Abstract
Human immune cells penetrate an endothelial barrier during their beneficial pursuit of infection and their destructive infiltration in autoimmune diseases. This transmigration requires Rap1 GTPase to activate Integrin affinity1. We define a new model system for this process by demonstrating with live imaging and genetics that during embryonic development, Drosophila melanogaster immune cells penetrate an epithelial, DE-Cadherin-based tissue barrier. A mutant in RhoL, a GTPase homolog that is specifically expressed in hemocytes, blocks this invasive step but not other aspects of guided migration. RhoL mediates Integrin adhesion caused by Drosophila Rap1 over-expression and moves Rap1 away from a cytoplasmic concentration to the leading edge during invasive migration. These findings indicate that a programmed migratory step during Drosophila development bears striking molecular similarities to vertebrate immune cell transmigration during inflammation and identify RhoL as a new regulator of invasion, adhesion and Rap1 localization. Our work establishes the utility of Drosophila for identifying novel components of immune cell transmigration and for understanding the in vivo interplay of immune cells with the barriers they penetrate.
INTRODUCTION
The ability of immune cells to penetrate tissue barriers is essential for normal and aberrant human physiology. Leukocytes patrol throughout the body in the blood, and to attack an infection must move through the blood vessel’s intervening endothelial wall2. Inappropriate activation of this transmigration can lead to damaging immune cell infiltrations that are involved in many medical conditions caused by autoimmunity3, 4. The ability to manipulate this process would have important medical implications.
Signals from infected tissue lead to changes in immune cell adhesion that underlie this specialized movement. Upon inflammation, endothelial cells in the vessel wall express selectins on their surface. Selectins transiently bind leukocyte surface proteins and promote an initial affinity of leukocyte Integrin for endothelial ligands, causing immune cells to roll along the vessel wall. Cytokine signaling from inflamed tissue and other immune cells then triggers Rap1 GTPase activation in leukocytes5 causing the Rap1 effectors RAPL and Mst1 to transit from a perinuclear region to the cell surface6, 7. This leads to increased Integrin affinity; leukocytes arrest rolling and actively migrate into the tissue, mostly at endothelial cell junctions3. How precisely Rap1 effectors are relocalized and lead to Integrin activation is unknown.
Hemocytes, Drosophila immune cells, have proven to be an excellent system to analyze guided cell migration. The predominant Drosophila immune cell, plasmatocytes, are macrophage-like, engulfing bacteria and viral invaders, phagocytosing dead cells8 and responding to wounds and tumors9-13. They are guided by PDGF/VEGFR homolog (PVR) and its ligands Pvf2 and 3 from their site of origin in head mesoderm along three main routes: towards the tail, along the ventral nerve cord (vnc) and along the dorsal vessel 12, 14, 15. Due to Drosophila’s open circulatory system, hemocytes need not penetrate an endothelial barrier to reach infections10. However, by examining plasmatocyte migration in live embryos during development, we discovered that Drosophila hemocytes invade an epithelial barrier as they move from the head region into the tail. We find that specific strategies for invasive immune cell transmigration such as the employment of Integrins and Rap1 are conserved with vertebrates. Thus Drosophila possesses a specific invasive migratory step which is now amenable to unbiased genetic analysis.
RESULTS
To determine if hemocytes, the Drosophila immune cells penetrate tissue barriers during their normal developmental migration, we examined fixed embryos expressing GFP from the hemocyte specific GAL4 driver srpHemo14. We visualized hemocytes with anti-GFP antibodies, nuclei with DAPI, and surrounding tissues with anti-DE-Cadherin antibodies. We observed hemocyte migration from the head region into the tail at stage (st) 11 and 12 from lateral (Supplementary Information, Fig.S1a-c) and dorsal directions (Fig. 1a-c’). Hemocytes initially encounter a barrier of tail epithelial cells (arrows in 1a, a’). However as hemocytes infiltrate they break the junctions that hold the cells together (arrows in 1b, b’) and stream into the tail (1c, c’).
FIGURE 1. Drosophila immune cells move into the tail during development by penetrating an epithelial barrier and carrying out chain migration.
Schematic drawings of embryos from a dorsal perspective with anterior to the left, showing hemocytes (green) migrating into the tail during st 11-12; boxed area corresponds to pictures below. Dashed box in rightmost schematic corresponds to (e). (a-c’) Dorsal confocal images of fixed wild type srpHemoGAL4 UAS-GFP embryos which express GFP in hemocytes. Embryos are from successively later stages as indicated above and were stained with anti-Cadherin (red) and anti-GFP (green) antibodies as well as DAPI to visualize nuclei (blue). Hemocytes moving into the tail are indicated with white arrows; examining these locations in the ‘ panels which show only the Cadherin channel reveals that a breach in the Cadherin barrier appears as hemocytes move into the tail. This is particularly evident when one compares the side of the hindgut (upper white arrow in b) where hemocytes have not yet entered and there is no break in the Cadherin staining (upper white arrow in b’) to the other side of the hindgut (lower arrow in b and b’) where they have. (d-e) Stills from 2-photon movies of ubi:shg-GFP; srpHemoGAL4 UAS-GFP embryos. Shg-GFP outlines all cells and hemocyte-expressed GFP is seen in cytoplasm. (d) Dorsal view of early st 11 single plane movie (Supplementary Information, Movie S1) showing the movement of hemocytes through surrounding cells into the tail (green arrow). (e) Dorsal view of a multi-plane movie (Supplementary Information, Movie S2) illustrating chain migration (green arrowhead) around the hindgut. Scale bars, (a-c) 25, (d) 10, or (e) 20 ⌈m. Anterior to left in all panels.
To directly observe hemocytes live while they are moving through surrounding tissues, we labeled all cell surfaces with ubi:DE-Cadherin-GFP and hemocytes with srpHemo>GFP. Seen in lateral views, hemocytes enter on the underside of the tail and push between ectodermal epithelia and neighboring mesoderm before germ band retraction commences (Supplementary Information, Fig. S1d, Movie S1). Movies of dorsal views revealed that hemocytes penetrated the epithelial barrier at the tail’s edge and then moved between the hindgut and surrounding cells (Fig. 1d) (Supplementary Information, Movie S2). Hemocytes displayed chain migration also seen in highly metastatic cancers16 (Fig. 1e) (Supplementary Information, Movie S3). We conclude that hemocytes migrate invasively into the tail during embryonic st 11 and 12.
To determine if this invasive movement into the tail utilizes a specialized cell biological program, we screened for mutants that specifically affect this step. We identified a viable transposon insert, pGawB10-161 (Supplementary Information, Fig. S2a) in rhoL17. In sequence alignments RhoL clusters within a branch of the Rho family that contains Rac, Cdc42, and less characterized Rhos (Supplementary Information, Fig. S2b). rhoL RNA is expressed specifically in anterior head mesoderm where hemocytes originate, starting at st 5 and continuing in all hemocytes through st 11 (Fig. 2a-c). Thereafter rhoL RNA ceases expression in hemocytes and begins expression in more posterior mesoderm (Fig. 2d). The rhoL10-161 insert causes a strong reduction in rhoL transcript levels (Supplementary Information, Fig. S2c-d). The phenotype is enhanced over Df(3R)by416, indicating that the insertion is a hypomorphic allele (data not shown). We generated a lethal excision, rhoLXA12, which behaves as a null allele and strongly reduces the movement of cells into the tail at st 11 and 12 (Fig. 2e-f, m). On average only 4±1 (n=41) hemocytes reached the tail compared to 31±1 (n=30) in wild type and 44% of embryos had no hemocytes in the tail, which is never observed in wild type. This causes an absence of hemocytes in the posterior nerve cord that persists through st 14 (Fig. 2h-i). We expressed the rhoL cDNA under the control of the hemocyte specific 8-163GAL4 driver (Materials and Methods) and achieved rescue (Fig. 2g, j) with 27±2 (n=24) hemocytes in the tail at st 12. We next assessed if RhoL affects hemocyte migration along their other non-invasive embryonic routes. Fixed rhoLXA12 mutant embryos displayed no defect in normal guided migration of hemocytes 12, 14, 15 along the anterior vnc (data not shown) or dorsal vessel (Fig. 2k-l, m). We conclude that RhoL, a member of the Rho GTPase family, is expressed in hemocytes at the time of their migration into the tail and is required specifically in hemocytes to permit this specialized movement. Thus RhoL defines a separate invasive migratory step.
FIGURE 2. Identification of the gene, rhoL, which is expressed and required in hemocytes to permit migration into the tail.
(a-d) In situ hybridizations with rhoL probe on wild type embryos show (a-c) hemocyte expression st 5-11 and (d) mesodermal expression st 12. Confocal images of fixed (e-g) st 12 and (h-l) st 14 embryos stained with anti-Cadherin (red) and anti-hemocyte expressed β-Gal (green) antibodies. In wild type embryos, hemocytes migrate (e) into the tail and (h) along the posterior vnc (indicated by white dashed oval). (f, i) In rhoLXA12 embryos, hemocytes do not. (g, j) In rhoLXA12 embryos expressing rhoL via the hemocyte driver 8-163GAL4, hemocyte migration (g) into the tail and (j) along the posterior vnc is restored. (k) Wild type and (l) rhoLXA12 mutant hemocytes migrate normally along the dorsal vessel. (m) Quantitation of hemocyte migration into tail and along dorsal vessel (** P value: 3×10−20, ns, not significant: P value: 0.3). Histograms throughout indicate mean ± s.e.m.. The number of embryos examined is recorded in this and all subsequent figures within the histogram. Error bars indicate s.e. throughout. (a-j) Scale bars in these and all other whole embryo pictures indicate 50 ⌈m. All embryos are oriented anterior to the left and dorsal up unless otherwise noted. (k-l) Scale bar, 20 ⌈m.
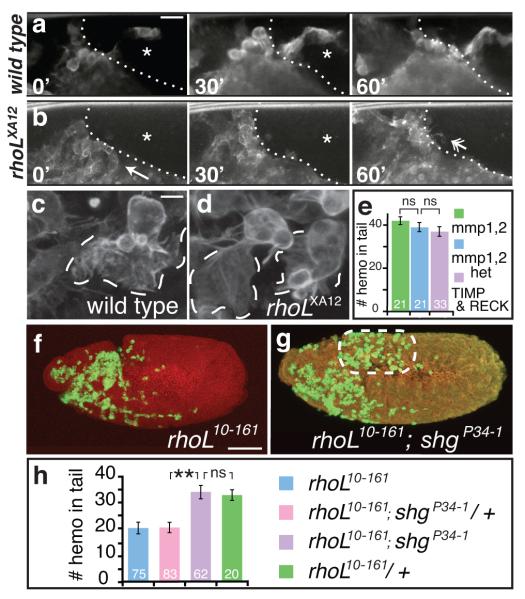
To analyse cellular aspects of hemocyte behavior, we observed hemocytes expressing the actin-binding protein Moesin fused to GFP in live embryos. In wild type, hemocytes entered the tail fluidly (Fig. 3a, Supplementary Information, Movie S4). Though hemocytes in rhoLXA12 mutants (Fig. 3b, Supplementary Information, Movie S5) moved up to the junction with the tail (arrow in 0′), and produced protrusions in its direction (arrow in 60′), they never entered the tissue, unlike in wild type (compare star in Fig. 3a-b). As suggested by fixed tissue analysis (Fig. 2i, l), rhoLXA12 hemocytes exhibited normal guided migration in anterior inner vnc movies. Pvfs guide hemocyte migration12 along the inner vnc and both wild type and rhoLXA12 hemocytes moved posteriorly in a directed manner (Supplementary Information, Movie S6 and S7, Fig. S3a-b). To determine if rhoLXA12 hemocytes had defects in actin structures that might prevent tail invasion, we created still images of live hemocytes. We observed no differences in hemocyte lamellipodia or filopodia between wild type and rhoLXA12 embryos (Fig. 3c-d). In contrast, Rac1 and Cdc42 are required in embryonic hemocytes to form broad and unipolar lamellae, respectively11, 18. Thus while Cdc42 and Rac1 affect migratory actin structures, RhoL does not. RhoL is also not required for guided migration, but is specifically required for the invasive migration of hemocytes.
FIGURE 3. RhoL does not affect hemocyte actin structures or guidance, but is required to penetrate a Cadherin-containing barrier for migration into the tail.
2-photon images of (a, c) wild type and (b, d) rhoLXA12 embryos expressing Moesin-GFP in hemocytes under the control of the 8-163GAL4 driver. (a, b) Stills from 2-photon movies (Supplementary Information, Movie S4, Movie S5). (a) Wild type hemocytes move into the tail; but (b) rhoLXA12 hemocytes move up to the tail (arrow in 0′) and produce protrusions towards it (arrow in 60′) but fail to enter (compare * in a and b). The elapsed number of minutes is indicated. (c, d) Single plane Z-sections show that RhoL is not required for the normal formation of lamellipodia or filopodia. (e) Matrix metalloproteases are not required for hemocyte movement into the tail (P values: left = 0.4, right = 0.5). (e, h) Quantitation of the number of hemocytes that move into the tail in the genotypes indicated. (f) Fewer hemocytes move into the tail in embryos carrying the rhoL10-161 hypomorphic allele. (g) This defect is rescued by the addition of the shgP34-1 mutant that decreases the expression of DE-Cadherin. (f-g) The tail is indicated by the white dashed oval. (h) P values: ** = 6×10−5, ns = 0.9. Scale bars, (a, b) 15 or (c, d) 5 ⌈m.
We wished to determine the nature of the barrier that hemocytes breach to move into the tail. The extracellular matrix (ECM) was one possibility; yet embryos lacking the two ECM metalloproteases in Drosophila, Mmp1 and Mmp219, or expressing inhibitors of ECM proteases displayed no migratory defects in hemocytes (Fig.3e, Supplementary Information, Fig. S3c-e). Next we tested if the barrier might consist of cellular junctions. Septate junctions form later, at st 14, but spot adherens junctions mature into zonula adherens by st 11 in the ectoderm20. If RhoL allows hemocytes to push apart adherens junctions to reach the tail, we reasoned that reducing the level of one junctional component, DE-cadherin, might rescue the rhoL phenotype. In the hypomorphic allele, rhoL10-161, 20±2 (n=75) hemocytes moved into the tail (Fig. 3f, h); however when rhoL10-161 was combined with an allele of DE-Cadherin, shgP34-1, that has reduced expression due to a transposon insertion in the 5′ UTR21, 22, the number of hemocytes moving into the tail increased to levels indistinguishable from rhoL10-161 heterozygotes (34±3 (n=46) versus 33±2 (n=23)) (Fig. 3g-h). An alternate interpretation of this rescue is that rhoL mutants display defects due to elevated levels of DE-Cadherin in hemocytes. However, we did not observe appreciable hemocyte expression of DE-Cadherin in either wild type or the null allele rhoLXA12 (Supplementary Information, Fig. S3f-g”). We therefore conclude that RhoL mediates penetration of hemocytes through a DE-Cadherin-containing barrier, most likely the zonula adherens.
Vertebrate leukocytes utilize Rap1 to regulate Integrin and move through endothelial barriers1, 5. To assess whether Drosophila hemocytes require Rap1 for invasive migration we examined mutants in dizzy, a GTP exchange factor (GEF), which is required for Rap1’s activation of Integrin-dependent adhesion in hemocytes23. The number of hemocytes that entered the tail decreased (Fig. 4a-b, d) in dizzyΔ1 null mutants. We did not assess Rap1 mutant embryos, due to their severe developmental defects23. Like rhoL, dizzyΔ1 is specific for invasive migration into the tail and does not affect general guided migration along the dorsal vessel (Supplementary Information, Fig. S4) or vnc (data not shown). To further examine the analogies between vertebrate leukocyte vessel transmigration and Drosophila hemocyte tail invasion, we assessed mutants in inflated, an α-Integrin subunit expressed in the mesoderm where hemocytes are specified24-26. We observed fewer hemocytes migrating into the tail in inflated null mutants (Fig. 4c-d), but no decrease in dorsal vessel migration (Supplementary Information, Fig. S4) or the vnc (data not shown). We conclude that hemocytes utilize the Rap1 pathway and Integrins for penetrating into the tail, and thus exhibit molecular parallels with vertebrate immune cells moving through blood vessel walls.
FIGURE 4. The Rap1 GEF Dizzy and the α-Integrin Inflated are required for tail invasion.

Confocal images of fixed early st 12 embryos stained with anti-Cadherin (red) and (a, c) anti-hemocyte-expressed β-Gal or (b) CD2 antibodies (green) showing that (a, b, d) the Rap1 GEF Dizzy and (a, c, d) the Integrin α-subunit Inflated are required for movement into the tail, but not for movement along the dorsal vessel (Supplementary Information, Fig. S4). (d) Quantitation of the number of hemocytes that move into the tail in the indicated genotypes. The deficiency utilized is Df(2L)BSC185 which removes dizzyΔ1 completely. P values: ** left = 5×10−5, right = 8×10-17, both ns = 0.6.
Our data suggest that RhoL acts with Dizzy and Rap1 to modulate Integrin function to permit invasive hemocyte migration. To determine where in the pathway RhoL functions, we utilized an Integrin-dependent hemocyte clustering assay23; adhesion is induced by expression of dominant active (DA) Rap1, Rap1V12, which bypasses the need for the dizzy GEF. Wild-type hemocytes do not congregate together in great numbers, (Fig. 5a, e), whereas hemocytes expressing Rap1DA form large tightly associated masses of cells (Fig. 5b, e). Rap1DA-induced hemocyte clustering is abolished in the presence of the rhoL10-161 allele (Fig. 5c, e). When we overexpressed RhoL, we observed the same numbers of hemocytes in masses as upon Rap1DA expression (Fig. 5d-e) with similar fractions of each hemocyte’s surface touching another hemocyte (Fig. 5 legend). We conclude that RhoL can regulate adhesion in this assay. To determine if RhoL functions in the same pathway with Dizzy and Rap1 during normal hemocyte movement, we created dizzyΔ1 rhoL10-161 double mutants and saw no additive effect (Fig. 4d). Our results show that Rap1 facilitates activation of Integrin during invasive migration and that this step is mediated by RhoL.
FIGURE 5. RhoL is required for Rap1 to induce adhesion and relocalize from an intracellular concentration to the cell surface in hemocytes.
(a, e) Wild type hemocytes located in lateral st 14 embryos do not form large clusters. (b, e) The expression of Rap1V12, a DA form, induces hemocyte adhesion and clustering which can be alleviated by the presence of (c, e) the rhoL10-161 hypomorphic allele. (d) The expression of RhoL by itself can lead to clustering. Quantitation of the fraction of hemocyte surfaces in contact with one another showed this was 42±3% in hemocytes overexpressing RapDA and 34±3% with RhoL (n .>17 hemocytes in 8 embryos each). (e) Quantitation of the largest number of hemocytes found in a cluster in embryos of indicated genotypes. P values: ** left = 4×10−10, right = 2×10−8, ns= 0.6. (f-g”) Confocal images from fixed embryos focusing on hemocytes at the junction with the tail or (h-i”) on a single hemocyte. (f-f’, h-h”) Rap1-GFP; srpHemoGAL4 UAS:CD2 and (g-g’, i-i”) Rap1-GFP; rhoL10-161-GAL4 UAS:lacZ embryos stained with antibodies against (f-f’, h-h”) the membrane marker CD2 or (g-g”, i-i”) cytoplasmic β-Gal to visualize hemocytes (red) and anti-GFP (green) to visualize Rap1. (f-g) Insets indicate % of hemocytes displaying a Rap1 concentration in each genotype (arrow), showing that mutating rhoL increases Rap1 concentrations three fold. (f-i”) Column heads indicate panels show Rap1-GFP staining (f, g, h, i), hemocyte marker staining (h”, i”) or a merge of the two (f’, g’, h’, i’). (h) In wild type hemocytes Rap1 is found at the leading edge (arrow), and in (i) rhoL10-161 mutants Rap1 is localized to an intracellular concentration that excludes cytoplasm (arrow). Scale bars, (a-d) 10, (f-g’) 20 or (h-i”) 5 ⌈m. (j) Model: To allow invasive migration and breach the DE-Cadherin barrier between epithelial cells, hemocytes require RhoL. The GEF Dizzy converts Rap1 to a GTP-bound form. RhoL causes Rap1-GTP to move from an intracellular concentration to the cell surface, where Rap1-GTP triggers increased Integrin affinity, permitting crossing of the DE-Cadherin barrier.
To determine how RhoL affects Rap1 activity, we examined a Rap1-GFP fusion expressed from the Rap1 promoter27. In wild type st 11 and 12 embryos we observed Rap1 in cytoplasmic concentrations in 11±1% of wild type hemocytes (n=33 embryos) (Fig. 5f-f’). The number of these concentrations was dramatically increased in rhoL10-161 embryos, to 30±2% (n=30 embryos) of hemocytes (Fig. 5g-g’). We examined Rap1 localization changes in hemocytes at higher magnification. In wild type embryos we observed Rap1 mostly at the leading edge during tail invasion (Fig. 5h-h”). In rhoL10-161 mutant embryos we found Rap1 located not at the membrane but in a large, roundish, intracellular concentration that excluded cytoplasm (Fig. 5i-i”). The Rap1 concentration is not due to phagocytosis of neighboring cells, because spectrin, which is expressed in the surrounding tissues but not in hemocytes, does not colocalize with Rap1 (data not shown). We conclude that RhoL mediates proper Rap1 localization to permit invasive migration into the tail.
Taken together our results show that the developmental movement of Drosophila hemocytes requires invasive migration. We observe hemocytes penetrating the epithelial barrier caused by the tail folding against the head during germband extension. Vertebrates also appear to require invasive migration of immune cells during development; recent studies show that hematopoietic stem cells are born in mesoderm underneath the dorsal aorta and must migrate into it to eventually colonize the fetal liver28. We propose that the ability of immune cells to move through an epithelial barrier evolved before closed circulation within blood vessels did.
We also identify a pathway in Drosophila that is required specifically for this invasive migratory step. We demonstrate a requirement for the Rap1 GEF Dizzy, the α-Integrin Inflated and the RhoL GTPase just in this type of migration; our work and that of others orders their function. We propose the following model based on our findings (Fig. 5j). When hemocytes encounter an epithelial barrier, a pathway for invasion is activated. The Dizzy GEF converts Rap1 to its active GTP form23. However, Rap1-GTP cannot induce Integrin activation unless RhoL is present to promote Rap1 localization to the leading edge, where Rap1 increases Integrin affinity as has been demonstrated in vertebrates29. The lack of basement membrane in EM studies30 argues that Inflated is not binding ECM during tail invasion. We posit that Inflated binds another membrane protein, akin to ICAM binding observed in vertebrates31, or DE-Cadherin itself, to facilitate transit between epithelial cells. While we cannot exclude that hemocytes exert force on epithelial cells to pry them apart, our observations (Movie S2) favor the model that hemocyte Integrin binding to an epithelial cell surface molecule induces a signaling pathway that decreases either the affinity or levels of Cadherin-containing junctions. During their migration into regions of the embryo other than the tail, hemocytes need not penetrate an epithelial barrier, and migrate along the surface of mesodermal or epithelial cell types.
In conclusion, we have provided the first evidence for modulation of Rap1 localization during epithelial penetration and for the involvement of a Rho GTPase homolog in controlling this shuttling. Chemokine signaling utilized during endothelial transmigration of vertebrate immune cells causes the Rap1 effectors RapL and Mst1 to leave an intracellular compartment in in vitro studies6, 7; however whether Rap1 localization changes is untested. T-cell receptor activation induces Rap1 trafficking from a perinuclear compartment to the cell surface to increase Integrin binding, yet how this shuttling is regulated is unclear29, 32. A Rho GTPase could control this process. In phylogenetic analyses (Supplementary Information, Fig. S2b) RhoL lies within a branch containing two Rhos involved in intracellular trafficking; RhoJ/TCL regulates early endocytosis and RhoQ/TC10 modulates exocytic vesicle fusion33, 34. This raises the possibility that RhoL acts as a switch to regulate trafficking of Rap1-containing vesicles in Drosophila and that RhoJ and/or RhoQ do the same in vertebrates.
Drosophila immune cells’ invasive migration displays striking similarity to vertebrate immune cells’ extravasation in pursuit of infection. Both require Rap1 and Integrin and do not appear to utilize MMPs30. Both must traverse a Cadherin-containing barrier31. In both vertebrate and Drosophila immune cell migration, Integrin is not needed for general interstitial migration35. We propose that during inflammation vertebrate immune cells move through an endothelial barrier by recapitulating the cell biological program originally evolved for developmental migration through epithelia. We have found many other mutants in our screens that affect tail invasive migration and have human homologs. We are thus encouraged that further examination of this new model for immune cell transmigration will provide novel insight into the cell biological controls that mediate this medically important process.
MATERIALS AND METHODS
Fly Stocks
U. Heberlein provided 1, 200 GAL4 enhancer trap lines containing the pGawB vector randomly inserted in the genome. R. Reuter provided srpHemoGAL4 UAS-GFP, dizzyΔ1/CyO; P(w+) srpHemoGAL4 UAS-CD2, UAS-rapV12, K. Brueckner and N. Perrimon provided pvr1 srpHemoGAL4/CyO ftz:lacZ; UAS-lacZnls, Pvr1 UAS-srcEGFP/CyO ftzlacZ; UAS-p35. N. Brown sent lines carrying a Rap1-GFP fusion under the control of the endogenous Rap1 promoter.
Flies were grown on an agar, cornmeal, molasses and yeast mixture containing Tegosept (Sigma) mold inhibitor, 1% methanol and 0.4% propionic acid. Embryos were collected on plates made from apple juice, sugar, agar and Tegosept.
The full genotype of the embryos shown were: Figure 1 (a-c) and Figure S1 (a-c): srpHemoGAL4 UAS-GFP. Figure 1 (d-e) and Figure S1 (d): ubi:shg-eGFP; srpHemoGAL4 UAS-GFP. Figure 2 (a-d) and Figure S2 (d): Oregon R. Figure 2 (e, h, k, m) and Figure 4 (a, d), Figure S3 (f-f”), and Figure S4 (a, d): 8-163GAL4 UAS-lacZ. Figure 2 (f, i, k, m), and Figure S3 (g-g”): 8-163GAL4 UAS-lacZ; rhoLXA12. Figure 2: (g, j, m) 8-163GAL4 UAS-lacZ; rhoLXA12 UAS-rhoL, Figure 2: (m) 8-163GAL4 UAS-lacZ; rhoLXA12/Df(3R)by416. Figure 3 (a, c) and Figure S3 (a): 8-163GAL4 UAS-Moe-GFP. Figure 3 (b, d) and Figure S3 (b): 8-163GAL4; rhoLXA12 UAS-Moe-GFP. Figure 3 (e) and Figure S3 (c): cn mmp2W307 bw sp mmp1Q112 /Cyo ftz:lacZ; srpHemoGAL4 UAS-CD2, Figure 3 (e) and Figure S3 (d): cn mmp2W307 bw sp mmp1Q112; srpHemoGAL4 UAS-CD2. Figure 3(e) and Figure S3 (e): srpHemoGAL4 UAS-GFP/ UAS-RECK; srpHemoGAL4 UAS-GFP/UAS-TIMP. Figure 3 (f, h) and Figure 4 (d): rhoL10-161GAL4 UAS-lacZ. Figure3 (g, h): shgP34-1; rhoL10-161GAL4 UAS-lacZ. Figure 4 (b, d) and Figure S4 (b, d): dizzyΔ1; srpHemoGAL4 UAS:CD2, Figure 4 (c, d) and Figure S4 (c, d): inflatedB2; 8-163GAL4 UAS:lacZ. Figure 4 (d) dizzyΔ1/ Df(2L)BSC185, dizzyΔ1; rhoL10-161GAL4 UAS-lacZ. Figure 5: (a, e) srpHemoGAL4 UAS:CD2, (b, e) UAS-rapV12; srpHemoGAL4 UAS:CD2, (c, e) UAS-rapV12; rhoL10-161GAL4 UAS:lacZ, (d, e) 9-1UAS-rhoL; srpHemoGAL4 UAS:CD2, (f-f’, h-h”) rap1-GFP; srpHemoGAL4 UAS:CD2, (g-g”, i-i”), rap1-GFP; rhoL10-161GAL4 UAS-lacZ. Figure S2 (c, d): rhoL10-161GAL4.
Hemocyte driver screen and 10-161 insert site localization
Males from GAL4 enhancer trap lines containing the pGawB vector were individually crossed to females carrying UAS:lacZ and the resulting embryos were stained with X-Gal to visualize expression. Lines were selected as possibly expressing in hemocytes based on the characteristic triangular early hemocyte pattern. Line 8-163 which showed normal hemocyte migration was utilized as a driver. Line 10-161 showed a defect in hemocyte migration into the tail. To determine the location of the 10-161 insert, the genomic region flanking the insertion site was amplified by inverse PCR (BDGP protocol) on MspI or Sau3A restricted genomic DNA (using GawB5’out and GawB5’in primers for the 5’end, and GawB3’int4 and Pry1 primers for the 3′ end), sequenced, and compared to the Drosophila genome. We mapped the insert to the promoter of the rhoL gene, 5 base pairs upstream of the transcript start.
P element excision and RhoL rescue
100 independent rhoL10-161 excision lines were created by crosses to the Δ2-3 transposase and balanced over TM3Sb. 16 lethal excisions were identified, 9 of which retained their lethality over Df(3R)by416 which removes RhoL. Primers (synthesized by IDT) located just next to the RhoL ORF and 2 kb upstream of the RhoL transcriptional start site were utilized to characterize the extent of the excision. Primer sequences were: RhoL ORF 5′ CTGTGGAGCGAGTACG; RhoL ORF 3′ TGACGGCGAACATAAC ; RhoL promoter 5′ GCGGAGCACTAAGGAG; RhoL promoter 3′ TGATTCGCTGTTCCAG. None of the excisions extended into the RhoL ORF, and all but one did not extend past the midpoint between the RhoL transcriptional start site and that of the neighboring gene. Genewiz Inc. did the sequencing.
To create a RhoL rescue construct containing the entire ORF (231bp to 803 bp after cDNA start) under the inducible control of UAS, the SD05212 RhoL cDNA was amplified with primers that encompassed the region from 51 bp to 890 bp. The primers contained an Eag1 site on the 5′ and an Xho1 site on the 3′ end to facilitate cloning into pUAST. Genetic Services Inc. created transgenes of these constructs. Rescue of the original rhoL10-161 insertion and the rhoLXA12 excision phenotype was tested with three independent UAS-RhoL insertions on the second chromosome. Insert 9-1 was utilized for the rescued embryos shown.
RT-PCR
RNA was prepared from 4-6 hr embryo collections using TRIzol (Invitrogen). We used a Superscript II Reverse Transcriptase Kit (Invitrogen) and Oligo dT primers to prepare cDNAs. PCR was conducted using the following primers which produce a 492 bp amplicon for RhoL RNA and a 126 bp amplicon for the RP49 control: RhoL 5′ GCGCCCGCTGAAAATAA; RhoL 3′ CTAACCGCCTCCTCGAAGAC; RP49 5′ GTATCGACAACAGAGTGCGT; RhoL 3′ CGTTGTGCACCAGGAAC.
In situ hybridization and immunobiochemistry
The RhoL cDNA, SD05212, was subcloned into BlueScript (Stratagene) utilizing Xho1 and EcoR1 to create DSPL15. T7 polymerase synthesized digoxigenin labelled anti-sense probes and in situ hybridization was performed using standard methods. Antibody staining of embryos was performed using a standard 5% formaldehyde/ heptane fix with methanol devitellinization except for anti-DE-Cadherin staining in Figure 1 for which we hand-devitellinized embryos after fixing them in 5% methanol free formaldehyde for 40′, anti-DE-Cadherin staining in the other Figures for which we fixed embryos with 1mM calcium and .925% formaldehyde and phalloidin for 15′, and anti-β-PS Integrin and anti-Rap1-GFP staining for which we devitellinized embryos with ethanol. The following antibodies or reagents were used (source, dilution): rabbit anti-β-Galactosidase (Cappel, 1:10,000), rabbit anti-GFP (A111-22) (Molecular Probes, 1:1000), mouse anti rat CD2 (MCA154G: Serotec, 1:50), rat anti-DE-Cadherin (DCAD2: T. Uemura, DSHB, 1:50), anti-dpERK (M8159: Sigma, 1:10), guinea pig anti-Hsc-3 antibody (D. Ryoo, 1:100), guinea pig anti-Hrs antibody (H. Bellen, 1:200), DAPI (Roche, 1:1,000), mouse anti-spectrin (3A9: D. Branton, R. Dubreuil, DSHB, 1:5), guinea pig anti-Spinster (G. Davis, 1:250), rabbit anti-GM130 --(MLO7: N. Lowe, 1:4,000). Cy2, Cy3 or Cy5 conjugated appropriate secondary antibodies were used at a 1:500 dilution (Invitrogen, Jackson ImmunoResearch Laboratories).
Bright field pictures were taken with a Zeiss Axiophot utilizing a 20X objective (Zeiss, Neofluor, air, NA 0.50) and a SPOT Insight 14.2 Color Mosaic CCD camera (Diagnostic Instruments) governed by SPOT 4.5.9.8 software; fluorescent images were visualized with a Zeiss LSM-510 confocal and a 20X (Zeiss, Plan Apochromat, air, NA 0.8), 40X (Zeiss, Plan Neufluor, oil, NA 1.3), and 63X (Zeiss, Plan Neufluor, dH2O, NA 1.3) objective. Sections spanning the entire depth of hemocyte fluorescence were taken every 3 μm and were merged to form the final images except in Figure 1 which are single 3 μm planes, Figure 5 (a-d, h-i”) which are single 1 μm planes.
Hemocyte movement quantitation
Embryos from early to mid st 12 were identified by having some separation of the germ band from the head, but not having extended past the midpoint of the embryo. The number of hemocytes in the tail were counted by visual examination on the LSM 510 Zeiss Confocal. Average numbers and standard error (SE) is reported. P values were determined using a 2-tailed equal variance T test in Microsoft Excel.
Live imaging
Bleach-dechorionated embryos were selected using a Leica MZH III fluorescent dissecting scope, lined up on an apple juice plate, glued to a 1.5 μm cover slip with heptane solubilized packing tape (Tesa 4124) (Beiersdorf AG, Germany) glue placed onto a custom made plastic frame holding a gas permeable standard membrane (YSL Inc.), and covered with Halocarbon oil 200 (Halocarbon). A series of sections every 3 μm were captured every 2 minutes. For Figure 1 (d, e), Figure S1 (d) and Figure S3 (a, b) we utilized a 40X objective (Olympus, UPlan FL, oil, NA 1.3), and for Figure 3 (c, d) a 63X objective (Olympus, UPlan FL, oil, NA 1.42) on a Prairie Technologies Ultima Multiphoton system with an Olympus microscope (BX51 W1), a Coherent Chameleon Ultra 1 80 MHz Ti:Saph laser controlled by Prairie View software v2.5.1.3. Figure 3 (c, d) were 1μm sections from Z-series acquisitions. For Figure 3 (a, b) we used a 40X objective (Nikon, Plan Fluor, water, NA 0.75) on a BioRad Radiance Multiphoton system with a Nikon microscope (Eclipse E600FN), a 10W pumped Tsunami laser (Spectra Physics) controlled by Laser Sharp software (BioRad)). Movies were assembled using Volocity 2.6.1 (Improvision) or Imaris 6.1.0 (Bitplane) software, and are shown at three frames per second. The movie in Figure 1d consists of only one plane.
Supplementary Material
Figure S1: Drosophila immune cells move into the tail during development by penetrating an epithelial barrier.
Schematic drawings show embryos from a lateral perspective with hemocytes (green) migrating into the tail during st 11-12. Hemocyte positions sketched from in situs of PVR. (a-c) Lateral confocal images of fixed wild type srpHemoGAL4 UAS-GFP embryos which express GFP in hemocytes; pictures correspond to the boxed area in the appropriate embryo schematic. Embryos are from successively later stages as indicated above and were stained with anti-Cadherin (red) and anti-GFP (green) antibodies as well as DAPI to visualize nuclei (blue). Hemocytes moving into the tail are indicated with white arrows; examining these locations in the ‘ panels which show only the Cadherin channel reveals that a breach in the Cadherin barrier appears as hemocytes move into the tail. Scale bars, 25 ⌈m.
(d) Stills from 2-photon movie of ubi:shg-GFP; srpHemoGAL4 UAS-GFP embryos. Shg-GFP outlines all cells and hemocyte-expressed GFP is seen in cytoplasm. (d) Lateral view starting st 11. Movie (Supplementary Information, Movie S1) corresponds to the dashed boxed area in the late st 11 schematic embryos above. Scale bars, (a-c) 25 ⌈m or (d) 20 ⌈m. Anterior to left and dorsal up in all panels.
Movie S6: Lateral view of hemocytes migrating along the ventral nerve cord in a wild type embryo, hemocytes labeled. Lateral movie of a wild type embryo expressing Moe-GFP in hemocytes showing hemocytes moving towards the posterior along the anterior most section of the ventral nerve cord. Stills shown in Supplementary Information, Figure S3a.
Movie S7: Lateral view of hemocytes migrating along the ventral nerve cord in a rhoLXA12 embryo, hemocytes labeled. Lateral movie of a rhoLXA12 embryo expressing Moe-GFP in hemocytes; hemocytes move normally towards the posterior along the anterior most section of the ventral nerve cord. Stills shown in Supplementary Information, Figure S3b.
FIGURE S2: Identification of the gene, rhoL, and an allele rhoL10-161 that strongly reduces rhoL expression. (a) Schematic of genomic region. The white box represents rhoL mRNA and the black box within it corresponds to the ORF; transposon insert site indicated with triangle and transposon excision mutant indicated above. (b) A Clustal W alignment of all human and Drosophila Rho GTPases; RhoL falls in a branch with Rac1, Cdc42 and 5 other Human Rhos. A box surrounds Drosophila RhoL; arrows indicate other Drosophila Rhos. (c) In situ hybridizations with rhoL probe on a rhoL10-161 embryo. (d) RT-PCR after 27 cycles utilizing rhoL and rp49 loading control primers on RNA from wild type and rhoL10-161. These results show severely reduced expression in the rhoL10-161 mutant.
FIGURE S3: RhoL is not required for migration along the anterior vnc or to alter Cadherin levels. Mmp1 and 2 are not required for invasive hemocyte migration into the tail.
(a, b) Stills from 2-photon multiplane movies of (a) wild type and (b) rhoLXA12 embryos expressing Moesin-GFP in hemocytes under the control of the 8-163 GAL4 driver. Movies focus on the solid boxed area in the embryo schematic above, and thus show hemocytes migrating along the anterior ventral nerve cord. Blue arrow in schematic indicates direction of hemocyte movement. Arrows in (a, b) indicate a single hemocyte moving over time. The elapsed number of minutes is indicated. (c-e) Confocal images of fixed early st 12 embryos stained with anti-Cadherin (red) and (c, d) hemocyte-expressed anti-CD2 or (e) anti-GFP antibodies to visualize hemocytes (green). Hemocytes move normally into the tail in (c) mmp1 Q112 mmp2 W307 heterozygous and (d) homozygous embryos as well as (e) embryos expressing the protease inhibitors TIMP and RECK. (f) Wild type and (g) rhoLXA12 embryos expressing lacZ in hemocytes under the control of the 8-163GAL4 driver. Embryos were stained with anti-DE-Cadherin antibodies (red, single channel shown in f, g) and anti-β-Gal antibodies (green, single channel shown in f’, g’) to visualize the level of DE-Cadherin in hemocytes. Merge shown in (f”, g”). No strong Cadherin staining was seen in hemocytes in either genotype.
Figure S4: inflated and dizzy mutants display normal dorsal vessel migration.
Confocal images of the dorsal vessel of fixed (a) wild type, (b) dizzyΔ1 and (c) inflated embryos stained with antibodies against (a, c) β-Gal or (b) CD2 to visualize hemocytes (green, white) and Cadherin (red) to visualize the surrounding tissue. (d) Quantitation of the % of embryos with at least one hemocyte in the central four segments of the dorsal vessel in the genotypes indicated.
Movie S1: Lateral view of hemocytes migrating in a wild type embryo, all cell outlines labeled. Lateral movie of an embryo expressing Cadherin-GFP ubiquitously to mark cell outlines and Moe-GFP in hemocytes, showing movement of hemocytes into tail before germband retraction. Stills shown in Figure S1d.
Movie S2: Single plane dorsal view of hemocytes invading the tail in a wild type embryo, all cell outlines labeled. Single plane dorsal movie of an embryo as in Movie S1 showing hemocytes moving into the tail, along the junction of the hindgut. Stills shown in Figure 1d.
Movie S3: Multiplane dorsal movie of hemocytes invading the tail with chain migration in a wild type embryo, all cell outlines labeled. Multiplane dorsal movie of an embryo as in Movie S1 showing hemocytes engaging in chain migration as they move around the hindgut. Still shown in Figure 1e.
Movie S4: Lateral view of hemocytes migrating into the tail in a wild type embryo, hemocytes labeled. Lateral movie of the tail of a wild type embryo expressing Moe-GFP in hemocytes; hemocytes move into the tail. Stills shown in Figure 3c.
Movie S5: Lateral view of hemocytes failing to migrate into the tail in a rhoLXA12 embryo, hemocytes labeled. Lateral movie of the tail of a rhoLXA12 embryo expressing Moe-GFP in hemocytes showing hemocytes moving up to the junction of the tail and following it during germband retraction, but failing to enter. Stills shown in Figure 3d.
Acknowledgements
We thank D. Jukam, P. Kunwar, A-M. Sudarov, and S. Wang for help during the GAL4 enhancer screen, and D. Demy for her results in our lab that influenced our thinking. We thank all the people who generously shared their reagents: U. Heberlein, K. Brueckner, N. Perrimon, D. Montell, R. Reuter, and A. Page-McCaw for stocks and T. Uemura, D. Branton, R, Dubreuil, H. Bellen, N. Lowe, D. Ryoo, and J. Treisman for antibodies. We thank the Developmental Studies Hybridoma Bank developed with NICHD support and maintained by the University of Iowa for antibodies and the Bloomington Stock Center for flies. We are grateful for the intellectual inspiration provided by conversations with P. Rangan, P. Kunwar and many other members of the lab. We thank M. Dustin, S. Schwab and lab members for comments on the manuscript. The initial screen for invasion mutants by DS was supported by RO1HD041900 to RL. RL is an investigator of the HHMI.
Abbreviations
- vnc
ventral nerve cord
- PVR
PDGF/VEGF Receptor homolog
- MMP
Matrix-metalloprotease
- DA
Dominant Active
- β-Gal
β-Galactosidase
- GEF
GTPase exchange factor
- UAS
Upstream activation sequence
- st
Stage
Footnotes
Competing interests statement The authors declare no competing financial interests.
BIBLIOGRAPHY
- 1.Abram CL, Lowell CA. The ins and outs of leukocyte integrin signaling. Annual review of immunology. 2009;27:339–362. doi: 10.1146/annurev.immunol.021908.132554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etzioni A, Doerschuk CM, Harlan JM. Of man and mouse: leukocyte and endothelial adhesion molecule deficiencies. Blood. 1999;94:3281–3288. [PubMed] [Google Scholar]
- 3.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nature immunology. 2005;6:1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 4.Kim JV, Kang SS, Dustin ML, McGavern DB. Myelomonocytic cell recruitment causes fatal CNS vascular injury during acute viral meningitis. Nature. 2009;457:191–195. doi: 10.1038/nature07591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimonaka M, et al. Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. The Journal of cell biology. 2003;161:417–427. doi: 10.1083/jcb.200301133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nature immunology. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 7.Katagiri K, Imamura M, Kinashi T. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nature immunology. 2006;7:919–928. doi: 10.1038/ni1374. [DOI] [PubMed] [Google Scholar]
- 8.Evans CJ, Hartenstein V, Banerjee U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Developmental cell. 2003;5:673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- 9.Wood W, Jacinto A. Drosophila melanogaster embryonic haemocytes: masters of multitasking. Nat Rev Mol Cell Biol. 2007;8:542–551. doi: 10.1038/nrm2202. [DOI] [PubMed] [Google Scholar]
- 10.Babcock DT, et al. Circulating blood cells function as a surveillance system for damaged tissue in Drosophila larvae. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10017–10022. doi: 10.1073/pnas.0709951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stramer B, et al. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. The Journal of cell biology. 2005;168:567–573. doi: 10.1083/jcb.200405120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood W, Faria C, Jacinto A. Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. The Journal of cell biology. 2006;173:405–416. doi: 10.1083/jcb.200508161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastor-Pareja JC, Wu M, Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Disease models & mechanisms. 2008;1:144–154. doi: 10.1242/dmm.000950. discussion 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruckner K, et al. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Developmental cell. 2004;7:73–84. doi: 10.1016/j.devcel.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Cho NK, et al. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 2002;108:865–876. doi: 10.1016/s0092-8674(02)00676-1. [DOI] [PubMed] [Google Scholar]
- 16.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 17.Murphy AM, Montell DJ. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. The Journal of cell biology. 1996;133:617–630. doi: 10.1083/jcb.133.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paladi M, Tepass U. Function of Rho GTPases in embryonic blood cell migration in Drosophila. Journal of cell science. 2004;117:6313–6326. doi: 10.1242/jcs.01552. [DOI] [PubMed] [Google Scholar]
- 19.Page-McCaw A, Serano J, Sante JM, Rubin GM. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Developmental cell. 2003;4:95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, Xu T. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2721–2726. doi: 10.1073/pnas.0611666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tepass U, et al. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes & development. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- 22.Pacquelet A, Rorth P. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. The Journal of cell biology. 2005;170:803–812. doi: 10.1083/jcb.200506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huelsmann S, Hepper C, Marchese D, Knoll C, Reuter R. The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development (Cambridge, England) 2006;133:2915–2924. doi: 10.1242/dev.02449. [DOI] [PubMed] [Google Scholar]
- 24.Bogaert T, Brown N, Wilcox M. The Drosophila PS2 antigen is an invertebrate integrin that, like the fibronectin receptor, becomes localized to muscle attachments. Cell. 1987;51:929–940. doi: 10.1016/0092-8674(87)90580-0. [DOI] [PubMed] [Google Scholar]
- 25.Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development (Cambridge, England) 1994;120:1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- 26.de Velasco B, Mandal L, Mkrtchyan M, Hartenstein V. Subdivision and developmental fate of the head mesoderm in Drosophila melanogaster. Development genes and evolution. 2006;216:39–51. doi: 10.1007/s00427-005-0029-4. [DOI] [PubMed] [Google Scholar]
- 27.Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science (New York, N.Y. 2002;295:1285–1288. doi: 10.1126/science.1067549. [DOI] [PubMed] [Google Scholar]
- 28.Bertrand JY, et al. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menasche G, Kliche S, Bezman N, Schraven B. Regulation of T-cell antigen receptor-mediated inside-out signaling by cytosolic adapter proteins and Rap1 effector molecules. Immunological reviews. 2007;218:82–91. doi: 10.1111/j.1600-065X.2007.00543.x. [DOI] [PubMed] [Google Scholar]
- 30.Tepass U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Developmental biology. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- 31.Choi EY, Santoso S, Chavakis T. Mechanisms of neutrophil transendothelial migration. Front Biosci. 2009;14:1596–1605. doi: 10.2741/3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mor A, Dustin ML, Philips MR. Small GTPases and LFA-1 reciprocally modulate adhesion and signaling. Immunological reviews. 2007;218:114–125. doi: 10.1111/j.1600-065X.2007.00538.x. [DOI] [PubMed] [Google Scholar]
- 33.de Toledo M, et al. The GTP/GDP cycling of rho GTPase TCL is an essential regulator of the early endocytic pathway. Molecular biology of the cell. 2003;14:4846–4856. doi: 10.1091/mbc.E03-04-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawase K, et al. GTP hydrolysis by the Rho family GTPase TC10 promotes exocytic vesicle fusion. Developmental cell. 2006;11:411–421. doi: 10.1016/j.devcel.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Drosophila immune cells move into the tail during development by penetrating an epithelial barrier.
Schematic drawings show embryos from a lateral perspective with hemocytes (green) migrating into the tail during st 11-12. Hemocyte positions sketched from in situs of PVR. (a-c) Lateral confocal images of fixed wild type srpHemoGAL4 UAS-GFP embryos which express GFP in hemocytes; pictures correspond to the boxed area in the appropriate embryo schematic. Embryos are from successively later stages as indicated above and were stained with anti-Cadherin (red) and anti-GFP (green) antibodies as well as DAPI to visualize nuclei (blue). Hemocytes moving into the tail are indicated with white arrows; examining these locations in the ‘ panels which show only the Cadherin channel reveals that a breach in the Cadherin barrier appears as hemocytes move into the tail. Scale bars, 25 ⌈m.
(d) Stills from 2-photon movie of ubi:shg-GFP; srpHemoGAL4 UAS-GFP embryos. Shg-GFP outlines all cells and hemocyte-expressed GFP is seen in cytoplasm. (d) Lateral view starting st 11. Movie (Supplementary Information, Movie S1) corresponds to the dashed boxed area in the late st 11 schematic embryos above. Scale bars, (a-c) 25 ⌈m or (d) 20 ⌈m. Anterior to left and dorsal up in all panels.
Movie S6: Lateral view of hemocytes migrating along the ventral nerve cord in a wild type embryo, hemocytes labeled. Lateral movie of a wild type embryo expressing Moe-GFP in hemocytes showing hemocytes moving towards the posterior along the anterior most section of the ventral nerve cord. Stills shown in Supplementary Information, Figure S3a.
Movie S7: Lateral view of hemocytes migrating along the ventral nerve cord in a rhoLXA12 embryo, hemocytes labeled. Lateral movie of a rhoLXA12 embryo expressing Moe-GFP in hemocytes; hemocytes move normally towards the posterior along the anterior most section of the ventral nerve cord. Stills shown in Supplementary Information, Figure S3b.
FIGURE S2: Identification of the gene, rhoL, and an allele rhoL10-161 that strongly reduces rhoL expression. (a) Schematic of genomic region. The white box represents rhoL mRNA and the black box within it corresponds to the ORF; transposon insert site indicated with triangle and transposon excision mutant indicated above. (b) A Clustal W alignment of all human and Drosophila Rho GTPases; RhoL falls in a branch with Rac1, Cdc42 and 5 other Human Rhos. A box surrounds Drosophila RhoL; arrows indicate other Drosophila Rhos. (c) In situ hybridizations with rhoL probe on a rhoL10-161 embryo. (d) RT-PCR after 27 cycles utilizing rhoL and rp49 loading control primers on RNA from wild type and rhoL10-161. These results show severely reduced expression in the rhoL10-161 mutant.
FIGURE S3: RhoL is not required for migration along the anterior vnc or to alter Cadherin levels. Mmp1 and 2 are not required for invasive hemocyte migration into the tail.
(a, b) Stills from 2-photon multiplane movies of (a) wild type and (b) rhoLXA12 embryos expressing Moesin-GFP in hemocytes under the control of the 8-163 GAL4 driver. Movies focus on the solid boxed area in the embryo schematic above, and thus show hemocytes migrating along the anterior ventral nerve cord. Blue arrow in schematic indicates direction of hemocyte movement. Arrows in (a, b) indicate a single hemocyte moving over time. The elapsed number of minutes is indicated. (c-e) Confocal images of fixed early st 12 embryos stained with anti-Cadherin (red) and (c, d) hemocyte-expressed anti-CD2 or (e) anti-GFP antibodies to visualize hemocytes (green). Hemocytes move normally into the tail in (c) mmp1 Q112 mmp2 W307 heterozygous and (d) homozygous embryos as well as (e) embryos expressing the protease inhibitors TIMP and RECK. (f) Wild type and (g) rhoLXA12 embryos expressing lacZ in hemocytes under the control of the 8-163GAL4 driver. Embryos were stained with anti-DE-Cadherin antibodies (red, single channel shown in f, g) and anti-β-Gal antibodies (green, single channel shown in f’, g’) to visualize the level of DE-Cadherin in hemocytes. Merge shown in (f”, g”). No strong Cadherin staining was seen in hemocytes in either genotype.
Figure S4: inflated and dizzy mutants display normal dorsal vessel migration.
Confocal images of the dorsal vessel of fixed (a) wild type, (b) dizzyΔ1 and (c) inflated embryos stained with antibodies against (a, c) β-Gal or (b) CD2 to visualize hemocytes (green, white) and Cadherin (red) to visualize the surrounding tissue. (d) Quantitation of the % of embryos with at least one hemocyte in the central four segments of the dorsal vessel in the genotypes indicated.
Movie S1: Lateral view of hemocytes migrating in a wild type embryo, all cell outlines labeled. Lateral movie of an embryo expressing Cadherin-GFP ubiquitously to mark cell outlines and Moe-GFP in hemocytes, showing movement of hemocytes into tail before germband retraction. Stills shown in Figure S1d.
Movie S2: Single plane dorsal view of hemocytes invading the tail in a wild type embryo, all cell outlines labeled. Single plane dorsal movie of an embryo as in Movie S1 showing hemocytes moving into the tail, along the junction of the hindgut. Stills shown in Figure 1d.
Movie S3: Multiplane dorsal movie of hemocytes invading the tail with chain migration in a wild type embryo, all cell outlines labeled. Multiplane dorsal movie of an embryo as in Movie S1 showing hemocytes engaging in chain migration as they move around the hindgut. Still shown in Figure 1e.
Movie S4: Lateral view of hemocytes migrating into the tail in a wild type embryo, hemocytes labeled. Lateral movie of the tail of a wild type embryo expressing Moe-GFP in hemocytes; hemocytes move into the tail. Stills shown in Figure 3c.
Movie S5: Lateral view of hemocytes failing to migrate into the tail in a rhoLXA12 embryo, hemocytes labeled. Lateral movie of the tail of a rhoLXA12 embryo expressing Moe-GFP in hemocytes showing hemocytes moving up to the junction of the tail and following it during germband retraction, but failing to enter. Stills shown in Figure 3d.