Abstract
A 26Å resolution map of the structure of human LDL was obtained from cryo-EM and single particle image reconstruction. The structure showed a discoidal shaped LDL particle with high-density regions mainly distributed at the edge of the particle and low-density regions at the flat surface that covers the core region. To determine the chemical components that correspond to these density regions and to delineate the distribution of protein and phospholipid located at the particle surface at the resolution of the map, we used Mono-Sulfo-NHS-Undecagold labeling to increase preferentially the contrast of the apoB protein component on the LDL particle. In the 3D maps from the image reconstruction of the undecagold labeled LDL particles, the high-density region from the undecagold label was distributed mainly at the edge of the particle and lower density regions were found at the flat surfaces that cover the neutral lipid core. This suggests that apoB mainly encircles LDL at the edge of the particle and the phospholipid monolayers are located at the flat surfaces, which are parallel to the cholesterol ester layers in the core and may interact with the core lipid layers through the acyl-chains.
Keywords: LDL structure, apoB, atherosclerosis, 3D image reconstruction, undecagold labeling
Introduction
Low Density Lipoprotein (LDL) plays an important role in lipid homeostasis in the body and serves as the primary mechanism for cholesterol transport from the liver to the peripheral tissues1; 2. Structurally, LDL is a molecular assembly with the major components consisting of a single copy of apolipoprotein B (apoB), phospholipid, cholesterol, cholesterol ester (CE) and triglyceride. The apoB and phospholipids distribute at the surface of LDL to solubilize and stabilize the hydrophobic lipid core of the CE, triglyceride and a small amount of cholesterol3. The composition in the lipid core may vary, and thus LDL particles may have different sizes. ApoB and the phospholipids at the surface must undergo rearrangement to accommodate changes in surface area and surface pressure resulting from compositional changes4. Thus, the distribution of the apoB and phospholipids at the particle surface is an important characteristic of the LDL structure. Furthermore, abnormally high levels of LDL are distinctive risk factors for coronary atherosclerosis5; 6 and it has been shown that LDL oxidation is an important step for the interaction of LDL with the macrophage6. Structural changes, especially at the surface, are likely to occur that favor such interaction. Therefore, understanding the arrangement of the apoB and phospholipid on the LDL particle surface is important to understand further the functional properties of LDL in both physiological and pathophysiological conditions.
Currently, the most informative evidence regarding the surface structure of LDL is from the mass distribution determined from cryo-EM and single particle reconstruction experiments7; 8. Recent studies have assigned the higher density regions at the particle surface to the protein content and lower density regions to the phospholipid content. A model of the apoB distribution has been proposed based on the contour of the high density regions in the 3D map 8. However, x-ray and neutron diffraction scattering studies have shown that at low resolution the density of a phospholipid phosphorylcholine head group is similar to that of protein9; 10. Furthermore the density of the phospholipid monolayer as a whole is unclear since it depends on the phospholipid packing, which is determined by the LDL surface pressure for which the values are not known4. Thus, at current resolution of the cryo-EM reconstruction of LDL, a definitive assignment of the protein to the highest density region may not be made. To delineate the location of the surface components more clearly, we labeled LDL with Mono-Sulfo-NHS-Undecagold to enhance preferentially the density of the region corresponding to apoB. The 3D map of undecagold labeled LDL showed high-density regions mainly at the edge of the particle. Thus, the reconstructions substantiate that the apoB protein, which corresponds to the high density, mainly resides at the edge of the discoidal shaped LDL particle, and the phospholipid monolayers that correspond to lower density regions reside at the flat surfaces. The structural implications of these surface components arrangements are discussed.
Results
Electron microscopy imaging and structure reconstruction of unlabeled LDL
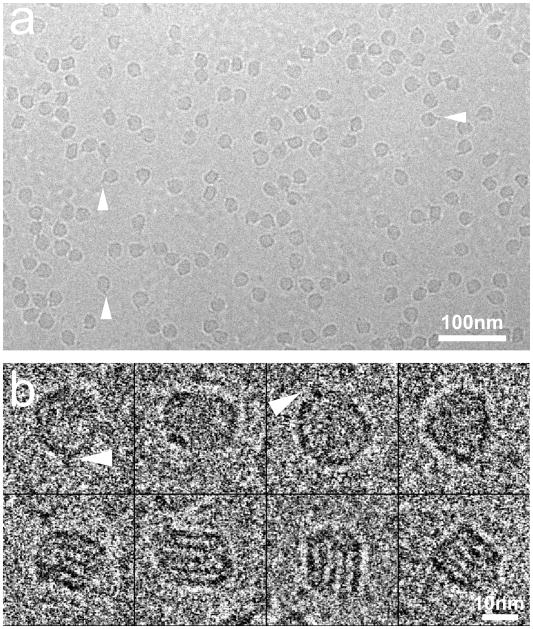
Samples were preserved in a stain free and hydrated state. The electron micrograph in Figure 1a shows a typical cryo-EM field of LDL particles that appear darker than the background. The LDL particles have random orientations as the projection images showed different shapes. Some particles show a pronounced “pointed feature”, which is consistent with the previous observations11 (triangles in Figure 1a, b). Representative individual particles are shown in Figure 1b. In some images, striped density features inside the particle were visible as described previously by us and others7; 8; 11. Such features were most clear at a defocus value ~ −1.8μm and only exist in the more “rectangular” shape projection views (Figure 1b).
Figure 1. Cryo-EM images of LDL.
a. Electron micrograph of LDL in vitreous ice, electron dense object appears as dark region on the micrograph. b. Typical LDL projection views in the electron micrograph. Triangles indicate the individual particles with the “pointed feature”.
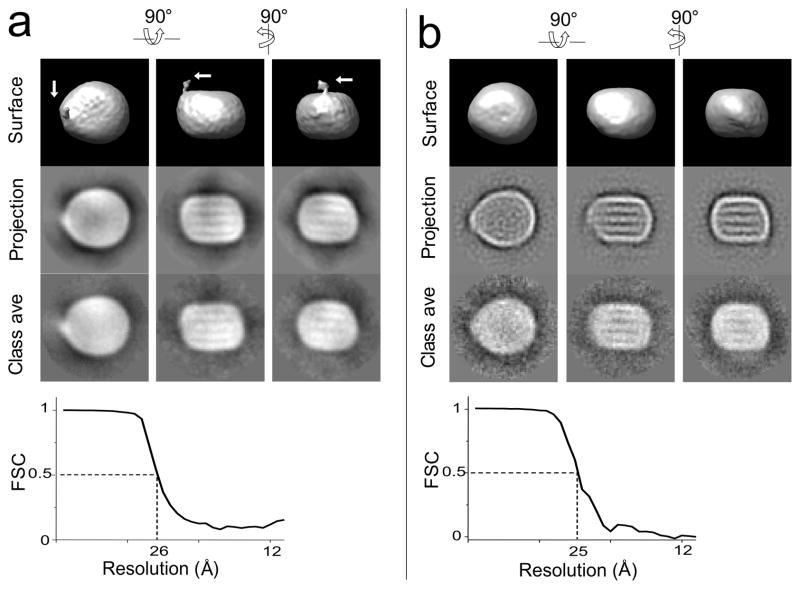
A ~31,000 particle dataset was used for the 3D structure reconstruction. The reconstructed volume showed an asymmetric discoidal shaped particle with the dimensions of approximately 240×210×165Å. The particle has a tip end along its long axis. A low-density region protruding from this tip is also observed (arrows in Figure 2a). The tip end and the protrusion of LDL particle correspond to the “pointed feature” seen in the 2D projection images in Figure 1 and in our previous studies11. The reconstruction agreed well with the data as the reconstructed surface, the projections of the structure and the corresponding class averages were clearly similar. The structure had a resolution of 26Å determined by the eotest program in the EMAN package (Fourier shell correlation = 0.5)12.
Figure 2. Refinement convergence of LDL and undecagold labeled LDL.
Comparison of the iso-surface structure volume with the projection map and the class averages. The first, second, and third columns are different views of the same structure rotated 90. The upper three images are the iso-surface display of the reconstructed LDL volume. The middle three images are the projection images corresponding to the same orientation as the iso-surface images; the lower three images are the average image from the class that match the projection view. Contrast is inverted from the micrograph as electron dense object appears as white region in panel Fourier Shell Correlation (FSC) curve calculated from structures reconstructed from odd and even numbered particle images. a. Reconstruction for LDL. The arrows indicate the position of the protrusion. b. Reconstruction for undecagold labeled LDL sample.
Undecagold labeling of LDL
The phospholipid composition of the LDL sample was quantified and the numbers of total phospholipid and the phosphatidyl-ethanolamine (PE) per LDL particle were calculated. The number of total phospholipid molecules (610) was in the range of a typical LDL subspecies4. The PE represented ~2% of total phospholipid that corresponds to ~12 molecules per particle, consistent with published results13. Mono-Sulfo-NHS-Undecagold was used to label the LDL.
An EYPC+CO emulsion control sample (no apoB) was prepared and incubated with Mono-Sulfo-NHS-Undecagold under identical conditions to the LDL sample. After dialysis, gold was only detectable in the ~5K MW eluate fraction of a size exclusion column, demonstrating that non-specific interaction with the emulsion was negligible. The amount of the unbound gold left in the control sample was ~ 8% of the original amount. The same amount of unbound gold was expected in the LDL labeling experiment and was subtracted during the quantitation of the LDL gold labeling
Based on the quantitation of the gold and the protein, the number of gold labels per particle was calculated to be ~175. Thus, if all the PE molecules had been labeled, the minimal number of gold labels bound to apoB was ~160. This number is within the maximum number of 225 lysine residues available for methylation on the surface of LDL14. Thus the number of gold labels on protein was 13 fold (160/12) higher than the number that labeled the PE, and the protein content of LDL was preferentially labeled. Furthermore, any unbound gold should distribute in the much larger background area of the image and be averaged out during the image reconstruction process. A ~31,000 particle dataset was used for image reconstruction, and a 25Å resolution map was obtained for the labeled LDL particle (Figure 2b).
The surface density of LDL
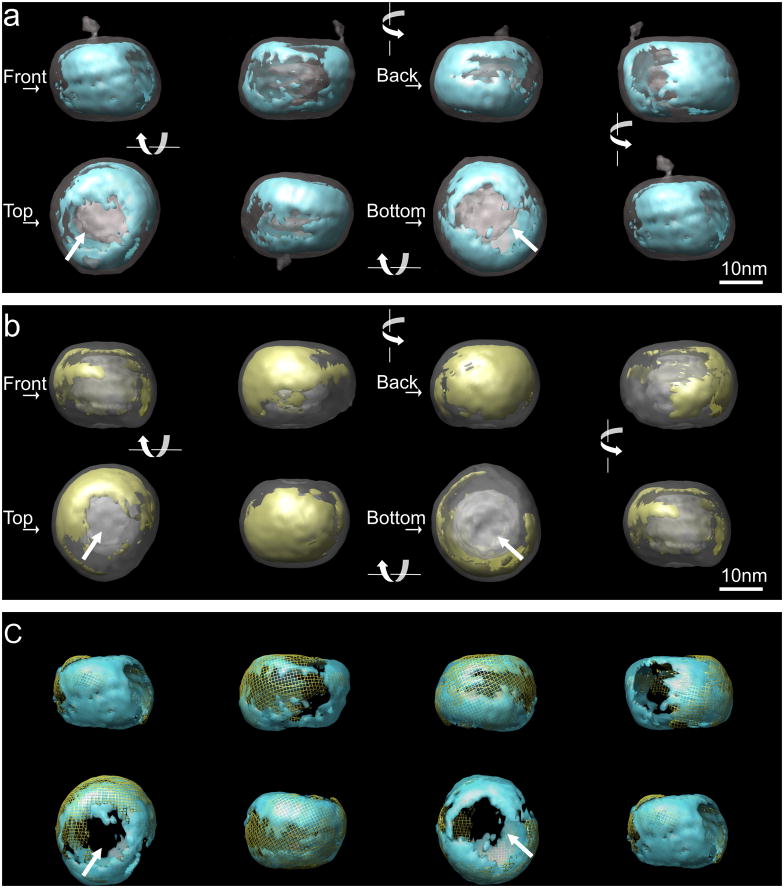
As shown in Figure 3a, in unlabeled LDL, a major structural feature of the LDL particle was the density distribution at the surface. There is a continuous high density region (thresholded to contain a volume of 6.2×105 Å3, corresponding to the volume calculated for apoB4) that mainly covers the edge of the particle. A lower density region is located on both of the two flat surfaces, which can be seen clearly in the “top” and “bottom” views. The reconstruction from undecagold labeled LDL showed similar features in terms of the overall shape, and the layered density in the core. Following alignment of the two reconstructed volumes and normalization of the density histograms (see Methods), the reconstructed volume was thresholded to include only the high density contributed from the gold labeling. This density was distributed mainly on the edge of the undecagold-LDL particle surface (Figure 3b) and largely overlaps with the high-density regions in unlabeled LDL (Figure 3c). Notably, the flat surfaces on the top and bottom remained as low density and did not contain contributions from the gold label.
Figure 3. Density distribution at the surface of LDL and undecagold labeled LDL.
Two volumes were aligned with EMAN program. For the underlying transparent grey structure, the threshold in the voxel density histogram was chosen to display the overall shape (~0.8σ and ~1.4σ for unlabeled and labeled LDL respectively). The overlaid structure was turned 90 in each frame and displayed from left → right in the first row and right → left in the second row. The orientations of the structure are arbitrary defined and indicated as “front”, “back”, “top” and “bottom”. In the “top” and “bottom” views, the arrows indicate the low density regions at the flat surfaces. a. Iso-surface display of LDL structure (transparent gray) overlaid with the higher density regions. The threshold contour level (~4σ) was chosen to include a volume corresponding to the volume of apoB (blue). b. The iso-surface display for undecagold labeled LDL structure volume overlaid with the higher density regions (threshold ~4.6σ) from gold contribution determined as described in Methods (yellow). c. Overlay of the high density regions in the structures of the two volumes shown in a and b. The high density regions from the unlabeled LDL are in transparent blue and the high density regions from undecagold labeled LDL are in yellow mesh.
Organization of the cholesterol ester core
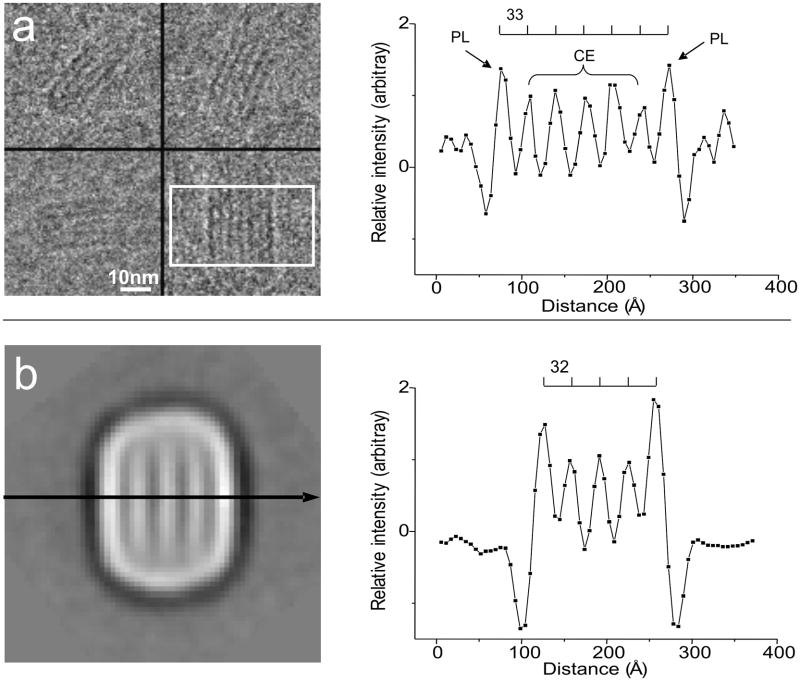
The projection views of LDL particles exhibiting striped features seen by us and others are thought to represent the organization of the CE core of the particle. To verify further this conclusion we compared LDL images to those of emulsion particles. Based on the hydrophobicity characteristics of the lipids, the EYPC+CO particles have a CO layered core and a monolayer of EYPC at the surface. Figure 4a shows the striped images of EYPC+CO emulsion particles. The EYPC monolayer at the surface exhibited higher density than the CO layers and the distance between the striped layers was ~33Å. The density profile in the direction perpendicular to the stripes from a class average image of unlabeled LDL (Figure 4b), was very similar to the density profile for the EYPC+CO emulsion particles in terms of the distance between the layers and the density of the two outer most layers compared to the CE layers (Figure 4a).
Figure 4. Comparing EYPC+CO emulsion and LDL particle striped images.
a. Selected views of the EYPC+CO emulsion in cryo-EM micrograph. Within the rectangular area, the pixel value in each column are summed and plotted with the distance between the columns. The electron dense object appears as dark region. The chemical compositions of the peaks are indicated. b. A class average image with the stripes. Contrast is inverted from the micrograph as electron dense object appears as white region. The pixel value is read along the arrow, the value of the pixels and the distance is plotted on the right. In both plots, the intensity is normalized (see methods section) and the distance between the peaks is indicated above the curve.
Discussion
LDL is a molecular assembly of the 4536 residue apoB protein and more than 2,000 lipid molecules13. As the lipid content varies, LDL has a dynamic composition and exhibits different subspecies4. Thus, LDL is a challenging subject for high-resolution structural studies using NMR or X-Ray crystallography. In this study, we separated LDL in an extremely narrow buoyant density range (1.035<d<1.04 g/ml) and examined the 3D structure by cryo-EM imaging and single particle image reconstruction. With this sample preparation, there was no large variation in LDL particle size (Figure 1) and the complete particle dataset was used for refinement rather than a sub-set of the data as in other studies8. Since protein was most concentrated in this buoyant density range, the LDL from this sample preparation represents the most typical LDL sub population. The reconstructed 3D volume showed the LDL to be a discoidal-shape particle with two flat surfaces on opposite sides and a layered structure in the core, consistent with previous electron microscopy studies by us and others7; 8; 11; 15; 16. A reliable feature of the reconstructed structure was the distribution pattern of the surface density. By thresholding the voxel density histogram, a continuous high-density region that roughly wraps around the discoidal shaped particle at the edge was clearly observed. (Figure 3a, Figure 5). Furthermore, this pattern of density distribution was reproducible in independent image reconstructions of LDL from different blood donors, and thus reflected intrinsic structural features of the LDL particle. The high-density distribution at the surface is similar but not identical to the structure recently described by Ren et. al.8 which may reflect differences in sample preparation. Interestingly, our reconstructed volume shows that the LDL particle has a “pointed feature” at one side of the long axis. This “pointed feature” contains a sharp curvature tip on the bulk of the particle and a lower density-value structure extruding from this tip towards the upper side of the LDL particle (Figure 2a). The low density of this protrusion may suggest a flexible nature, but the feature has been observed reproducibly in several independent image reconstructions of LDL from different donors. This “pointed feature” is consistent with the individual LDL particle 2D projection images (Figure 1) and our previous observations11, and thus reflects an intrinsic feature of LDL particle. The reconstruction of the LDL-LDL receptor complex by Ren et. al.8 showed a density feature similar to this “pointed feature” that was thought to represent the bound receptor domain.
Figure 5. Proposed lipid packing model in LDL.
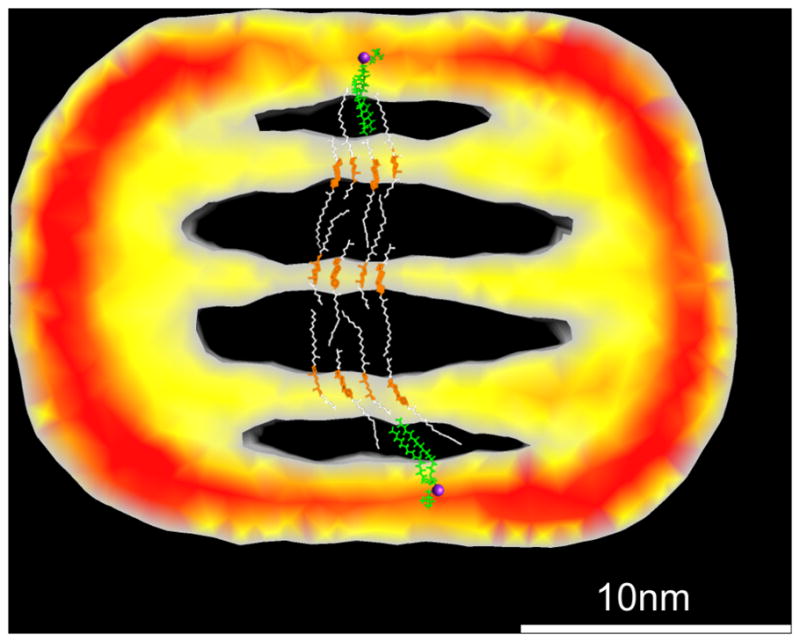
Cholesteryl palmitate and dipalmitoyl-phosphatidylcholine (green) were used to build the model. The sterol rings are colored in orange and the acyl chains from cholesteryl palmitate are colored in white. The phosphorus atoms are colored in purple. A slice of the unlabeled LDL structure with the inside alternating density feature is shown in the background at the same scale. The color indicates the intensity values within the slice as Red > Yellow > Grey.
The solvent exposed surface of LDL, based on current understanding, is composed of apoB, and a monolayer of phospholipids. ApoB has amphipathic α-helix and β-sheet structures3, and the phospholipids are orientated with the polar head group exposed to the aqueous surroundings17. Since the LDL particle surface is covered by either the protein or the monolayer of phospholipids, the density distribution at the surface should reflect the distribution of the surface components. However, it is not entirely clear as to whether the protein corresponds solely to the highest density region and can be clearly differentiated from the phospholipid head group containing region, since at the resolution of the reconstruction, the density of the phospholipid head groups may be close to that of protein. Hydration of the phospholipid head group and protein 18 and the packing of the phospholipid monolayer19 may further complicate a density comparison between the two components on the basis of theoretical calculations. To delineate clearly the protein-containing regions, we used Mono-Sulfo-NHS-Undecagold to label LDL. This gold derivative specifically labels primary amine groups. Thus, at the surface of LDL the available lysine residues of the protein and the phosphatidyl ethanolamine groups of the phospholipid are both labeled. However, based on our quantification, the number of gold labels on apoB was ~13 fold that of the PE molecules per LDL particle. Also, the PE molecules are more mobile than the protein and more likely to be averaged out in the reconstruction. Thus, this labeling agent should preferentially enhance the density of the lysine residues to distinguish clearly the regions corresponding to the protein. An image reconstruction of the undecagold-LDL particle was obtained with a 25Å resolution and showed similar overall shape to the unlabeled LDL (Figure 2b, Figure 3b). The low-density protrusion was not seen in this reconstruction probably because of its flexible nature and the addition of undecagold particles, which makes the protrusion less distinguishable from the background. An individual undecagold cluster has a diameter of 8Å, and, at 25Å resolution, individual gold clusters cannot be visualized nor can their positions be determined. However, the addition of ~175 gold clusters on LDL surface is expected to enhance the density at the particle surface and the contrast of the particle in the images. Indeed, the surface of undecagold labeled LDL has higher contrast than the unlabeled LDL as seen in the class averages and the projection images from the two reconstructions (Figure 2b), demonstrating that the labeling was reflected in the 3D reconstructed volume. Selecting a threshold level to include only the additional density from the gold labeling demonstrated that this additional density is found at the edge of the particle not at the flat surfaces that cover the core lipid layers on the top and the bottom of the particle (Figure 3b). Thus the distribution of density attributable to the gold label corresponds to features seen in unlabeled LDL although the high density distributes slightly differently on the edge of the LDL particle (Figure 3c). Since the undecagold labeling preferentially enhanced the contrast of apoB, the highest density regions at the edge of the discoidal shaped particle surface correspond to the distribution of the apoB protein and the regions at the flat surfaces of LDL particle may correspond to the phospholipid monolayers. This lipid organization is further supported by the imaging of the lipid emulsion particles.
As shown in Figure 4, EYPC + CO emulsion particles that have a CO core solublized by a monolayer of EYPC exhibited similar stripes in the projection images. When the averaged intensities of the stripes are plotted along the line perpendicular to the stripes, the monolayer of EYPC at the surface shows a higher value than the CO layers, and the distance between the stripes is ~33Å. The class average view of LDL particles with the striped feature shows a very similar profile to the emulsion particle in terms of the distance between the stripes and the intensity values of the two outer most stripes compared to the CE. This suggests that the LDL particle and the emulsion particles have the similar molecular arrangement producing the striped features.
As illustrated in Figure 5, the position of the phospholipid monolayer suggests an alternative lipid packing model to that recently proposed by Ren et al8. In this model the major surface component that covers the acyl-chains of the cholesterol ester layers is the phospholipid monolayer as opposed to the β-sheet domain from the apoB. Thus, in this model, the acyl-chain moieties of the phospholipids and CE are largely disordered, and the sterol ring moieties and the phospholipid polar regions are well ordered and aligned. When the angle of the electron beam is parallel with the planes, the pixel value profile of these three regions is phospholipid polar region > sterol ring > acyl-chain as seen in Figure 4b. The acyl-chain interactions between lipid layers is important to stabilize the structure, as evidenced from the EYPC+CO emulsion particles where the layered lipid packing can exist without the presence of the apoB protein(Figure 4a) and the distance between the adjacent layers may be maintained by the overlapping acyl-chain moieties. The reconstruction of undecagold labeled LDL also revealed that the location of gold labeling was mainly at the high-density regions of unlabeled LDL. Thus, the high-density regions in the unlabeled LDL structure largely correspond to the apoB protein as opposed to phospholipid head groups. Our reconstruction of Mono-Sulfo-NHS-Undecagold labeled LDL is a unique way to circumvent the difficulties in clear assignment of molecular components and provide a more reliable structural interpretation at the current resolution limit.
Materials and methods
LDL preparation and characterization
Human plasma was prepared from blood of healthy non-fasting donors and treated with EDTA. LDL was separated from the plasma by ultracentrifugation floatation with the buoyant density range of 1.02–1.063 g/ml 20. This broad buoyant density range LDL was further fractionated with two rounds of KBr density gradient ultracentrifugation to provide a highly homogeneous LDL particle distribution. In brief, LDL separated over the broad buoyant density range was transferred into five SW55 centrifuge tubes. In each tube, 1ml LDL, 1ml 1.040g/ml, 1ml 1.030g/ml, 1.5ml 1.020g/ml KBr solutions were carefully overlaid sequentially. One blank tube was overlaid in the same way, except that the LDL was replaced by 1.050g/ml KBr. The tubes were centrifuged using a BECKMAN Optima L-90 K Ultracentrifuge at 50k rpm, 20°C with “no brake” setting. After 20 hours, the density gradients of KBr solution reached equilibrium. A dense color band observed by eye was found at almost the same height in all tubes. The contents of each tube were aliquoted to take 200μl LDL from the dense color region at the same height in each tube. The same aliquots were taken from the blank tube for density determination by refractive index. The fractions from the most color dense region were mixed and adjusted to 1.05g/ml by adding solid KBr. The sample was then transferred to a new centrifuge tube, and overlaid with 3ml 1.03g/ml KBr solution for a second round of density gradient centrifugation to fractionate the LDL further. At the end of the second run, 200μl LDL from the middle of the most color dense region was taken and dialyzed overnight against 10mM phosphate, 150mM sodium chloride, pH 7.4 buffer. This provided the final fractionated sample with the buoyant density between 1.035—1.04g/ml and was used for biochemical characterization and frozen for cryo-electron microscopy with or without undecagold labeling. Protein concentrations for all LDL samples were measured with a Markwell adapted Lowry procedure21. Phospholipid components were extracted from LDL by Folch extraction with CHCl4/MeOH, followed by thin layer chromatographic separation. The concentration of total phospholipid and PE were measured with the Bartlett assay 22.
Undecagold labeling
Mono-Sulfo-NHS-Undecagold was purchased from Nanoprobes Inc, Yanphank, NY, USA. The labeling with undecagold was done by following the manufacture’s protocol 23. Fifty nmol Mono-Sulfo-NHS-Undecagold and 0.08nmol LDL (by apoB) was reacted in 20mMol HEPEs buffer (pH7.5, apoB=0.15mg/ml). The reaction solution was placed at ambient temperature for 3hrs and at 4 C overnight. The reaction solution was dialysized against 10mM phosphate and 150mM sodium chloride buffer pH7.4. An EYPC+CO emulsion solution with similar amount of CO was used as a labeling and dialysis control. After dialysis, the undecagold concentration (C) in the LDL and emulsion control were determined by measuring the absorbance at UV420nm as described in the vendor’s instructions24. The amount of gold label on LDL was obtained by subtracting the C(control) from C(LDL) concentrations and normalized to the protein concentration to derive the number of gold per LDL particle.
EYPC+CO emulsions
The emulsion preparation was carried out with similar procedures to those described previously25. Five mg/ml EYPC and 5mg/ml CO in PBS buffer was intensively sonicated for 5hrs, under a N2 atmosphere and the solution was kept above the CO phase transition temperature, 52 C, at all times. After the sonication, the solution was briefly centrifuged and the top milky color region was separated as the emulsion solution.
Cryo-Electron microscopy and image processing
The cryo-preservation was performed on Quantifoil® Cu-400 mesh holey carbon grids with the Vitrobot® plunging system. The incubation conditions were set to 22 C, 100% humidity. First, the grid was air glow discharged. 4μl sample solution was applied to the grid surface. The grid was blotted and plunged into the liquid ethane. The frozen grids were transferred into a storage box and kept in liquid N2 until cryo-imaging26. Images were collected at 120Kv on a Tecnai TF20 electron microscope with a Gatan or Oxford cryo holder using low dose conditions. The defocus ranged from −0.8 μm to −4.5 μm. For the images shown in Figure 1, the data was collected at 50,000X magnification and recorded on KODAK SO163 film (calibrated with vermiculite). The films were digitized with a Creo EVERSMART® scanner and binned by a factor of 3×3. After binning, the image file was at 2.73 Å/pix. For the large particle number data sets for image reconstructions (LDL and undecagold labeled LDL) and the EYPC+CO emulsion data sets, the data were collected at 29,000X magnification and recorded on a 4K TVIPS® CCD camera with an image sampling of 2.9Å/pix (calibrated with Tobacco Mosaic Virus).
For image reconstruction, particle images were picked with e2boxer from the EMAN2 program27, and contrast transfer function (CTF) correction was performed with ctfit from the EMAN program package. The CTF corrected images were further 2X binned to give 5.8Å/pix for refinement with the EMAN image processing package28. The resolutions of the reconstructed structures were determined by the eotest program from EMAN. The 3D reconstruction image results were displayed with the EMAN package. A low pass filter to the resolution of the reconstruction was applied to the volume before the 3D iso-surface display. Lipid model building was achieved and displayed with the Chimera program29.
Pixel value determination
The values were read with the SPIDER LI command30. For EYPC+CO emulsion particles, the pixel value in each column within the rectangular area (Figure 4a) was summed and plotted against the distance between the columns. For the selected 2D class average image, only the pixels along the indicated line was plotted (Figure 4b). In both plots, the inside peaks (except the two outside peaks) were considered as cholesterol ester layer peaks. The pixel values in the plots were normalized so that the average of the minimum value for the cholesterol peaks had a value of 0, and the average of the maximum for the cholesterol ester peaks had a value of 1.
Structure volume histogram normalization
The two structure volumes were normalized so that the minimum density had a value of 0 and the mean density had a value of 1. The extra density range in undecagold labeled structure histogram was considered to be the contribution from the gold labeling.
Acknowledgments
We thank Cheryl England and Michael Gigliotti for their help in preparing LDL sample; We thank Dr. Jean-François Ménétret for the help during the experiments and the discussion about this manuscript.
This work was supported by grant NIH/NHLBI P0126335
Abbreviations
- LDL
low density lipoprotein
- EM
electron microscopy
- EYPC
egg yolk phosphatidylcholine
- CE
cholesteryl ester
- CO
cholesteryl oleate
- PE
phosphatidyl-ethanolamine
- EDTA
ethylenediaminetetraacetic acid
- PBS
phosphate buffered saline
Footnotes
Accession numbers
The 3D density maps of LDL and undecagold labeled LDL are accessible from EMDB entry IDs of EMD-5239 and EMD-5241 respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldstein JL, Brown MS. The LDL pathway in human fibroblasts: a receptor-mediated mechanism for the regulation of cholesterol metabolism. Curr Top Cell Regul. 1976;11:147–81. doi: 10.1016/b978-0-12-152811-9.50011-0. [DOI] [PubMed] [Google Scholar]
- 2.Mahley RW, Innerarity TL, Rall SC, Jr, Weisgraber KH. Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res. 1984;25:1277–94. [PubMed] [Google Scholar]
- 3.Segrest JP, Jones MK, De Loof H, Dashti N. Structure of apolipoprotein B-100 in low density lipoproteins. J Lipid Res. 2001;42:1346–67. [PubMed] [Google Scholar]
- 4.McNamara JR, Small DM, Li Z, Schaefer EJ. Differences in LDL subspecies involve alterations in lipid composition and conformational changes in apolipoprotein B. J Lipid Res. 1996;37:1924–35. [PubMed] [Google Scholar]
- 5.Goldstein JL, HHH, Brown MS. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, ALB, Sly WS, Valle D, editors. McGraw-Hill; New York: 2001. pp. 2863–2913. [Google Scholar]
- 6.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–41. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orlova EV, Sherman MB, Chiu W, Mowri H, Smith LC, Gotto AM., Jr Three-dimensional structure of low density lipoproteins by electron cryomicroscopy. Proc Natl Acad Sci U S A. 1999;96:8420–5. doi: 10.1073/pnas.96.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren G, Rudenko G, Ludtke SJ, Deisenhofer J, Chiu W, Pownall HJ. Model of human low-density lipoprotein and bound receptor based on CryoEM. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0908004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson D, Shipley GG. Structural studies of plasma lipoproteins. In: Schoenborn BP, editor. Neutrons in biology. Plenum press; New York: 1984. pp. 211–226. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson D, Smith HM, Dickson J, Austin JP. Interaction of apoprotein from porcine high-density lipoprotein with dimyristoyl lecithin. 1. The structure of the complexes. Eur J Biochem. 1976;64:541–7. doi: 10.1111/j.1432-1033.1976.tb10334.x. [DOI] [PubMed] [Google Scholar]
- 11.Spin JM, Atkinson D. Cryoelectron microscopy of low density lipoprotein in vitreous ice. Biophys J. 1995;68:2115–23. doi: 10.1016/S0006-3495(95)80392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy. Structure. 2004;12:1129–36. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Hevonoja T, Pentikainen MO, Hyvonen MT, Kovanen PT, Ala-Korpela M. Structure of low density lipoprotein (LDL) particles: basis for understanding molecular changes in modified LDL. Biochim Biophys Acta. 2000;1488:189–210. doi: 10.1016/s1388-1981(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 14.Lund-Katz S, Ibdah JA, Letizia JY, Thomas MT, Phillips MC. A 13C NMR characterization of lysine residues in apolipoprotein B and their role in binding to the low density lipoprotein receptor. J Biol Chem. 1988;263:13831–8. [PubMed] [Google Scholar]
- 15.Coronado-Gray A, van Antwerpen R. Lipid composition influences the shape of human low density lipoprotein in vitreous ice. Lipids. 2005;40:495–500. doi: 10.1007/s11745-005-1409-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman MB, Orlova EV, Decker GL, Chiu W, Pownall HJ. Structure of triglyceride-rich human low-density lipoproteins according to cryoelectron microscopy. Biochemistry. 2003;42:14988–93. doi: 10.1021/bi0354738. [DOI] [PubMed] [Google Scholar]
- 17.Laggner P, Kostner GM, Rakusch U, Worcester D. Neutron small angle scattering on selectively deuterated human plasma low density lipoproteins. The location of polar phospholipid headgroups. J Biol Chem. 1981;256:11832–9. [PubMed] [Google Scholar]
- 18.Atkinson D, Davis MA, Leslie RB. The structure of a high density lipoprotein (HDL3) from porcine plasma. Proc R Soc Lond B Biol Sci. 1974;186:165–80. doi: 10.1098/rspb.1974.0044. [DOI] [PubMed] [Google Scholar]
- 19.Ibdah JA, Lund-Katz S, Phillips MC. Molecular packing of high-density and low-density lipoprotein surface lipids and apolipoprotein A-I binding. Biochemistry. 1989;28:1126–33. doi: 10.1021/bi00429a029. [DOI] [PubMed] [Google Scholar]
- 20.Schumaker VN, Puppione DL. Sequential flotation ultracentrifugation. Methods Enzymol. 1986;128:155–70. doi: 10.1016/0076-6879(86)28066-0. [DOI] [PubMed] [Google Scholar]
- 21.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–10. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett GR. Phosphorus assay in column chromatography. J Biol Chem. 1959;234:466–8. [PubMed] [Google Scholar]
- 23.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci U S A. 1997;94:9556–61. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hainfeld JF, Furuya FR. A 1.4-nm gold cluster covalently attached to antibodies improves immunolabeling. J Histochem Cytochem. 1992;40:177–84. doi: 10.1177/40.2.1552162. [DOI] [PubMed] [Google Scholar]
- 25.Ginsburg GS, Small DM, Atkinson D. Microemulsions of phospholipids and cholesterol esters. Protein-free models of low density lipoprotein. J Biol Chem. 1982;257:8216–27. [PubMed] [Google Scholar]
- 26.Menetret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6:1219–32. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 27.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 29.Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–82. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]