Abstract
Here we describe the cloning of a sequenced WUMS isolate of murine gammaherpesvirus-68 (MHV-68, γHV-68, also known as MuHV-4) as a bacterial artificial chromosome (BAC). We engineered the insertion of the BAC sequence flanked by loxP sites into the left end of the viral genome before the M1 open reading frame. The infectious viruses were reconstituted following transfection of the MHV-68 BAC DNA into cells. The MHV-68 BAC-derived virus replicated indistinguishably from the wild-type virus in cultured cells. Excision of the BAC insert was efficiently achieved by coexpressing the Cre recombinase. Although the BAC insertion did not significantly affect acute productive infection in the lung, it severely compromised the ability of MHV-68 to establish splenic latency. Removal of the BAC sequence restored the wild-type level of latency. Site-specific mutagenesis was carried out by RecA-mediated recombination to demonstrate that this infectious BAC clone can be used for genetic studies of MHV-68.
1. Introduction
Two human gamma-herpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV or HHV-8), and Epstein-Barr virus (EBV), are implicated in multiple malignancies. Like other herpesviruses, they establish life-long persistent infections, interchanging between lytic replication and latency, while being subjected to the control of the host immune system. An enormous amount of knowledge has been built on the molecular biology of KSHV and EBV. However, due to the lack of a small animal model, the studies on the in vivo interplay between the virus and the host during persistent infections have been limited.
Murine gammaherpesvirus 68 (MHV-68, γHV-68, also known as MuHV-4), originally isolated from wild rodents, was found to be closely related to KSHV/HHV-8 and EBV [1–4]. Following intranasal inoculation into laboratory mice, MHV-68 first replicates in lung epithelial cells to produce infectious viruses and then establishes latency in B cells [5, 6]. Remarkably, massive lymphoproliferation that occurs at about two weeks after infection resembles infectious mononucleosis seen in some individuals experiencing primary EBV infection [7, 8]. Persistent infections of MHV-68 also lead to B-cell lymphomas in a small fraction of mice and the tumor incidence increases in immuno-deficient mice [9]. The genome of MHV-68 was completely sequenced [1] and the majority of the identified open reading frames (ORFs) share homology to those of KSHV and EBV. However, unlike human gamma-herpesviruses, MHV-68 readily infects a variety of cell lines and undergoes lytic replication. Therefore, MHV-68 represents a valuable model to study not only the virus-host interactions but also viral lytic replication of gamma-herpesviruses.
In virology, genetic mutagenesis is the primary approach to examine the role of a viral gene. Mutations are introduced into a specific gene on the viral genome and the phenotype of the resultant mutant virus is studied. Herpesviruses have rather large DNA genomes up to 250 kb and thus the mutant virus is generated by homologous recombination between the viral genome and the DNA fragment carrying the desired genetic alterations. This method is time consuming because the mutant virus needs to be separated from the parental virus by multiple rounds of plaque purifications or limiting dilutions. It would be even more cumbersome to generate the mutants that are defective in viral replication. A breakthrough in genetic mutagenesis of herpesviruses came from the method that was established to clone the entire herpesviral genome into a bacterial artificial chromosome (BAC) [10–16]. BAC, a vector derived from F factor of E. coli., has the capacity to accommodate a large insert (up to 300 Kb) and stably maintain it in bacterial hosts. Once the viral genome is cloned and propagated as a BAC plasmid in E. coli, it can be efficiently modified using the well-developed bacterial genetic methods. The cloning method exploits the fact that the linear herpesviral DNA within an infectious viral particle becomes circularized once entering a cell. For cloning of the herpesviral genome, the first step is to construct a recombinant virus that contains the insertion of the BAC sequence at a tolerable site, and then this BAC-containing virus is used to infect permissive cells. Next, the circularized BAC-containing viral DNA is harvested from the infected cells and used to transform E. coli to obtain colonies, each of which harbors a viral BAC. Two major recombination systems have been used for site-specific mutagenesis of viral BACs. One is RecA-mediated allelic exchange [17], relying on the shuttle plasmid to transfer the mutation flanking by at least 350-bp homologous sequences into the viral BAC plasmid. The other is carried out by recombinases E and T between linear DNA fragment and the BAC plasmid, which only requires a 50-bp homologous region on either site of the mutation [18]. However, ET-recombination might suffer from a higher risk of illegitimate recombination.
In order to efficiently and systematically study the functions of individual viral genes encoded by MHV-68, we decided to clone the sequenced WUMS isolate [1] as a BAC plasmid. After obtaining the MHV-68 BAC plasmid, we conducted transposon-mediated random insertional mutagenesis to systematically identify the viral ORFs essential for viral replication [19] as well as to functionally profile a genetic locus in great details [20]. Due to the extensive use of our MHV-68 BAC plasmid in the field, we describe here the construction and characterization of this MHV-68 BAC. The BAC sequence flanked by the loxP sites was inserted into upstream of the leftmost open reading frame, M1. The infectious viruses were reconstituted after transfection of the MHV-68 BAC plasmid and shown to replicate indistinguishably compared to the wild-type virus. However, we found that the excision of the BAC sequence from the reconstituted viruses by Cre/loxP recombination is necessary for the in vivo study. After verifying the MHV-68 BAC clone we generated, mutagenesis was carried out to explore the role of ORF11. ORF11 is conserved among gamma-herpesviruses, but it does not have any assigned function. Nonsense mutations were inserted near the N-terminus of ORF11 on the MHV-68 BAC by a RecA-mediated mutagenesis system. Disruption of ORF11 had no effect on viral replication in vitro or on viral infection in vivo following intranasal inoculation into Balb/c mice.
2. Materials and Methods
2.1. Virus and Cells
MHV-68 was originally obtained from the American Type Culture Collection (VR1465). The working virus stocks were grown by infecting BHK-21 cells (ATCC CCL-10) at a multiplicity of infection (MOI) of 0.05 PFU/cell. BHK-21 and 293T cells were cultured in Dulbecco's-modified Eagle's medium (DMEM) plus 10% fetal bovine serum and NIH3T3 cells were cultured in DMEM plus 10% bovine calf serum. The plaque assays were performed on BHK-21 cells as previously described [21].
2.2. Plasmids
The pBAC(loxP) plasmid was constructed from pBeloBAC11 by inserting a second copy of loxP sequence. This was achieved by annealing two oligonucleotides (loxP4 and loxP5, listed in Table 1) and cloning into the unique BamHI and SfoI sites of pBeloBAC11. The puromycin expression cassette derived from pTW39 which contains the puromycin coding sequence driven by the CMV promoter was inserted into pBAC(loxP) to create pBAC(loxP+PUR). pBAC(loxP+PUR) was then linearized at the unique ClaI site between two loxP sites, blunted and cloned into the unique StuI site in the MHV-68 genome region (nt 106–4340) [1] to generate the targeting plasmid pTW64 for homologous recombination.
Table 1.
Sequences of the primers.
| Primer name | Sequence | Position (MHV-68 sequence) |
|---|---|---|
| loxP4 | GATCCATAACTTCGTATAATGTATGCTATA | |
| CGAAGTTAT TTAAATATCGATGCTAGCG | ||
| loxP5 | CCGCTAGCATCGATTTAAATAACTTCGTAT | |
| AGCATACATTATACGAAGTTATG | ||
| AF | TCTCCTCGAGggttgaatgcatcttgtagg | 59095–59114 |
| AR | GGATCCTTAATTAATTGACCGGgcctcaccgctcatctgaaa | 59479–59498 |
| BF | CCGGTCAATTAATTAAGGATCCtgggtagatccaggagagat | 59499–59518 |
| BR | CTATGCTAGCgatctagcaaacctctggga | 59948–59929 |
The non-MHV-68 sequences of primers are in caps. The italic indicates the engineered restriction enzymes. For loxP4 and loxP5, NheI, ClaI, and SwaI were added next to the loxP sequence (underlined) for cloning purposes. For AF and BR, XhoI and NheI sites were introduced into the primers, respectively, for cloning into pGS284. For AR and BF, BamHI and PacI were included in the primers for screening purposes.
The shuttle plasmids used for allelic exchange are derived from pGS284 [15]. The translational stop codons were introduced into the targeted ORF by PCR. The PCR reactions were carried out as suggested by the manufacturer using a proof-reading enzyme, Vent DNA polymerase (Biolab) and the cycle number was limited to 25. The sequences upstream of the stop codons (A fragments) were amplified by primers of AF and AR while the downstream sequences (B fragments) were by primers of BF and BR using wild-type MHV-68 virion DNA as the template. The A and B fragments were designed to have a 21 bp overlapping sequence containing restriction enzyme sites BamHI and PacI for later screening purposes. In a subsequent PCR reaction, the A and B fragments were used as templates and amplified by primers of AF and BR. The final PCR products were digested with appropriate enzymes and cloned into pGS284. The resultant shuttle plasmid pORF11S(GS284) was sequenced before used for allelic exchange. All the primers used to construct the shuttle plasmid were listed in Table 1.
2.3. Virus Construction
The MHV-68(BAC) was generated by cotransfecting pTW64 linearized with AvrII and the MHV-68(EGFP) virion DNA into BHK-21 cells. At 5 days after transfection, the pool of MHV-68(BAC) and MHV-68(EGFP) was subjected to limiting dilution. The recombinant MHV-68(BAC) candidates were selected by the loss of enhanced green fluorescent protein expression. Their genomic structures were analyzed by Southern blots and the correct recombinant was identified.
2.4. Isolation of Viral DNA and BAC Plasmids
Virion DNA was harvested from viral particles released from cells. Generally four 10-cm plates of BHK-21 cells were infected at MOI of 0.05 PFU/cell. At the time when 50–80% cells were detached from the plate, the supernatants were harvested and clarified twice by spinning down cell debris for 15 min at 1500 rpm, followed by digestion of DNaseI (5 μg/ml) and RNaseA (5 μg/ml) in the presence of 3 mM of MgCl2 and CaCl2 at 37°C for 30 min. The treated supernatants were subjected to high-speed centrifugation in a SA-600 rotor (Sovall) at 12,000 rpm for 2 hr at 4°C. The pellets were resuspended in the buffer (200 mM Tris [pH 8] and 100 mM EDTA) and digested with 1 mg/ml of proteinase K (Sigma) in the presence of 0.5% SDS at 60°C for overnight. The digested samples were extracted twice with phenol : chloroform (1:1) and the DNA was precipitated with ammonium acetate and ethanol.
For isolating circular viral DNA, the Hirt extraction method was used [22]. Briefly, BHK-21 cells were infected at MOI of 100 PFU/cell and cells were harvested at 3 hr after infection in PBS. The cell pellet was resuspended in the buffer containing 10 mM Tris (pH 8), 10 mM EDTA, 0.5% SDS, and 1 M NaCl. The sample was then incubated at 4°C for overnight and spun down at 14,000 rpm for 20 min at 4°C. The supernatant was digested with proteinase K (60 μg/ml) and RNaseA (30 μg/ml) at 37°C for 30 min. The DNA was extracted twice with phenol : chloroform (1 : 1) and precipitated with ammonium acetate and ethanol. The pellet was resuspended in TE and 1 μL was used to electroporate E.coli DH10B.
All the BAC plasmids were isolated by the Midi-Prep kit (Qiagen) according the modifications suggested by the manufacture.
2.5. DNA Transfection and Electroporation
For transfection of BHK-21 cells, Lipofectamine Plus (Invitrogen) was used according to the manufacture's recommendations. To remove the BAC sequence, a Cre recombinase expression plasmid was co-transfected with the viral BAC plasmid. For electroporation of DH10B (Invitrogen), a Gene Pulser II system (Bio-rad) with 0.1-cm cuvettes (Fisher) was used.
For reporter assays, 10 ng of a firefly luciferase construct driven by the ISRE (interferon-stimulated response element) promoter and 1 ng of a Renilla luciferase construct driven by the housekeeping PGK (phosphoglycerate kinase) promoter were transfected into 293T cells seeded in 48-well plates together with 100 ng of a protein expression plasmid of a viral gene. One day later, the transfected cells were treated with interferon α (IFN-α) and 24 hours (24 hrs) later, the total cell lysates were harvested for Dual Luciferase Assay (Promega). The normalized value was obtained by dividing the firefly luciferase reading with the Renilla luciferase activity of each sample.
2.6. Analysis of Viral DNA
To analyze virion DNA and BAC plasmids, digestion of restriction enzymes was performed and the products were separated on 0.7% agarose gel. The gel was then transferred onto a Hybond N+ membrane (Amersham). The membrane was then hybridized with isotope-labeled probes and the signals were developed as previously described [21]. To quantify the copy number of viral DNA, quantitative real-time PCR was performed using primers and probes derived from the M9 gene as previously described [23].
2.7. Mice, Lung Titers, and Reactivation Assays
Female Balb/c mice (Charles River laboratories, Wilmington, Mass.), 5-6 weeks-old, were first anesthetized by intraperitoneal injection of a mixture of Ketamine and Xylazine (100 mg/Kg and 2 mg/Kg) and then inoculated intranasally with 20 PFU of each virus in 20 μl of DMEM. At the times indicated, lungs and spleens were harvested and weighed. To determine the viral titers in the lungs, serial dilutions of the tissue homogenates were incubated with BHK-21 cells for plaque assays. Single-cell suspensions were obtained from the spleens and subjected to reactivation assays. Serial numbers of splenocytes were laid onto BHK-21 cells and after 6 days BHK-21 cells were fixed to determine the numbers of plaques. The plaques are referred to as infectious centers and each infectious center arises as a result of a virus reactivating from latency. Preformed viruses in splenocytes were examined by freeze-thawing cells once prior to incubating with BHK-21 cells and no plaques were detected. The viral copy numbers present in splenocytes were determined by quantitative real-time PCR described in the previous section.
2.8. Allelic Exchange
The pORF11S(BAC) plasmid was generated by RecA-mediated allelic exchange in E. coli., as described by [15]. Briefly, the donor strain is GS111 carrying the shuttle plasmid, pORF11S(GS284), and the recipient strain is GS500(recA+) harboring pMHV-68(BAC). The shuttle plasmid contains an ampicillin-resistant marker and the negative selection marker SacB, which causes the toxicity to bacteria in the presence of sucrose. A chloramphenicol-resistance marker is located within the BAC sequence. Conjugation was done by cross-streaking the donor and the recipient strains. Cointegrates were selected by growing in the presence of chloramphenicol (34 μg/ml) and ampicillin (100 μg/ml) and resolved by 8-hr growth in the presence of chloramphenicol only. After resolution, negative selection against the bacteria retaining the shuttle plasmid was done by growing in the presence of 5% sucrose and chloramphenicol on LB plates lacking NaCl. The resulting colonies were streaked on LB plates containing either chloramphenicol or ampicillin. The recombinants that lost the integrated shuttle plasmid were chloramphenicol resistant and ampicillin sensitive. The incorporation of the stop codons was determined by PCR and restriction enzyme digestion to look for the insertion of new sites engineered next to the stop codons.
3. Results and Discussion
3.1. Construction of a Full-Length MHV-68 BAC
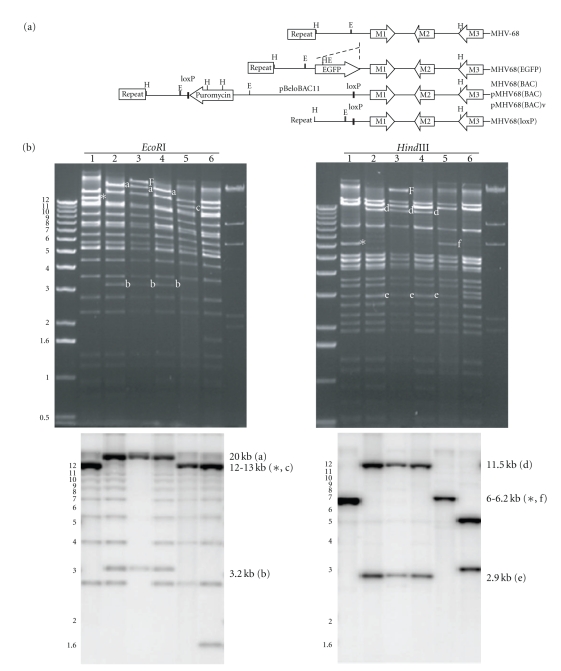
To facilitate genetic manipulation of MHV-68, the viral genome was cloned as a BAC plasmid and mutagenized in E. coli, based on a strategy used for mutagenesis of MCMV [15]. First, a recombinant MHV-68 bearing the BAC sequence was constructed. The locus chosen for the insertion of the BAC sequence is nt 1891 [1]. The site has been used for insertion in our laboratory to construct several recombinant viruses which replicated comparably to wild-type MHV-68 in vitro and in vivo. The plasmid pBeloBAC11 was modified to contain two tandem loxP sites for the excision of the BAC sequence mediated by the Cre recombinase. To promote the selection of the BAC-inserted recombinant virus, an expression cassette of the puromycin resistance gene was also inserted into pBeloBAC11. The resultant modified plasmid designated as pBAC(loxP+puro), was linearized between two loxP sites and inserted into a StuI site of a 4.2 kb MHV-68 sequence such that the BAC sequence was flanked by regions of 1.8 kb (nt 106–1891) and 2.4 kb (nt 1892–4340). This final construct was co-transfected into BHK-21 cells with purified virion DNA from MHV-68(EGFP), containing an EGFP insertion at nt 1891. The recombinant virus was screened for the loss of green fluorescence. After several rounds of purification by limiting dilutions, the recombinant virus, MHV-68(BAC), was obtained and its genomic structure was analyzed by restriction enzyme digestion (Figure 1(b), lane 2). The virion DNA of MHV-68(BAC) exhibited an expected pattern of restriction enzyme digestions. As a result of the insertion of the BAC sequence at nt 1891, the wild-type 12.7-kb EcoRI fragment (labeled with an asterisk in lane 1) were replaced by the 3.2-kb and 20 kb fragments in the digested MHV-68(BAC) DNA (labeled with a and b in lane 2). Similarly, the wild-type 6.1-kb HindIII fragment (labeled with an asterisk in lane 1) was replaced by the 2.9- and 11.5-kb fragments (labeled with d and e in lane 2) in the digested MHV-68(BAC) DNA.
Figure 1.
The genomic structures of the MHV-68 recombinant viruses and the MHV-68 BAC plasmid. (a) Schematic diagram of the construction of the MHV-68 BAC virus. Shown is the left end of the viral genome encompassing terminal repeats and ORFs M1–M3. The BAC vector sequence and a puromycin expression cassette was recombined into the locus where the EGFP expression cassette was inserted in MHV-68(EGFP). MHV-68(BAC) was selected for the loss of the EGFP expression. The pMHV-68(BAC) plasmid was obtained by electroporating the circular genome of MHV-68(BAC) into DH10B cells. Following transfection of pMHV-68(BAC) into BHK-21 cells, the reconstituted virus, pMHV-68(BAC)v, was generated. The structures of pMHV-68(BAC) and pMHV-68(BAC)v are identical to MHV-68(BAC), except that pMHV-68(BAC) is circular with termini fused. To remove the BAC insert flanking by loxP sites, pMHV-68(BAC)v was co-transfected with a Cre recombinase expression plasmid and MHV-68(loxP) was isolated. The sites of HindIII are indicated with H and the ones of EcoRI were indicated with E. (b) Restriction enzyme analysis of the genome of MHV-68 recombinant viruses and the MHV-68 BAC plasmid. Each DNA sample was digested with either EcoRI or HindIII. Lane 1 is virion DNA of wild-type MHV-68; lane 2 is virion DNA of MHV-68(BAC); lane 3 is the plasmid DNA of pMHV-68(BAC); lane 4 is virion DNA of pMHV-68(BAC)v; lane 5 is virion DNA of MHV-68(loxP) and lane 6 is virion DNA of MHV-68(EGFP). The wild-type fragments which were eliminated due to the insertion of the BAC sequence were marked with asterisks. The fragments marked with a, b, d, and e, were generated because of the BAC insert. The fragments marked with c and f, resulted from the removal of BAC, 200 bp larger than the fragments indicated with asterisks. The fragments marked with F were from the fused termini of the circular pMHV-68(BAC). On the bottom of each agarose gel picture is the corresponding Southern blot using the probe spanning nt 51–6298. The laddering patterns in EcoRI-digested samples were due to various number of repeats on the linear virion DNA but not present in pMHV-68(BAC) (lane 3).
Next, the circular viral genomes of MHV-68(BAC) were harvested from infected BHK-21 cells by the Hirt extraction method and then electroporated into E. coli DH10B. The plasmids from several colonies were prepared and digested with HindIII to identify the clone that contains the full-length MHV-68 genome. One clone, pMHV-68(BAC), was analyzed by EcoRI and HindIII digestions (Figure 1(b), lane 3). Compared to MHV-68(BAC) virion DNA (Figure 1(b), lane 2), the only difference is the appearance of a larger fragment (labeled with F in lane 3) derived from the fused terminal fragment due to the circular nature of the BAC plasmid.
3.2. Characterization of the BAC-Derived Viruses
Transfection of the pMHV-68(BAC) into BHK-21 initiated viral lytic replication and led to the formation of plaques. A similar amount of wild-type MHV-68 virion DNA was transfected as a control and the appearance of plaques was about at the same time, 3 days after transfection. The supernatant containing the reconstituted virus, pMHV-68(BAC)v, was harvested to infect fresh BHK-21 cells to produce more viruses and obtain virion DNA for the analysis of restriction enzyme digestions. As expected, the restriction pattern of virion DNA of pMHV-68(BAC)v (Figure 1(b), lane 4) is identical to MHV-68(BAC) (Figure 1(b), lane 2). In order to remove the rather large insert of the BAC sequence (10 kb), pMHV-68(BAC)v was co-transfected with a protein expression plasmid of Cre recombinase. The resultant pool of viruses was subjected to limiting dilution and screened by PCR for the absence of the BAC sequence. The virus, MHV-68(loxP), lacking the BAC sequence but retaining one copy of the loxP site, was then amplified in BHK-21 cells to obtain virion DNA for analyses. After Cre-mediated recombination, there is ∼200-nt BAC sequence left on the genome of MHV-68(loxP), which causes a shift in the 6.1-kb HindIII wild-type fragment (labeled with an asterisk in lane 1) to a larger fragment (labeled with f in lane 5).
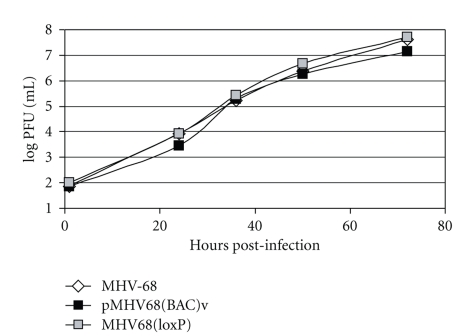
Next, the growth kinetics of pMHV-68(BAC)v and MHV-68(loxP) were examined by determining their multiple-growth curves on BHK-21 cells. These two viruses replicated indistinguishably from each other and comparably to wild-type MHV-68 (Figure 2). Thus, the 10-kb insertion of the BAC sequence did not significantly affect the replication of MHV-68 in cultured cells.
Figure 2.
pMHV-68(BAC)v and MHV-68(loxP) displayed similar growth curves to wild-type MHV-68 in vitro. BHK-21 cells were infected with each virus at an MOI of 0.05. The viral titers in cells and the supernatants harvested at various times after infection were determined by plaque assays.
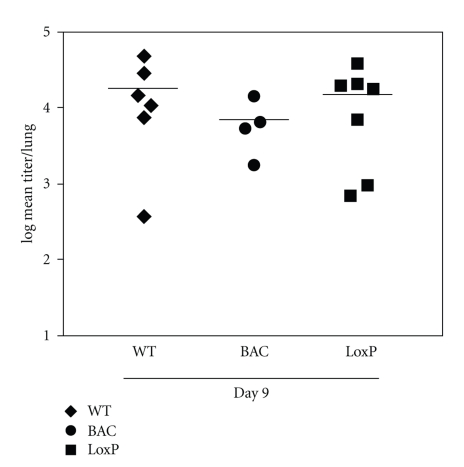
One major advantage of studying MHV-68 is the availability of an in vivo infection mouse model. Thus, we needed to determine whether the reconstituted virus behaved similarly to the wild-type virus in mice. Balb/c mice were intranasally inoculated with 20 PFU of wild-type MHV-68, pMHV-68(BAC)v, or MHV-68(loxP). At the peak of acute lytic infection in the lungs, mice infected with MHV-68(loxP) generated comparable viral titers to those infected with wild-type MHV-68 (Figure 3). The viral titers in mice infected with pMHV-68(BAC)v were slightly lower (2.6-fold) than those infected with wild-type MHV-68 but the difference did not reach the statistical significance (p > 0.1). At day 11 after infection, no infectious viruses were recovered from any group of mice. The results indicate that the virus derived from our BAC clone infects and replicates in the lungs as well as wild-type MHV-68 and the presence of the BAC insert does not significantly interfere with viral productive infection in the lungs.
Figure 3.
Infection of pMHV-68(BAC)v and MHV-68(loxP) produced similar viral titers to wild-type MHV-68 in lungs of mice. Balb/c mice were infected with 20 PFU of each virus via intranasal inoculation. At day 9 after infection, the lungs were harvested and homogenized for determining viral titers using plaque assays. Each symbol represents the lung titer of individual mice.
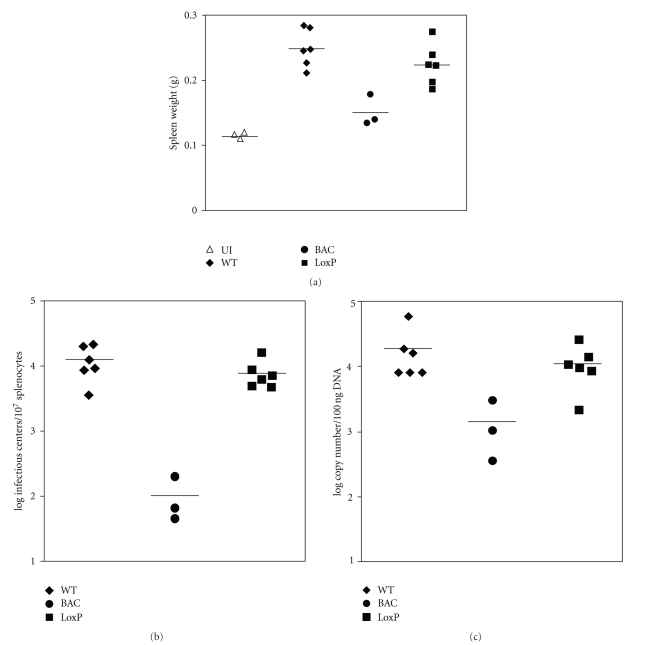
Following acute infection in the lungs, MHV-68 establishes latency in the spleen accompanied with splenomegaly. The spleens of mice infected either with wild-type MHV-68 or MHV-68(loxP) enlarged to similar extents compared to uninfected mice (Figure 4(a)). The levels of latently-infected cells determined by infectious center assays were also comparable between mice infected with wild-type virus and those with MHV-68(loxP) (Figure 4(b)). Similar results were obtained from measuring the viral DNA copy numbers present in the spleen (Figure 4(c)). In contrast, the virus retaining the BAC insert failed to establish the wild-type level of latency in the spleen. There were 100-fold fewer infectious centers and 10-fold lower viral DNA copies in the spleen compared to those from mice infected with wild-type MHV-68. Taken together, the BAC insertion in the viral genome severely attenuated viral splenic latency and its removal restored the ability of MHV-68 to establish the wild-type level of latency.
Figure 4.
The removal of the BAC insert restored the ability of MHV-68 to establish viral latency in the spleens of mice. Balb/c mice were infected with 20 PFU of each virus via intranasal inoculation. At day 14-15 after infection, the spleens were weighed and harvested for reactivation assays and quantification of viral DNA copy numbers. Each symbol represents the weight of the spleen (a), the number of infectious centers (b), and the viral DNA copies (c) of individual mice.
The present study indicates that our MHV-68 BAC clone is suitable for genetic mutagenesis to investigate the function of individual viral genes. In fact, random mutagenesis by transposon insertions was already conducted on this MHV-68 BAC plasmid, and the resultant mutant viruses were used to identify genetic requirements for productive replication in vitro and in vivo [19]. Moreover, we also constructed a firefly luciferase-expressing virus using the MHV-68 BAC genetic engineering system described here and examined viral infection via the whole body bioluminescence imaging [24]. It was shown that the virus derived from our MHV-68 BAC clone mainly replicates in the lung following intranasal inoculation and produces infectious virus that can be recovered from it, which is similar to what has been observed for wild-type MHV-68 [5].
Although a different MHV-68 BAC plasmid was constructed and published previously by Adler et al. [25], we independently carried out the cloning process and inserted the BAC sequence at a different location. Our BAC sequence insertion is at 1.8 kb from the terminal repeats of the genome while Adler et al. inserted the BAC sequence adjacent to the repeats [25]. We showed that the presence of the 10-kb BAC sequence did not significantly reduce the replication ability of the virus in cultured cells or in the lungs of mice, unlike the previous MHV-68 BAC clone constructed in the other laboratory [10].
Despite that the BAC insertion does not interfere with viral lytic replication, it severely attenuates viral ability to establish latency in the spleen. We observed less splenomegaly, 10-fold lower viral DNA copy numbers and 100-fold reduction in viral reactivation from splenocytes. This result is similar to the MHV-68 BAC clone made by Adler et al. [10], but not surprising, since several studies have shown that the left end of the MHV-68 genome is important for viral latency [26–29]. In addition, several microRNA genes were identified on the left end of the genome [30]. It is possible that the insertion of a large BAC sequence alters the expression of the neighboring genes. Thus, it is necessary to remove the BAC sequence using Cre recombinase prior to the study of viral latency.
3.3. Site-Specific Mutagenesis of ORF11
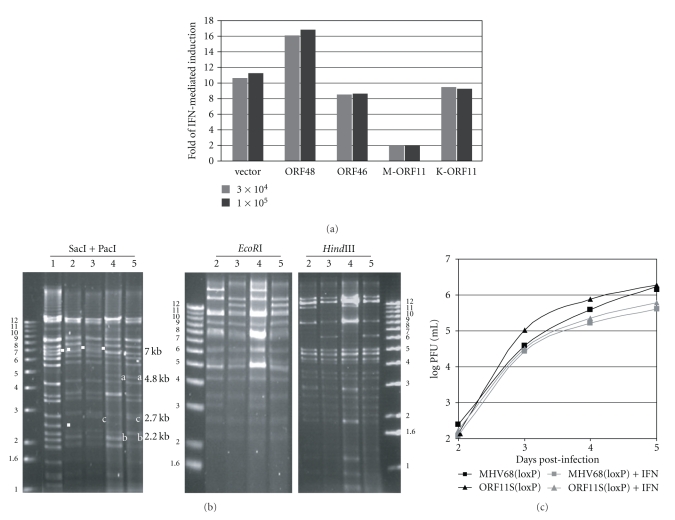
ORF11 is one of MHV-68 genes whose functions remain unknown. Previously, ORF11 was shown to be a virion component but dispensable for viral replication in vitro [31]. A recent study on the EBV ORF11 homologue (also known as LF2) indicates that this viral protein play a role in counteracting type I interferon (IFN) responses [32]. Here, we used transient transfection assays to examine whether MHV-68 ORF11 (M-ORF11) affected the IFN-mediated induction of an ISRE (interferon-stimulated response element) reporter. We also included transfection of a KSHV ORF11 (K-ORF11) expression plasmid as a comparison. The induction by IFN-α was 10~12-fold in the cells transfected with an empty vector but reduced to 2-fold in the cells receiving the M-ORF11 plasmid (Figure 5(a)). Interestingly, K-ORF11 did not have the same inhibitory effect as M-ORF11. Neither did expression of other MHV-68 genes, such as ORF48 and ORF46, inhibited the induction of ISRE reporter activity. The reporter assays indicate that M-ORF11 can reduce the IFN responses but not K-ORF11.
Figure 5.
ORF11 is not required for MHV-68 to resist IFN-α. (a) ORF11 inhibits the IFN-mediated induction of the ISRE reporter activity. 293T cells were transfected with the ISRE-driven firefly luciferase reporter and the protein expression plasmid of a viral ORF indicated at the bottom of the graph. For normalization, a Renilla luciferase construct driven by the housekeeping PGK promoter was included in each transfection. One day after transfection, the cells were either mock treated or treated with 3 × 104 units/ml of IFN-α and 24 hrs later, the total lysates were harvested for Dual Luciferase assays. For another experiment, the cells were treated with 105 units/ml of IFN-α. The fold of induction was derived by dividing the normalized valuses of treated samples with those of mock treated samples. (b) Restriction enzyme analysis of the genomes of pORF11S(BAC)v and ORF11S(loxP). Each virion DNA sample was digested with either combination of SacI and PacI, EcoRI or HindIII. Lane 1 is virion DNA of wild-type MHV-68; lane 2 is virion DNA of pMHV68(BAC)v; lane 3 is virion DNA of MHV68(loxP); lane 4 is virion DNA of pORF11S(BAC) and lane 5 is virion DNA of ORF11S(loxP). The wild-type fragments marked with solid squares were eliminated in lanes 4 and 5 due to the insertion of stop codons. The a and b fragments were generated as a result of the insertion of the stop codons. The c fragments were resulted from the removal of the BAC sequence. (c) Lack of ORF11 does not increase the sensitivity of MHV-68 to IFN-α. NIH3T3 cells were pretreated with 100 units/ml of IFN-α for 16 hrs and then infected with MHV-68(LoxP) or ORF11S(LoxP) at an MOI of 0.01. The whole cultures were harvested at various times post-infections for plaque assays to determine the viral titers.
In order to determine the biological significance of the anti-IFN activity of M-ORF11 in the context of viral replication, we constructed an ORF11-deficient mutant virus using the pMHV-68 BAC plasmid. The recA-mediated allelic exchange method was used to introduce translational stop codons after N-terminal 41 amino acids of the predicted 388 amino-acid ORF11 gene product. The corresponding shuttle plasmid to transfer the stop codons into the pMHV-68 BAC plasmid was constructed and verified by sequencing. After allelic exchange in RecA+E. coli as described in Section 2, the recombinant BAC plasmid, pORF11S(BAC), was isolated and examined by SacI and PacI digestion. To reconstitute the virus, the pORF11S(BAC) plasmid was transfected into BHK-21 cells together with a Crerecombinase expression plasmid. The pool of viruses containing both ORF11S(BAC) and ORF11S(loxP) was subjected to limiting dilution and screened for the absence of the BAC sequence. One isolate, ORF11S(loxP), was selected and its genomic structure was examined by restriction analysis. As expected, the virion DNA of ORF11S(BAC) and ORF11S(loxP) have the similar restriction patterns of SacI and PacI altered in the fragment containing the ORF11 coding sequence (Figure 5(b), lanes 4 and 5) compared to wild-type pMHV68(BAC)v and MHV68(loxP). The 7.0-kb SacI wild-type fragment (labeled with squares in lanes 1–3 of Figure 5(b)) containing the ORF11 coding sequence is replaced by the 4.7- and 2.2-kb fragments (labeled with a and b in lanes 4-5) as a result of the new PacI site introduced along with the stop codons into the ORF11. The removal of the BAC sequence led to the appearance of the 2.7-kb SacI in ORF11S(loxP) (labeled with c in lane 5 of Figure 5(b)), which is also detected in MHV68(loxP) (Figure 5(b), lane 3). This 2.7-kb fragment is 200 bp larger than the 2.5-kb wild-type fragment (Figure 5(b), lane 1) as a result of the remaining loxP and BAC vector sequence. Restriction digestions by HindIII and EcoRI displayed similar patterns compared to the wild-type pMHV68(BAC) plasmid, indicating no detectable illegitimate DNA rearrangements during recombination in E. coli.
To determine whether ORF11 contributes to the resistance of MHV-68 to type I interferons, we studied the multiple-step growth curves of MHV68(loxP) and ORF11S(loxP) with or without the treatment of IFN-α. For this experiment, we used NIH3T3 cells and mouse IFN-α. The ORF11S(loxP) virus replicated comparably to MHV68(loxP) in the absence of interferons, indicating that ORF11 is not required for viral replication in vitro (Figure 5(c)), consistent with the previous results [19, 31]. Furthermore, the lack of ORF11 did not increase the sensitivity of MHV-68 to the treatment of IFN-α. There were about 3∼4-fold reductions by IFN-α in the peak viral titers of both MHV68(loxP) and ORF11S(loxP) (Figure 5(c)). There are two major interpretations for this result. One is that the anti-IFN activity of ORF11 observed in the transient transfection reporter assay is a nonspecific effect due to overexpression of the protein. This is unlikely since overexpression of other viral proteins, especially K-ORF11, does not have the same inhibitory effect. The other interpretation is that there are additional viral proteins with similar anti-IFN functions to complement the loss of ORF11. It is not unusual for large DNA viruses, such as herpesviruses, to utilize multiple viral proteins to counteract type I interferon responses. As reviewed elsewhere [33–35], several KSHV proteins can inhibit the functions of IRF3 and IRF7, the two important transcriptional factors that regulate type I interferon responses. Therefore, it is likely that interferons play an important role in the control of gamma-herpesvirus infection and the viruses utilize multiple mechanisms to counteract the host responses for their survival.
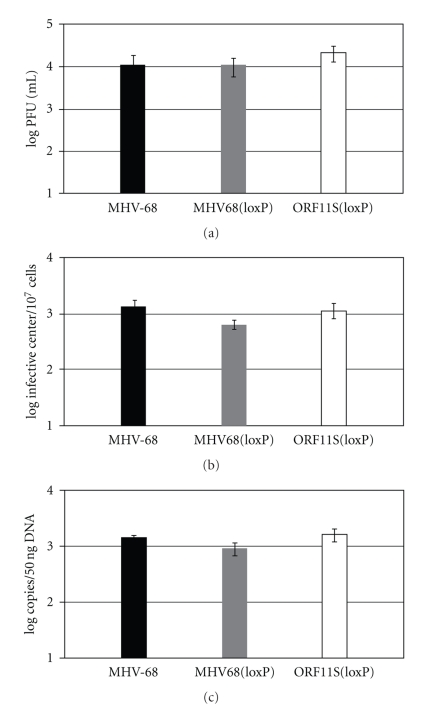
To examine the role of ORF11 in MHV-68 infection in vivo, Balb/c mice were intranasally inoculated with 20 PFU of wild-type MHV-68, MHV68(loxP), or ORF11S(loxP). The peak viral titers in the lungs were similar among groups of mice at day 9 after infection (Figure 6(a)). At day 11, no infectious viruses were detected in any mice (data not shown). The viral latency at day 15 after infection in the spleens of mice infected with ORF11S(loxP) was comparable to that in the mice infected with wild-type MHV-68 (Figure 6(b)). Although MHV68(loxP) seemed to establish lower levels of latency \ than the other two groups of infected mice, this difference did not reach statistical significance. Similar results were obtained from the analysis of DNA copy numbers measured by real-time PCR. In conclusion, ORF11 is not required for MHV-68 to establish lytic and latent infections in mice, consistent with the previous studies [19, 31].
Figure 6.
ORF11 is not required for viral infection in Balb/c mice. (a) The acute lung viral titers. Balb/c mice were intranasally inoculated with 20 PFU of each virus. The lungs were harvested at day 9 after infection and the titers were determined by plaque assays. Shown are the means of viral titers from 3 mice per virus infection and the standard errors of the means. (b) The viral latency in spleens. 20 PFU of each virus was inoculated into Balb/c mice. The spleens were harvested at day 15 after infection and the amounts of latent virus were determined by infective center assays. (c) The copy numbers of viral genomes in the spleens harvested at day 15 after infection. 50 ng of splenic DNA was used for real-time PCR to determine the viral DNA copy numbers. Shown are the means from 3 mice per virus infection and the standard errors of the means.
One notable piece of information about ORF11 is that ORF11 as well as ORF10 were previously postulated to be derived from the viral dUTPase gene, ORF54, based on sequence similarities and structural predictions [36]. However, ORF10 and ORF11 do not retain the critical residues at the catalytic site of dUTPase and thus they are not expected to have any enzymatic activity. Since these three proteins, ORF54, ORF10, and ORF11, have some degrees of homology, it may be that they have the same unknown functions, such as anti-IFN activity, and can complement each other; thus, elimination of ORF11 alone is not sufficient to generate a phenotype. Nevertheless, it is also possible that ORF11 is only required under specific circumstances. For example, the virus deficient in the M3 gene only exhibited a different phenotype from the wild type when infecting CNS [37]. We have not tested other aspects of virus infection besides replication in the lungs and latency in the spleens.
4. Conclusions
The MHV-68 BAC plasmid generated in our laboratory has been extensively used to construct various mutant viruses. In this report, we provide the details for the cloning and characterization of our MHV-68 BAC. In comparison to the wild-type MHV-68, the virus reconstituted from the BAC plasmid replicated comparably in cultured cells and produced similar titers in the lung of infected mice. However, removal of the BAC insertion is necessary for the studies of viral splenic latency. In summary, our MHV-68 BAC plasmid is a valid and useful resource for genetic mutagenesis.
Acknowledgments
The authors thank Vaithilingaraja Arumugaswami and Seungmin Hwang for their helpful comments. T. T. Wu is supported by the Leukemia and Lymphoma Society and NIH (R21 DE018337). R. Sun is supported by NIH (DE015752, DE19085), STOP Cancer Foundation and Burroughs Wellcome Fund.
References
- 1.Virgin HW, IV, Latreille P, Wamsley P, et al. Complete sequence and genomic analysis of murine gammaherpesvirus 68. Journal of Virology. 1997;71(8):5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. Journal of General Virology. 1990;71(6):1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 3.Efstathiou S, Ho YM, Minson AC. Cloning and molecular characterization of the murine herpesvirus 68 genome. Journal of General Virology. 1990;71(6):1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 4.Mackett M, Stewart JP, Pepper SDV, et al. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. Journal of General Virology. 1997;78(6):1425–1433. doi: 10.1099/0022-1317-78-6-1425. [DOI] [PubMed] [Google Scholar]
- 5.Sunil-Chandra NP, Efstathiou S, Arno J, Nash AA. Virological and pathological features of mice infected with murine gammaherpesvirus 68. Journal of General Virology. 1992;73(9):2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 6.Sunil-Chandra NP, Efstathiou S, Nash AA. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. Journal of General Virology. 1992;73(12):3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 7.Doherty PC, Tripp RA, Hamilton-Easton A-M, Cardin RD, Woodland DL, Blackman MA. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Current Opinion in Immunology. 1997;9(4):477–483. doi: 10.1016/s0952-7915(97)80098-2. [DOI] [PubMed] [Google Scholar]
- 8.Tripp RA, Hamilton-Easton AM, Cardin RD, et al. Pathogenesis of an infectious mononucleosis-like disease induced by a murine γ-herpesvirus: role for a viral superantigen? Journal of Experimental Medicine. 1997;185(9):1641–1650. doi: 10.1084/jem.185.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunil-Chandra NP, Arno J, Fazakerley J, Nash AA. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. American Journal of Pathology. 1994;145(4):818–826. [PMC free article] [PubMed] [Google Scholar]
- 10.Adler H, Messerle M, Koszinowski UH. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. Journal of Virology. 2001;75(12):5692–5696. doi: 10.1128/JVI.75.12.5692-5696.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adler H, Messerle M, Koszinowski UH. Cloning of herpesviral genomes as bacterial artificial chromosomes. Reviews in Medical Virology. 2003;13(2):111–121. doi: 10.1002/rmv.380. [DOI] [PubMed] [Google Scholar]
- 12.Borst E-M, Hahn G, Koszinowski UH, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. Journal of Virology. 1999;73(10):8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delecluse H-J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(14):8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delecluse H-J, Kost M, Feederle R, Wilson L, Hammerschmidt W. Spontaneous activation of the lytic cycle in cells infected with a recombinant Kaposi’s sarcoma-associated virus. Journal of Virology. 2001;75(6):2921–2928. doi: 10.1128/JVI.75.6.2921-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith GA, Enquist LW. Construction and transposon mutagenesis in Escherichia coli of a full- length infectious clone of pseudorabies virus, an alphaherpesvirus. Journal of Virology. 1999;73(8):6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou F-C, Zhang Y-J, Deng J-H, et al. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. Journal of Virology. 2002;76(12):6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244(4910):1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Buchholz F, Muyrers JPP, Francis Stewart A. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genetics. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 19.Song MJ, Hwang S, Wong WH, et al. Identification of viral genes essential for replication of murine γ-herpesvirus 68 using signature-tagged mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3805–3810. doi: 10.1073/pnas.0404521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arumugaswami V, Sitapara R, Hwang S, et al. High-resolution functional profiling of a gammaherpesvirus RTA locus in the context of the viral genome. Journal of Virology. 2009;83(4):1811–1822. doi: 10.1128/JVI.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T-T, Usherwood EJ, Stewart JP, Nash AA, Sun R. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. Journal of Virology. 2000;74(8):3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. Journal of Molecular Biology. 1967;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 23.Rickabaugh TM, Brown HJ, Martinez-Guzman D, et al. Generation of a latency-deficient gammaherpesvirus that is protective against secondary infection. Journal of Virology. 2004;78(17):9215–9223. doi: 10.1128/JVI.78.17.9215-9223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang S, Wu T-T, Tong LM, et al. Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. Journal of Virology. 2008;82(24):12498–12509. doi: 10.1128/JVI.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adler H, Messerle M, Wagner M, Koszinowski UH. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. Journal of Virology. 2000;74(15):6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clambey ET, Virgin HW, IV, Speck SH. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. Journal of Virology. 2000;74(4):1973–1984. doi: 10.1128/jvi.74.4.1973-1984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clambey ET, Virgin HW, IV, Speck SH. Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency. Journal of Virology. 2002;76(13):6532–6544. doi: 10.1128/JVI.76.13.6532-6544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacoby MA, Virgin HW, IV, Speck SH. Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation. Journal of Virology. 2002;76(4):1790–1801. doi: 10.1128/JVI.76.4.1790-1801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macrae AI, Usherwood EJ, Husain SM, et al. Murid herpesvirus 4 strain 68 M2 protein is a B-cell-associated antigen important for latency but not lymphocytosis. Journal of Virology. 2003;77(17):9700–9709. doi: 10.1128/JVI.77.17.9700-9709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeffer S, Zavolan M, Grässer FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 31.Boname JM, May JS, Stevenson PG. Murine gammaherpesvirus 68 open reading frame 11 encodes a nonessential virion component. Journal of Virology. 2005;79(5):3163–3168. doi: 10.1128/JVI.79.5.3163-3168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Fossum E, Joo CH, et al. Epstein-Barr virus LF2: an antagonist to type I interferon. Journal of Virology. 2009;83(2):1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang C, Lee J-S, Jung JU. Immune evasion in Kaposi’s sarcoma-associated herpes virus associated oncogenesis. Seminars in Cancer Biology. 2008;18(6):423–436. doi: 10.1016/j.semcancer.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aresté C, Blackbourn DJ. Modulation of the immune system by Kaposi’s sarcoma-associated herpesvirus. Trends in Microbiology. 2009;17(3):119–129. doi: 10.1016/j.tim.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Coscoy L. Immune evasion by Kaposi’s sarcoma-associated herpesvirus. Nature Reviews Immunology. 2007;7(5):391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 36.Davison AJ, Stow ND. New genes from old: redeployment of dUTPase by herpesviruses. Journal of Virology. 2005;79(20):12880–12892. doi: 10.1128/JVI.79.20.12880-12892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Berkel V, Levine B, Kapadia SB, Goldman JE, Speck SH, Virgin HW., IV Critical role for a high-affinity chemokine-binding protein in γ-herpesvirus-induced lethal meningitis. Journal of Clinical Investigation. 2002;109(7):905–914. doi: 10.1172/JCI14358. [DOI] [PMC free article] [PubMed] [Google Scholar]