equilibrium Langevin dynamics (LD) runs with and without the drug molecule (AMP or METH) bound to scFv. For the case when scFv is without a drug molecule, 21 ns LD has been conducted. However, significant changes are only present in the first 5 ns. For the case when a drug molecule is bound to scFv, 5 ns LD has been conducted. The results are discussed in the previous sections of this paper.
Waters in-between drug and scFv
To determine how many waters are at the binding site of the protein, we remove the drug molecule (AMP or METH) and place it outside the binding pocket of scFv. We run equilibrium LD for 1 ns while holding the drug's center-of-mass fixed. Then we release the drug's center-of-mass and run equilibrium LD without any constraint to the system. We observe that a drug molecule falls into the binding pocket along with a number of waters as discussed in the previous section.
Absolute binding energies of METH and AMP
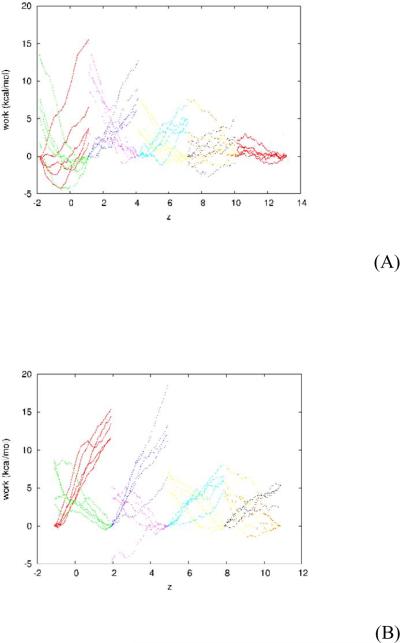
We perform SMD10, 20 and use BD-FDT17, 18 to compute the absolute binding energies of AMP and METH. Specifically, we pull (steer) the center-of-mass z-coordinate of the drug molecule (AMP or METH) at a constant speed v (v = 50Å / ns, 25Å / ns, or 12.5Å / ns) to sample transition paths between the dissociated state (zd ≥ 9.5Å) and the bound state (zb ~ −0.5Å). We use the infinitely stiff spring to pull the center-of-mass z-degree of freedom17 while all the other degrees of freedom of the system follow the equilibrium LD. We divide the whole separation along the z-axis between the dissociated state and the bound state into multiple sections of 3Å each. Over each section, we sample five forward pulling paths (in the direction towards the bound state) and five reverse paths (in the direction away from the bound state) and measure the work of pulling along them. The work curves are shown in Fig. 6. Note that the pulling paths are irreversible, WA→B + WB→A ≠ 0.
Fig. 6.
Work measurements along the pulling paths between the bound state and the dissociated state of scFv:drug complexes: (A) scFv:AMP and (B) scFv:METH. Over each section of 3Å, the five forward paths diverge toward the negative z-direction and the five reverse paths diverge toward the positive z-direction. The curves are for WA→B(z) along forward paths and WB→A(z) along reverse paths. The pulling speed is 50 Å / ns.
Over each section, we pull the center-of-mass z-coordinate of the drug molecule from the right end (zA) to the left end (zB) along a forward path and measure the work WA→z ≡ WA→B(z), the amount of work done to the system when it is pulled from the right end of the section to a point in the section whose z-coordinate is z. Along the reverse path from the left end of a section to the right end of the same section, we measure the work WB→A(z) as the amount of work done to the system when it is pulled from the left end to a point in the section whose z-coordinate is z. Therefore along the partial path from a point in the section whose z-coordinate is z back to the right end of the section, the work is Wz→A ≡ WB→A(zA) − WB→A(z). We apply the BD-FDT to each section to map the free-energy landscape over the section as follows17:
| (1) |
where GA and G(z) are the free energies of the system when the drug's center-of-mass z-coordinate are at the right end of a section and at z, respectively. β=1/kBT with kB and T being the Boltzmann constant and the absolute temperature respectively. The statistical average over the forward paths is
| (2) |
where the index p indicates the p-th of the NF forward paths from State A to State B.
The statistical average over NR reverse paths, from State B to State A, is
| (3) |
In this way, we map out the free-energy landscape over all the sections between the dissociated state and the bound state. The overall free-energy landscapes of AMP and METH in complex with scFv are shown in Fig. 5 and discussed in the previous section..
In silico experiments of single-chain antibody fragment against drugs of abuse.
Solution structures of scFv found to deviate from its crystallographic form
Extra water molecules found at binding site in addition to the crystallographic waters
Absolute binding energies computed in agreement with in vitro experimental measurements
Monotonous descend in the free-energy landscapes of scFv:drug complexes along the binding pathway from the dissociated state to the bound state
ACKNOWLEDGEMENTS
The authors acknowledge support from an NIH SC3 grant (GM084834), the UTSA Computational Biology Initiative, and the Texas Advanced Computing Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.NDIC . National Drug Intelligence Center; Johnstown, PA: 2008. [Google Scholar]
- 2.Celikel R, Peterson EC, Owens SM, Varughese KI. Protein Science. 2009;18(11):2336–2345. doi: 10.1002/pro.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson EC, Laurenzana EM, Atchley WT, Hendrickson HP, Owens SM. Journal of Pharmacology and Experimental Therapeutics. 2008;325(1):124–133. doi: 10.1124/jpet.107.134395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gentry WB, Ruedi-Bettschen D, Owens SM. Clinical Pharmacology & Therapeutics. 2010;88(3):390–393. doi: 10.1038/clpt.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isomura S, Hoffman TZ, Wirsching P, Janda KD. Journal of the American Chemical Society. 2002;124(14):3661–3668. doi: 10.1021/ja012376y. [DOI] [PubMed] [Google Scholar]
- 6.Lape M, Paula S, Ball WJ. European Journal of Medicinal Chemistry. 2010;45(6):2291–2298. doi: 10.1016/j.ejmech.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurenzana EM, Gunnell MG, Gentry WB, Owens SM. Journal of Pharmacology and Experimental Therapeutics. 2003;306(3):1092–1098. doi: 10.1124/jpet.103.053140. [DOI] [PubMed] [Google Scholar]
- 8.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. Journal of Computational Chemistry. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 10.Isralewitz B, Baudry J, Gullingsrud J, Kosztin D, Schulten K. J. Mol. Graph. Model. 2001;19(1):13–25. doi: 10.1016/s1093-3263(00)00133-9. [DOI] [PubMed] [Google Scholar]
- 11.Itsuo H, et al. Journal of Physics: Condensed Matter. 2008;20(25):255238. [Google Scholar]
- 12.Burendahl S, Danciulescu C, Nilsson L. Proteins. 2009;77(4):842–856. doi: 10.1002/prot.22503. [DOI] [PubMed] [Google Scholar]
- 13.Mascayano C, Nunez G, Acevedo W, Rezende MC. Journal of Molecular Modeling. 2010;16(5):1039–1045. doi: 10.1007/s00894-009-0616-9. [DOI] [PubMed] [Google Scholar]
- 14.Parravicini C, Abbracchio MP, Fantucci P, Ranghino G. Bmc Structural Biology. 2010;10 doi: 10.1186/1472-6807-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strzelecki J, Mikulska K, Lekka M, Kulik A, Balter A, Nowak W. Acta Physica Polonica A. 2009;116:S156–S159. [Google Scholar]
- 16.Yang LJ, Zou J, Xie HZ, Li LL, Wei YQ, Yang SY. PLoS ONE. 2009;4(12) doi: 10.1371/journal.pone.0008470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen LY, Bastien DA, Espejel HE. Phys. Chem. Chem. Phys. 2010;12(25):6579–6582. doi: 10.1039/b926889h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LY. J. Chem. Phys. 2008;129(14):144113–144114. doi: 10.1063/1.2992153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J. Comput. Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Schulten K. J. Chem. Phys. 2004;120(13):5946–5961. doi: 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14(1):33–&. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]