Abstract
The gene expression changes produced by moderate hypothermia are not fully known, but appear to differ in important ways from those produced by heat shock. We examined the gene expression changes produced by moderate hypothermia and tested the hypothesis that rewarming after hypothermia approximates a heat-shock response. Six sets of human HepG2 hepatocytes were subjected to moderate hypothermia (31°C for 16 h), a conventional in vitro heat shock (43°C for 30 min) or control conditions (37°C), then harvested immediately or allowed to recover for 3 h at 37°C. Expression analysis was performed with Affymetrix U133A gene chips, using analysis of variance-based techniques. Moderate hypothermia led to distinct time-dependent expression changes, as did heat shock. Hypothermia initially caused statistically significant, greater than or equal to twofold changes in expression (relative to controls) of 409 sequences (143 increased and 266 decreased), whereas heat shock affected 71 (35 increased and 36 decreased). After 3 h of recovery, 192 sequences (83 increased, 109 decreased) were affected by hypothermia and 231 (146 increased, 85 decreased) by heat shock. Expression of many heat shock proteins was decreased by hypothermia but significantly increased after rewarming. A comparison of sequences affected by thermal stress without regard to the magnitude of change revealed that the overlap between heat and cold stress was greater after 3 h of recovery than immediately following thermal stress. Thus, while some overlap occurs (particularly after rewarming), moderate hypothermia produces extensive, time-dependent gene expression changes in HepG2 cells that differ in important ways from those induced by heat shock.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-010-0181-2) contains supplementary material, which is available to authorized users.
Keywords: Cold stress, Cell stress response, DNA microarray, Heat shock proteins
Introduction
Hypothermia affects human tissues in physiologically and therapeutically important ways. Clinically, moderate hypothermia (33°C) improves neurological outcome after cardiac arrest (Bernard et al. 2002), and there may be benefits to induced hypothermia in other clinical conditions, including some cases of closed head injury (Marion et al. 1997; McIntyre et al. 2003). However, some have suggested that patients exposed to hypothermia may also be at increased risk for infection (Remick and Xioa 2006). Because of the clinical risks and benefits of hypothermia, the wider clinical use of this technique has increased interest in identifying how it affects cells and tissues. Proposed mechanisms include reductions in oxygen consumption and metabolic activity, as well as protective alterations in critical oxidation–reduction molecules (Safar and Kochanek 2002). Additionally, in vitro, cultured mammalian cells respond to hypothermia through alterations in the expression of many genes outside of metabolic and redox pathways, including pathways with the potential to influence inflammatory responses and tissue survival characteristics (Sonna et al. 2002a).
In vitro, moderate hypothermia has been reported to trigger increases in the expression of RNA-binding proteins such as cold-induced RNA-binding protein (CIRBP) and RNA-binding motif 3 (RBM3) (Danno et al. 1997; Nishiyama et al. 1997a, b). It is noteworthy that the times and temperatures that typically produce clinical benefit in trials of systemic hypothermia (32–34°C for 12–24 h) are very similar to those which have been found to induce a gene expression response in vitro. This suggests that some of the effects of moderate hypothermia might be reflected in, or mediated by, altered expression of genes that affect tissue recovery and survival. The full extent to which moderate hypothermia affects gene expression in humans is unknown, but previous work with the THP-1 human cell line (Sonna et al. 2006) suggests that the response is in fact extensive.
Animal models suggest that many tissues, including brown adipose tissue (Boss et al. 1997; Denjean et al. 1999; Yamashita et al. 1999), white adipose tissue (Yamashita et al. 1999), heart (Boss et al. 1997), soleus muscle (Boss et al. 1997), spinal cord (Mizuno et al. 2000), and spleen (Ganta et al. 2006) do indeed display at least some changes in gene expression in response to cold. Hypothermia can also modify gene expression responses to ischemia/reperfusion of brain (Fukui et al. 2006; Kobayashi et al. 2008; Ohta et al. 2007; Yenari and Han 2006) and liver (Niemann et al. 2010). In the spleen, changes in cytokine and chemokine expression occur even after sympathetic denervation, suggesting that cold itself, and not the increase in sympathetic activity that occurs during hypothermic exposure, may be responsible for some of the changes seen (Ganta et al. 2006). The importance of examining different tissues is illustrated by the observation that in rat (Boss et al. 1997; Mizuno et al. 2000), UCP-2 responses to hypothermia appear to vary somewhat as a function of tissue type.
Recent work has reported that expression of the cytoprotective HSPs-70 and -32 proteins after ischemia–reperfusion of rat liver was substantially augmented by hypothermia applied during the period of ischemia (Niemann et al. 2010), and these animals had improved survival, reduced elevation of transaminases, less histological evidence of liver injury, and reduced induction of both TNF-alpha and macrophage inflammatory protein-2. Thus, it appears that hypothermia can affect rat liver gene expression in the context of ischemia–reperfusion. However, this study was specifically designed to examine the effects of hypothermia on responses to ischemia–reperfusion, not the effects of hypothermia per se; neither was it designed to determine the breadth of the effects of hypothermia and rewarming on liver gene expression.
In humans, therapeutic hypothermia is undertaken with the understanding that the patient will eventually be rewarmed. Accordingly, a full understanding of the effects of hypothermia on gene expression requires studies that include both hypothermia and rewarming. At least some important cold-induced genes, including HSPs, typically demonstrate changes in expression only after a period of recovery at normothermic conditions (typically, 37°C; Fuller 2003; Sonna et al. 2002a). However, the full extent of the human gene expression response to rewarming is unknown. Furthermore, we do not know whether the broad effects on human gene expression observed in a blood cell line (THP-1 cells) also occur in a human cell line derived from a solid organ, such as liver.
Here we report the use of DNA microarrays to examine in greater detail the effects of moderate hypothermia and rewarming on mRNA expression by human hepatocytes (HepG2 cells). HepG2 cells were selected because this cell line is derived from a solid organ that is involved in a wide variety of metabolic processes (and as such, expresses a large number of genes that are potentially inducible or repressible) and in our hands has demonstrated robust changes in gene expression in response to environmental stressors such as hypoxia and heat shock (Sonna et al. 2003). Furthermore, unlike our previous microarray study in THP-1 cells, we chose to look at the gene expression changes that occur both during hypothermia itself and after a period of recovery at 37°C. The hypothermic exposure of 31°C for 16 h is comparable to those achieved by therapeutic hypothermia protocols (Bernard et al. 2002; Marion et al. 1997). As controls, we examined cells maintained at 37°C as well as cells subjected to heat shock at 43°C for 30 min and allowed to recover at 37°C. We chose heat shock as a positive control because it produces a broad, intense gene expression response in many experimental systems that has been well characterized (Sonna et al. 2002a), is known to produce congruent changes in mRNA and protein expression, and is reliably detectable by microarray analysis (Sonna et al. 2002b). Our study had two principal goals: (1) to identify novel genes and pathways affected by hypothermia and rewarming in a human hepatocellular cell line and (2) to test the hypothesis that moderate hypothermia would reliably produce an mRNA expression response that is distinct from that of heat shock, with changes in expression upon rewarming that include a heat-shock response.
Materials and methods
Cell culture
Human hepatocellular carcinoma HepG2 cells were obtained from the American Type Culture Collection (catalog number HB-8065) and grown to 90–95% confluence in Minimum Essential Medium Eagle (MEME) (ATCC catalog number 30-2003) supplemented with 10% Fetal Bovine Serum (Gibco/Invitrogen), and 2% Penicillin–Streptomycin (Gibco/Invitrogen).
Heat and cold exposure
The experiments were performed in serum-free media. Prior to the beginning of each experiment, cells were allowed to equilibrate at 37°C for 4 h in serum-free media. Heat and cold stresses were delivered by placing flasks into thermostatic water baths contained within 5% CO2 tissue culture incubators, with both water baths and incubators maintained at the desired temperature (31°C for cold stress, 43°C for heat shock). Control cells were maintained in a 37°C incubator. The thermal stress conditions tested were: 31°C for 16 h (henceforth referred to as cold stress or moderate hypothermia), or 37°C for 15.5 h, followed by 43°C for 0.5 h (heat shock), or 37°C for 16 h (control). Temperatures were stable as measured with mercury thermometers during the experiments.
The hypothermic exposure time was chosen based on preliminary experiments in HepG2 cells that confirmed literature observations, made in other cell lines that increased expression of CIRBP as a result of moderate hypothermia requires 12 to 24 h of exposure to cold (Nishiyama et al. 1997a, b). The heat shock conditions were experimentally conventional (6°C over normal culture temperatures for 30 min) and have been shown to produce a vigorous heat shock response in HepG2 cells (Sonna et al. 2003).
The time point at which cells were removed from thermal stress conditions was defined as time 0 (T = 0). Cells were studied at time 0 or allowed to recover at 37°C for either 3 h (microarray experiments) or for 1, 3, and 6 h (confirmatory reverse transcription PCR experiments).
RNA extraction and quality assurance
At each time point, the culture media were aspirated off, the cells were briefly rinsed once with cold 1% phosphate buffered saline, 5 ml of pre-warmed trypsin solution (Gibco, catalog number 25200-056) was added to facilitate the removal of the cells from each flask, and each flasks were then returned to the control incubator for several minutes to allow time for the cells to detach. The trypsin was inactivated by addition of 5 ml of MEME, and Trypan Blue Exclusion measurement of cell viability was performed on an aliquot of the resulting cell suspension.
Extracted cells were then pelleted by centrifugation, and total RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen, catalog number 74104). RNA quality was assessed by spectrophotometry and by examination of an aliquot of RNA by gel electrophoresis. All samples analyzed met prospectively defined quality criteria (Farrell 1998; Sonna et al. 2002b).
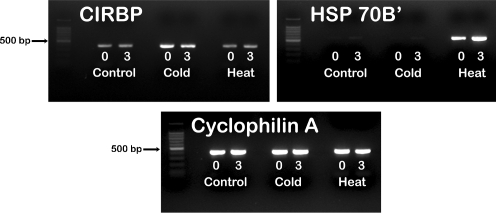
Additionally, each sample set submitted for microarray analysis was required to demonstrate appropriate differential expression of well-established positive control genes (CIRBP for samples subjected to cold stress, HSP70B’ for samples subjected to heat shock) and stable expression of a control gene (cyclophilin A).
DNA microarray analysis
Expression analysis was performed using Affymetrix Gene Chip™ U133A arrays, as described in detail elsewhere (Sonna et al. 2002b). Pre-processing was performed using MAS 5.0 software, with the clipped average signal intensity set to 500. Signal intensities at each condition/time point combination represented the data input used for the statistical analysis.
Statistical analysis
Statistical analyses were performed using SPSS version 10, R version 1.9, and a two-way repeated-measures software algorithm that was written in Java and whose output was verified by comparison to that of SPSS (Khatri et al. 2006). Where appropriate, we also used Microsoft Excel and Sigma Stat 2.03.
Sequences that had a statistically significant change in expression were identified by two-way repeated-measures analysis of variance (ANOVA) performed on the signal intensities reported by MAS 5.0. A P value of 0.05 or less was taken as an indication of statistical significance.
To identify the source(s) of the differences detected by two-way ANOVA, signal intensities at each time point were subsequently analyzed by one-way ANOVA. For sequences that displayed homogeneity of variance as defined by Levene’s test, ANOVA was performed using the equal variance assumption with the R statistical package; otherwise, the Oneway procedure in R was used. Post-hoc multiple group comparisons were then performed with SPSS using Tukey’s HSD for sequences that met the equal variance assumption and Dunnett’s C for sequences that did not.
The purpose of this study was to identify gene expression sequences that were affected by thermal stress. Accordingly, sequences were excluded from consideration if there was a statistically significant change over time that was entirely independent of thermal exposure condition (i.e., no main effect of exposure condition and no interactive effect between time and exposure condition). Only 5.7% of sequences (see “Results” section) were excluded based on this criterion.
Additionally, where noted in this manuscript, we excluded sequences from further consideration if they did not demonstrate a twofold or greater change in expression at one or both of the time points studied.
Ontological analysis
We performed ontological analysis of the genes affected by thermal stress by both manual and computer-assisted methods.
In the first method, each sequence of interest was assigned a functional class manually using resources available at the NCBI web site (http://www.ncbi.nlm.nih.gov), such as Entrez Gene and Online Mendelian Inheritance in Man.
In a second (computer-assisted) method, we used Onto-Express (Draghici et al. 2003; Khatri et al. 2002) to look for functional categories specifically affected by hypothermia and heat shock. Onto-Express assigns Gene Ontology Annotations (the European Bioinformatics Institute; Ashburner et al. 2000) to each sequence significantly affected by the stress of interest and to each sequence represented on the microarray. It then looks for functional categories that are statistically overrepresented in the dataset. Because we were interested in a side-by-side comparison of genes affected immediately after thermal stress (T = 0) and after recovery at 37°C (T = 3 h), a corrected P value of 0.025 or less was considered statistically significant.
As in other automated ontology tools, it is possible for an individual gene to be represented in more than one high-level ontological category. Also, because Onto-Express does not cluster categories (the way tools such as DAVID does), it is possible for several top-level categories to be functionally similar (e.g., “RNA splicing” and “RNA splicing via spliceosome”). Because each individual top-level category is assigned a unique P value by the algorithm (for reasons noted above), we chose not to manually combine categories for purposes of this report.
Third, we performed KEGG pathway analysis using DAVID 6.0 (http://david.abcc.ncifcrf.gov/home.jsp) on all genes significantly affected by cold stress at each of the time points under study. In this analysis, as in the Onto-Express Analysis, gene sequences were included regardless of magnitude of effect or direction of change. We used the default settings to identify affected pathways (EASE P value ≤0.1, two or more genes affected per pathway).
Confirmation of key findings by reverse transcription PCR
Reverse transcription PCR was performed as described previously (Sonna et al. 2002b) using the Retroscript First-Strand Synthesis Kit (Ambion, Austin, TX, USA), Taq polymerase and buffers obtained from Promega (Madison, WI, USA), and primer sequences as listed in Supplemental Table 1.
Results
Confirmation of gene expression responses to cold stress and heat shock
Samples from six independently performed experiments were submitted for microarray analysis (a total of 36 gene chips; 3 conditions per experiment × 2 time points per experiment × 6 experiments = 36 chips). The temperatures measured during these six experiments were (mean ± SD): cold stress, 31.0 ± 0.2°C (n = 12, or two measurements per experiment); heat shock, 43.1 ± 0.1°C (n = 12); control, 37.0 ± 0.1°C (n = 18, or three measurements per experiment); and recovery, 37.0 ± 0.1°C (n = 6). Cell survival was measured by the Trypan blue assay both at the end of the thermal stress period and again after recovery and ranged from 94.5% to 97.4%.
Figure 1 provides a representative example of the heat- and cold-induced changes in the expression of control genes (as ascertained by RT-PCR) that occurred in the sample sets submitted for microarray analysis. There was a noticeable increase in expression of CIRBP after cold stress but not after heat shock. By contrast, HSP70B’ showed a substantial increase in expression after heat shock but not hypothermia and furthermore, demonstrated maximal expression after a period of recovery at 37°C. Expression of cyclophilin A appeared to be uniform under all conditions tested.
Fig. 1.
Reverse transcription PCR showing the expression responses to heat shock and cold stress from one of the independent experiments submitted for microarray analysis. The numbers (0, 3) denote the recovery time at 37°C, in hours, after heat shock (43°C × 30 min) or cold stress (31°C × 16 h). CIRBP cold-induced ribonuclear binding protein, HSP heat shock protein
Thermal stress produced extensive gene expression changes that included a large component of downregulation
Of 22,283 sequences on the U133A array, 6,609 (29.7%) showed changes in expression that were both statistically significant and relevant to the experimental hypothesis, as determined by two-way repeated-measures ANOVA. Of these, 3,572 (54%) showed an effect of thermal stress condition only (a “main effect” of heat, cold, or control temperature), 1,407 (21%) showed both a main effect and an interactive effect between time and thermal stress condition, and 1,630 (25%) showed only an interactive effect. Another 1,275 (5.7%) of the 22,283 sequences on the array showed only a significant effect of time (i.e., no main effect and no interactive effect). This last number corresponds closely to what would be expected by random chance alone and represents changes that occurred independently of thermal stress (and therefore not of direct relevance to the experimental hypothesis). Accordingly, these 1,275 sequences were excluded from further consideration.
One-way ANOVA was performed at each of the time points to help identify the source(s) of the differences found by two-way ANOVA. This analysis revealed that, of the 6,609 sequences identified by two-way ANOVA, 5,379 also showed a statistically significant effect of temperature (heat, cold, control) at one or both of the time points examined. Post-hoc analysis found a significant difference between controls and at least one of the thermal stress conditions (heat, cold) for 4,253 sequences, of which 3,256 were affected by cold stress at one or both time points and 1,749 by heat shock.
Tables 1 and 2 and Fig. 2 present complimentary displays of how the 4,253 sequences in this experiment were affected by time and thermal stress; Table 1 lists affected sequences by time and thermal condition, Table 2 parses these sequences into those with increased and decreased expression, and Fig. 2 presents a graphical display that allows a visual comparison of the number of sequences affected by the alternative forms of thermal stress. Because many sequences fell into more than one category (for example, some were affected both by heat and by cold at a given time point or were affected by a given stress at both time points), the totals in the tables and in the figure add up to more than 4,253. Unless otherwise specified, the magnitude of change was not specifically accounted for in these presentations of the data; so, for example, a gene whose expression was significantly increased both by heat and by cold at T = 0 would be listed as “similarly affected by heat and cold,” regardless of the actual magnitude of the changes.
Table 1.
Pattern of expression of the sequences affected by cold stress and heat shock
| Pattern of expression | During thermal stress (T = 0 h) | After recovery (T = 3 h) | |||||
|---|---|---|---|---|---|---|---|
| SignificantaN = 2,987 | Significantly affected ≥twofold | High certaintyb | SignificantaN = 2,632 | Significantly affected ≥twofold | High certaintyb | ||
| Affected only by cold | Total | 2,357 | 381 | 323 | 1,310 | 162 | 98 |
| Increased | 1,222 | 138 | 114 | 748 | 73 | 51 | |
| Decreased | 1,135 | 243 | 209 | 562 | 89 | 47 | |
| Affected only by heat | Total | 389 | 52 | 24 | 975 | 195 | 128 |
| Increased | 144 | 30 | 14 | 362 | 126 | 87 | |
| Decreased | 245 | 22 | 10 | 613 | 69 | 41 | |
| Similarly affected by cold and heat | Total | 160 | 12 | 296 | 13 | ||
| Increased | 21 | 2 | 50 | 8 | |||
| Decreased | 139 | 10 | 246 | 5 | |||
| Oppositely affected by cold and heatc | Total | 81 | 3 | 3 | 51 | 0 | 0 |
| Cold up–heat down | 54 | 0 | 0 | 48 | 0 | 0 | |
| Cold down–heat up | 27 | 3 | 3 | 3 | 0 | 0 | |
| Cold totals | Significantly different from controls | 2,598 | 409d | 1657 | 192d | ||
| Increased | 1,297 | 143d | 846 | 83d | |||
| Decreased | 1,301 | 266d | 811 | 109d | |||
| Heat totals | Significantly different from controls | 630 | 71d | 1322 | 231d | ||
| Increased | 192 | 35d | 415 | 146d | |||
| Decreased | 438 | 36d | 907 | 85d | |||
aImplies that a significant difference was found both by two-way and one-way ANOVA and that post-hoc comparisons showed a significant difference between control and one or both of the thermal stress conditions
bImplies that there was also a significant difference between responses to heat and cold in the post-hoc comparisons
cAll of these showed, in the post-hoc analysis, a significant difference between control conditions and each of the thermal conditions tested, as well as a significant difference between heat and cold
dComputed without regard to the magnitude of the effect of the opposite thermal stimulus and therefore can add up to more than the sum of the previous categories
Table 2.
Effect of time on gene expression responses to moderate hypothermia and heat shock
| Direction of response | Timing of response | Hypothermia | Heat shock | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Increased | Thermal stress only (T = 0) | 828 | 25.4% | 149 | 8.5% |
| Recovery only (T = 3) | 366 | 11.2% | 373 | 21.3% | |
| Sustained (T = 0 and T = 3) | 464 | 14.3% | 40 | 2.3% | |
| Subtotal | 1,658 | 50.9% | 562 | 32.1% | |
| Decreased | Thermal stress only (T = 0) | 771 | 23.7% | 278 | 15.9% |
| Recovery only (T = 3) | 292 | 9.0% | 746 | 42.7% | |
| Sustained (T = 0 and T = 3) | 514 | 15.8% | 158 | 9.0% | |
| Subtotal | 1,577 | 48.4% | 1,182 | 67.6% | |
| Subtotal | 3,235 | 99.4% | 1,744 | 99.7% | |
| Bidirectional | Increased at T = 0, then decreased at T = 3 | 5 | 0.2% | 3 | 0.2% |
| Decreased at T = 0, then increased at T = 3 | 16 | 0.5% | 2 | 0.1% | |
| Subtotal | 21 | 0.6% | 5 | 0.3% | |
| Total | 3,256 | 100% | 1749 | 100% | |
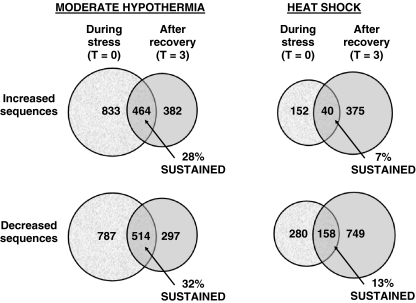
Fig. 2.
Venn diagrams illustrating the time dependence of gene expression after moderate hypothermia and heat shock. Only a minority of sequences at each time point showed sustained increases or decreases in expression
Cold stress and heat shock produced different expression patterns
Table 1 compares the number of sequences affected by hypothermia to those affected by heat shock. The gene expression response to cold stress was somewhat more extensive than the response to heat shock, as evidenced by the observation that hypothermia affected more sequences than did heat shock at both time points examined. The difference in the number of sequences affected was greater at T = 0, the end of the thermal stress period, than after 3 h of recovery at 37°C (Table 1).
Additionally, the total number of sequences affected by heat shock increased during recovery at 37°C (as expected), whereas the number of sequences affected by hypothermia decreased over the same time period. These differences held true even when we excluded sequences that showed changes in expression of less than twofold relative to controls (Table 1).
The gene expression responses to both thermal stress conditions also included substantial components of decreased expression. About half of the sequences affected by cold stress and about two thirds of the sequences affected by heat shock showed decreased expression relative to controls (Table 1).
The sequences affected by each thermal stress varied over time
Cold stress and heat shock produced time-dependent changes in expression. As noted previously, slightly less than half of the sequences identified by the two-way ANOVA analysis showed evidence of an effect of time on gene expression. Similar conclusions were reached in the one-way ANOVA analysis, as illustrated in detail in Table 2 and Fig. 2. Of the 4,253 sequences identified as showing expression that was significantly different from controls at one or both time points, 3,256 were affected by cold stress and 1,749 by heat shock. However, less than one third (978 of 3,256) of the sequences affected by hypothermia and less than one eighth (198 of 1,749) of the sequences affected by heat shock showed sustained changes in expression (i.e., significant changes in expression in the same direction at both of the time points studied, Fig. 2). Very few sequences (<1%) exhibited a bidirectional response to thermal stress (i.e., significantly decreased at one time point, increased at the other, Table 2).
These expression patterns showed greater overlap during the recovery period
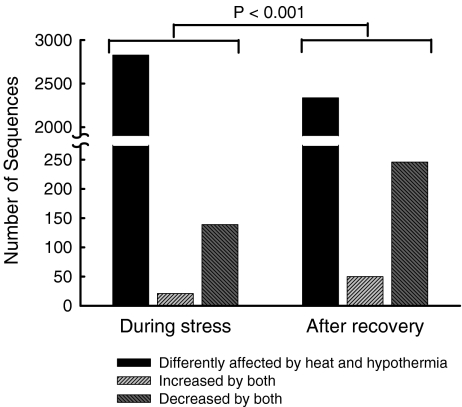
Some sequences showed changes in the same direction (increased or decreased) in response to both cold stress and heat shock (Table 1 and Fig. 3). The proportion of sequences displaying congruent changes in expression roughly doubled during recovery at 37°C. Specifically, 5% (160 of 2,987) of the significantly affected sequences showed changes in the same direction immediately after cold stress and heat shock, as compared to 11% (296 of 2,632) after 3 h of recovery at 37°C.
Fig. 3.
Effect of heat shock and hypothermia on gene expression. The ordinate represents the number of sequences affected in each category. Sequences were said to be similarly affected by the two thermal stress conditions if both hypothermia and heat shock produced statistically significant differences from control expression in the same direction (increased or decreased) The P-value was computed by Chi-square
Effect of thermal stress on negative and positive control sequences
Several important negative control sequences corresponding to genes that are generally unaffected by a wide variety of different stimuli (“housekeeping genes”) were likewise unaffected by thermal stress in this experiment. Supplemental Table 2 details the effect of moderate hypothermia and heat shock on a list of such control sequences, including beta-actin, GAPDH, cyclophilin A, a number of other sequences found in previous gene chip array experiments to demonstrate stable expression after heat shock (Sonna et al. 2002b) and hypoxia (Sonna et al. 2003), and several of the ribosomal proteins most highly expressed in this cell line as determined by signal intensity on the microarrays. Of the 19 genes represented by the 25 sequences in this table, only three (beta-actin, GAPDH, and ribosomal protein L30) showed statistically significant changes in expression by two-way ANOVA. However, the magnitudes of the expression changes were very small; none of the geometric mean expression ratios in these three genes exceeded 1.2-fold.
Additionally, sequences corresponding to genes known to respond to thermal stress (positive controls) showed significant changes in expression. Specifically, sequences corresponding to the cold-responsive genes CIRBP and RBM3 were increased in response to hypothermia. Two sequences corresponding to CIRBP were increased 4.8-fold and 3.5-fold, respectively, during cold stress; these sequences were increased 3.5-fold and 2.7-fold after recovery. A sequence corresponding to RBM3 was increased 2.5-fold during cold stress and 2.4-fold after recovery. None of these sequences were significantly affected by heat. Likewise, as discussed below, many sequences corresponding to heat shock proteins (including HSP70B’) showed increases in expression in response to heat shock.
Effect of thermal stress and recovery on expression of heat shock proteins
The two-way ANOVA analysis identified 76 sequences corresponding to heat shock proteins (HSPs), chaperonins, and co-chaperonins that showed statistically significant changes in expression. Subsequent one-way ANOVA with post-hoc analysis revealed a source of the difference(s) for 67 of these sequences.
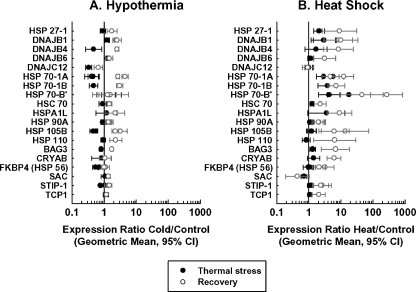
Heat shock and hypothermia produced different effects on the expression patterns of sequences corresponding to HSPs and related proteins. Whereas heat shock typically produced increases in HSP mRNA expression that became larger and more extensive after 3 h of recovery at 37°C, hypothermia often had an inhibitory response on HSP expression, followed by a mild increase in HSP expression during the recovery period. Specifically, heat shock produced increases in the expression of 15 sequences during thermal stress and of 37 sequences after recovery. One HSP sequence (corresponding to sacsin) was slightly (though significantly) decreased by heat shock during thermal stress (expression ratio 0.70) and two (corresponding to sacsin and DNAJA3) decreased during recovery (expression ratios, 0.44 and 0.75, respectively). By contrast, moderate hypothermia led predominantly to decreased HSP expression at the end of thermal stress (16 decreased sequences, as compared to five increased sequences), followed by increases in expression relative to controls after recovery (a total of 27 increased sequences, as compared to three decreased sequences). A total of 16 sequences were significantly increased both by moderate hypothermia and heat shock after 3 h of recovery at 37°C.
Figure 4 illustrates these effects in the subset of HSPs most strongly affected by heat shock or hypothermia (i.e., sequences, which, among other criteria, showed expression ratios ≥2 or ≤0.5).1
Fig. 4.
Effect of hypothermia and heat shock on the expression of selected HSPs, chaperonins, and co-chaperonins. For illustrative purposes, the sequences shown were significantly affected at one or more time points in the ANOVA analysis had a geometric mean expression ratio of twofold or greater (0.5 or less for decreased sequences) at one or more of the significant time point(s) and had at least one set of 95% confidence intervals that excluded unity. Some genes were represented by more than one sequence and thus have more than two data points per line
Effect of thermal stress on expression of heat shock factors
Hypothermia also had an effect on the expression of a sequence corresponding to heat shock factor-1 (HSF-1), the major transcription factor responsible for increased expression of HSPs during thermal stress (Cotto and Morimoto 1999). Hypothermia produced a small but statistically significant decrease in expression of a sequence corresponding to heat shock factor-1 (expression ratio 0.74) during thermal stress that was no longer significantly different from control after recovery. By contrast, heat shock did not produce any changes in HSF-1 RNA expression.
A sequence corresponding to heat shock factor-2 was significantly decreased by a small amount (expression ratio 0.63) after recovery from heat shock but not affected at either time point by moderate hypothermia.
We did not identify any sequences corresponding to heat shock factor-4 that were significantly affected by heat shock.
Biological processes affected by thermal stress
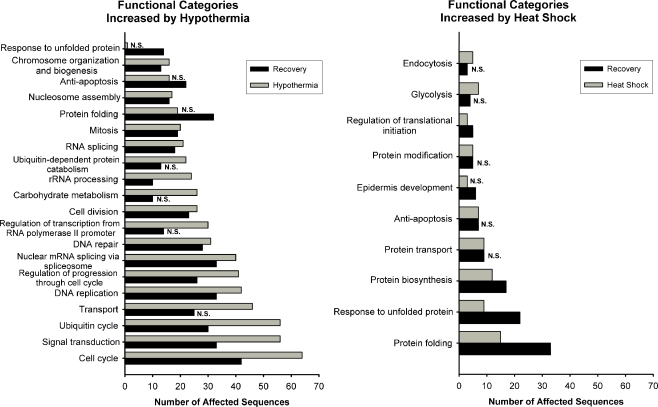
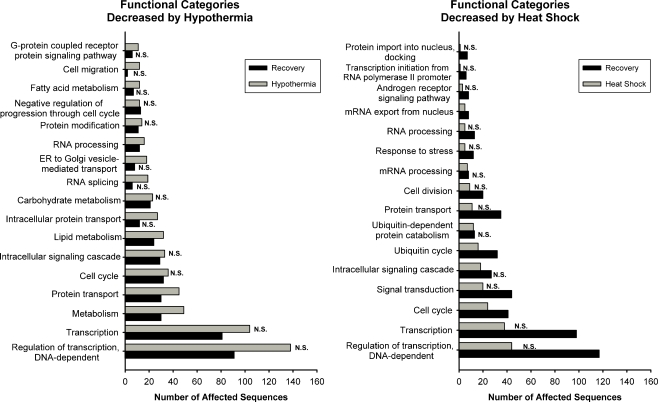
We used the Onto-Express software package (Draghici et al. 2003) to gain additional insight into the biological processes affected by hypothermia and rewarming. Figure 5 presents the biological processes most strongly affected by genes whose expression were increased during or after thermal stress, and Fig. 6 presents those which were decreased. These figures only depict pathways that are statistically overrepresented at one or both of the time points examined. Because the total number of increased sequences was different for each combination of thermal stress and time (Tables 1 and 2), it is possible for two time points to have the same number of affected sequences in a given biological process and yet for one not to be statistically significant. Furthermore, because the categories were generated by a statistical algorithm, some of them are overlapping or are subsets of others (e.g., “lipid metabolism” and “metabolism”). Because of this, it is possible for a gene to be represented in more than one category and as might be expected, it is also possible for multifunctional genes to be included in more than one of the categories presented. The Gene Ontology Annotation numbers corresponding to the categories shown, and their consensus definitions, are listed in Supplemental Table 3.
Fig. 5.
Functional categories most highly affected by increases in gene expression after heat shock and hypothermia. N.S. the number of affected sequences in that category at the time point in question does not differ from what would be expected from a random sampling of sequences from the microarray (i.e., P > 0.025)
Fig. 6.
Functional categories most highly affected by decreases in gene expression after heat shock and hypothermia. N.S. the number of affected sequences in that category at the time point in question does not differ from what would be expected from a random sampling of sequences on the microarray (i.e., P > 0.025)
As might be expected, the sequences increased by heat shock affected biological processes known to be involved in protein biosynthesis, folding, and in the response to unfolded protein; furthermore, the largest effect was seen after a period of recovery at 37°C (Fig. 5). By contrast, and consistent with our manual analysis of HSPs (above), hypothermia did not significantly affect the response to unfolded protein until after recovery at 37°C. Hypothermia-induced genes heavily affected pathways involved in cell replication (including cell cycle, DNA replication, mitosis, etc.), signal transduction, and the ubiquitin cycle. We also detected substantial effects of hypothermia on pathways involved in RNA splicing and processing.
A similar analysis was performed on genes whose expression was decreased by heat shock and hypothermia (Fig. 6). Pathways involved in transcription and the regulation of transcription were the ones most extensively affected both by heat shock and hypothermia, but this association was only significant after 3 h of recovery at 37°C. A gene expression signature of the metabolic response to hypothermia was clearly detected. Hypothermia led to decreased expression of sequences of key metabolic pathways, including those of lipid metabolism and carbohydrate metabolism (Fig. 6).
An analysis of KEGG pathways affected by hypothermia was performed using DAVID version 6 (Table 3). Consistent with the results of our Onto-Express analysis, a strong effect of hypothermia was noted on pathways involved in cell cycle, signal transduction (including the p53 signaling pathway, the insulin signaling pathway, and the phosphatidylinositol signaling system), ubiquitin-mediated proteolysis, and several metabolic pathways.
Table 3.
KEGG Pathways affected by hypothermia
| KEGG pathway number | KEGG pathway name | During cold stress (T = 0) | After recovery (T = 3) | ||
|---|---|---|---|---|---|
| No. of affected genes | P | No. of affected genes | P | ||
| hsa04110 | Cell cycle | 42 | <0.001 | 28 | <0.000 |
| hsa04120 | Ubiquitin-mediated proteolysis | 37 | <0.001 | 25 | 0.001 |
| hsa00230 | Purine metabolism | 27 | 0.072 | 22 | 0.018 |
| hsa04520 | Adherens junction | 20 | 0.012 | 17 | 0.003 |
| hsa05222 | Small cell lung cancer | 21 | 0.044 | 15 | 0.066 |
| hsa04115 | p53 signaling pathway | 17 | 0.026 | 14 | 0.010 |
| hsa04910 | Insulin signaling pathway | 29 | 0.033 | ||
| hsa05131 | Pathogenic Escherichia coli infection (EPEC) | 15 | 0.012 | 13 | 0.003 |
| hsa05130 | Pathogenic Escherichia coli infection (EHEC) | 15 | 0.012 | 13 | 0.003 |
| hsa00240 | Pyrimidine metabolism | 23 | 0.001 | 17 | 0.003 |
| hsa04070 | Phosphatidylinositol signaling system | 19 | 0.019 | ||
| hsa05212 | Pancreatic cancer | 18 | 0.054 | ||
| hsa05220 | Chronic myeloid leukemia | 17 | 0.087 | ||
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 17 | 0.034 | ||
| hsa03320 | PPAR signaling pathway | 17 | 0.039 | ||
| hsa01030 | Glycan structures (biosynthesis 1) | 16 | 0.034 | ||
| hsa00330 | Arginine and proline metabolism | 9 | 0.084 | 7 | 0.087 |
| hsa00562 | Inositol phosphate metabolism | 15 | 0.007 | ||
| hsa05214 | Glioma | 11 | 0.083 | ||
| hsa05110 | Cholera (infection) | 11 | 0.053 | ||
| hsa03050 | Proteasome | 10 | 0.004 | ||
| hsa00020 | Citrate cycle (TCA cycle) | 9 | 0.039 | ||
| hsa00860 | Porphyrin and chlorophyll metabolism | 8 | 0.067 | ||
| hsa00220 | Urea cycle and metabolism of amino groups | 8 | 0.018 | ||
| hsa00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 4 | 0.043 | ||
| hsa00130 | Ubiquinone biosynthesis | 4 | 0.053 | ||
The number of affected genes includes genes whose expression was significantly increased or decreased at each of the time points studied relative to controls
Effect of thermal stress on other sequences
Supplemental Tables 4–7 present lists of genes not traditionally considered heat shock proteins (“non-HSPs”) whose expression was maximally affected by moderate hypothermia and by heat shock. The lists of genes in these tables again suggest that heat shock and moderate hypothermia produced contrasting effects on gene expression. Importantly, some of the expression changes in non-HSPs observed have been previously reported in studies of thermal stress. As noted previously, hypothermia produced changes in expression of genes widely acknowledged to be cold responsive, including CIRBP and RBM3. Likewise, the non-HSPs affected by heat stress included several genes previously found to be highly heat responsive in a previous study in peripheral blood mononuclear cells (Sonna et al. 2002b), including Rad (Supplemental Table 7), the phosphatidylserine receptor (Supplemental Table 7), and NF-kappa B repressing factor (increased after recovery from heat stress by 2.3-fold but unaffected by moderate hypothermia).
Comparison to the responses of THP-1 cells
It is generally acknowledged that cellular responses to heat involve a number of changes that are likely shared by many different cell types (Kregel 2002; Lindquist 1986; Parsell and Lindquist 1993; Sonna et al. 2002a). However, the extent to which cold-induced changes in expression are similarly independent of cell type is not well described. We compared the findings in HepG2 cells to sequences that were identified in previous work as being affected by moderate hypothermia (32°C × 24 h) in the acute monocytic leukemia cell line THP-1 (Sonna et al. 2006). Because the experiments with THP-1 cells did not involve rewarming, only the changes in expression at the end of cold exposure in HepG2 cells were used for comparison.
Of the 1,297 sequences that showed statistically significant increases in expression by moderate hypothermia in HepG2 cells at the end of thermal stress (Table 1), 456 (35%) were also significantly increased in THP-1 cells (irrespective of post-hoc filter criteria). Of the 143 sequences that showed significant expression ratios of twofold or greater in HepG2 cells as a result of moderate hypothermia, 51 were also significantly increased in THP-1 cells (irrespective of magnitude), of which 12 also met all post-hoc expression call filter criteria in the THP-1 cells (a change of twofold or greater, expression calls of “present” or “marginal” in at least half of the cells exposed to hypothermia). By contrast, of these 143 sequences, only three showed statistically significant decreases in expression in THP-1 cells (P < 0.001 by z test of proportions with Yates’ correction).
Of the 1,301 sequences that showed statistically significant decreases in expression at the end of moderate hypothermia in HepG2 cells, 484 (37%) were also significantly decreased in THP-1 cells (irrespective of post-hoc filter criteria). Of the 266 sequences in HepG2 cells that showed significant expression ratios of 0.5 or less, 96 were also significantly decreased in THP-1 cells (irrespective of magnitude), of which 13 also met the post-hoc filter criteria in THP-1 cells outline above. By contrast, of these 266 sequences, only 11 were increased by moderate hypothermia in THP-1 cells (P < 0.001 by z test of proportions with Yates’ correction).
Thus, 25 sequences (12 increased, 13 decreased) representing 23 genes showed changes in expression in both cell lines that were statistically significant and that also met the post-hoc filter criteria of the respective studies, including geometric mean changes in expression of at least twofold, Table 4. Importantly, the sequences in this consensus list include both CIRBP and RBM3, which are known to be increased by moderate hypothermia (Danno et al. 1997; Nishiyama et al. 1997a, b; Sonna et al. 2002a). Additionally, HSP70-1 was significantly decreased during the period of cold exposure in both experiments (though as noted previously, HSP70-1 showed increases in expression upon rewarming).
Table 4.
Nonspecific cold response genes
| Functional Class | Common name(s) | Identifier | Expression ratio cold/control | ||
|---|---|---|---|---|---|
| HepG2 Cells (geometric mean) | THP-1 Cells (geometric mean, 95% CI) | ||||
| Decreased by cold | |||||
| Apoptosis | TIA1; TIA1 cytotoxic granule-associated RNA-binding protein | AL046419 | 0.46 | 0.48 | (0.39–0.59) |
| DNA conformation and repair | RAD50; RAD50 homolog (Saccharomyces cerevisiae) | NM_005732 | 0.33 | 0.49 | (0.40–0.59) |
| HSPs, chaperonins, and co-chaperonins | HSPA1A; Heat shock 70 kD protein 1A; HSP 70-1 | NM_005345 | 0.41 | 0.37 | (0.26–0.53) |
| NM_005345 | 0.45 | 0.36 | (0.25–0.50) | ||
| HSPH1; Heat shock 105/110 kDa protein 1; HSP105B; heat shock 105 kD; HSP 105; KIAA 0201 | NM_006644 | 0.45 | 0.29 | (0.25–0.33) | |
| Membrane protein | TM7SF3; transmembrane 7 superfamily member 3 | NM_016551 | 0.32 | 0.41 | (0.27–0.64) |
| Membrane traffic and receptor sorting | SORL1; Sortilin-related receptor, L(DLR class) A repeats-containing | AV728268 | 0.29 | 0.49 | (0.40–0.62) |
| Metabolism and biosynthesis | HIBCH; 3-hydroxyisobutyryl-coenzyme A hydrolase | NM_014362 | 0.50 | 0.44 | (0.23–0.85) |
| GART; phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase; PGFT; PRGS | NM_000819 | 0.43 | 0.46 | (0.36–0.60) | |
| Unknown | CHORDC1; cyteine and histidine-rich domain (CHORD)-containing, zinc binding protein 1; CHP1 | NM_012124 | 0.45 | 0.37 | (0.27–0.50) |
| KIAA0436; putative prolyl oligopeptidase; FLJ16627 | AW000954 | 0.48 | 0.44 | (0.31–0.63) | |
| BTN3A3; butyrophilin, subfamily 3, member A3 | NM_006994 | 0.48 | 0.45 | (0.29–0.68) | |
| UBPH; similar to ubiquitin binding protein; ubiquitin binding protein homology | NM_019116 | 0.46 | 0.49 | (0.36–0.67) | |
| Increased by cold | |||||
| Cell growth, proliferation, and differentiation | PPM1B; protein phosphatase 1B (formerly 2C), magnesium-dependent, beta-isoform | AJ271832 | 2.12 | 2.20 | (1.63–2.96) |
| CTNNBIP1; Catenin, beta-interacting protein 1; ICAT | NM_020248 | 2.24 | 4.16 | (2.41–7.16) | |
| Cytoskeleton | TMEM4; transmembrane protein 4; MSAP; MIR-interacting saposin-like protein | BC001027 | 2.56 | 2.06 | (1.69–2.51) |
| Protein degradation | PSMD11; proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | BF432873 | 16.20 | 5.66 | (2.37–13.54) |
| RNA binding | RBM3; RNA binding motif (RNP1, RPM) protein 3 | NM_006743 | 2.50 | 2.10 | (1.49–2.97) |
| CIRBP; cold inducible RNA binding protein; CIRP | NM_001280 | 3.54 | 2.72 | (2.22–3.35) | |
| NM_001280 | 4.76 | 3.72 | (2.91–4.76) | ||
| Signal transduction | NUDT3; nudix (nucleoside diphosphate linked moiety X)-type motif 3 | AK025759 | 2.08 | 2.13 | (1.86–2.45) |
| Unknown | C6orf62; chromosome 6 open reading frame 62 | AW972292 | 2.54 | 3.63 | (2.29–5.75) |
| DKFZP434C171; DKFZP434C171 protein | AL080169 | 4.60 | 2.57 | (1.63–4.07) | |
| CDNA FLJ11898 fis, clone HEMBA1007322 | AK021960 | 4.46 | 4.90 | (2.51–9.56) | |
| Homo sapiens (clone B3B3E13) Huntington’s disease candidate region mRNA fragment | L37198 | 5.31 | 8.07 | (3.81–17.12) | |
Included are all genes that showed statistically significant, twofold or greater, changes in expression in both HepG2 and THP-1 cells during cold exposure and that also met the post-hoc filter criteria of the respective studies
Confirmatory reverse transcription PCR
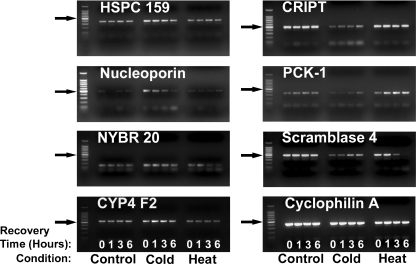
Figure 7 presents qualitative confirmatory experimental data that illustrate the time course of the change in expression of several of the genes identified that were affected by moderate hypothermia and heat shock in our microarray studies. Our objective here was to confirm only change in direction and to confirm qualitative differences in the responses to heat shock and hypothermia, not to perform a formal correlation between magnitudes of change observed on the microarray and by PCR (which would require qrPCR).
Fig. 7.
Confirmation of selected microarray findings by reverse transcription PCR. Cells were subjected to heat shock or cold stress and allowed to recover at 37°C for 0, 1, 3, or 6 h. Arrows point to the bright 500 bp ladder band
Cells in culture were exposed to thermal stress conditions as described previously and allowed to recover at 37°C for varying amounts of time, at which point RNA was isolated and subjected to expression analysis by conventional reverse transcription PCR. The directions of the changes observed were congruent with the findings made by microarray. Additionally, as predicted by the microarray, the effects of heat shock on the expression of these genes differed noticeably from the effects of moderate hypothermia in the direction of change, the time course, or both.
Discussion
We found that moderate hypothermia followed by rewarming led to extensive alterations in the mRNA expression signature of human HepG2 cells as compared to normothermic controls. The changes included a component that differs substantially from the heat shock response, most evident during the period of hypothermia itself, and a mild heat shock signature response that occurred after rewarming. The changes also included a large component of decreased gene expression; however, the actual genes affected and the functional pathways in which they were involved differ substantially from those that are decreased by heat shock. These findings suggest that cellular thermal history can be deduced from the resulting temporal changes in expression signature.
There are mechanistic implications to the similarities and differences between the gene expression responses observed after moderate hypothermia and heat shock. For example, although both heat shock and hypothermia produced widespread changes in gene expression during the period of thermal stress, the specific genes affected differed substantially between the two thermal conditions, as did the functional pathways identified. These findings suggest that the mechanisms involved in gene expression during thermal stress likely differ substantially between heat shock and moderate hypothermia. By contrast, the observation that the gene expression response to rewarming includes a mild heat shock response raises the possibility that there may be some common mechanisms involved in gene expression during recovery from hypothermia and heat shock. This concept is supported by studies of hypothermia in the cryopreservation range (Fuller 2003) that have reported increases in HSP and stress protein expression after periods of profound hypothermia too brief to trigger cold-specific changes in expression. It thus seems plausible that heat shock and hypothermia affect gene expression by largely distinct mechanisms during the period of thermal stress itself but share potentially important common mechanistic elements during the period of recovery. This leads to the hypothesis that some of the beneficial effects of therapeutic hypothermia might be related to the ability of rewarming to produce a cytoprotective heat shock response without subjecting the individual to the potentially deleterious elevations in core temperature that would otherwise be required. Studies in animal models of hypothermia/rewarming will be required to test this therapeutically relevant possibility.
The differing effects of hypothermia/rewarming and heat shock on gene expression become even more apparent when the analysis is confined to HSPs (Fig. 3). As noted, moderate hypothermia led to an initial decrease in expression of many HSPs. It is interesting to note that expression of mRNA encoding the transcription factor heat shock factor-1 was also decreased during hypothermia. Because heat shock factor-1 is known to be responsible for increased expression of many HSPs during thermal stress (Cotto and Morimoto 1999), this observation suggests one of several candidate mechanisms by which hypothermia might produce coordinated, decreased expression of multiple HSPs. Increased HSP-1 expression during rewarming after moderate hypothermia represents a coordinating candidate mechanism for promoting the cytoprotective effects of heat shock proteins in the context of therapeutic hypothermia. These findings imply that studies of heat shock factor-1 expression and activity during hypothermia might yield important insights into the mechanism by which mild hypothermia affects gene expression.
It has recently been reported, in a rat liver model of ischemia–reperfusion, that recovery from hypothermia in the absence of ischemia does not trigger increases in protein levels of HSP-70 or -32 (Niemann et al. 2010). However, the total duration of hypothermia used in this animal model (<2 h) was much shorter than what is typically used to trigger changes in expression of the hypothermia-induced genes CIRBP and RBM3 (Danno et al. 1997; Nishiyama et al. 1997a, b) and was also much shorter than the duration of hypothermia used to produce gene expression changes in both our previous study of THP-1 cells (24 h) and in the present study (16 h). It may be that a relatively short period of hypothermia is sufficient to modify the effects of other stressors (such as ischemia–reperfusion) but not sufficient in and of itself to trigger changes in gene expression.
A comparison of the findings in HepG2 cells to the effects of 24 h of moderate hypothermia (without rewarming) in THP-1 cells identified 23 genes whose response to moderate hypothermia is substantial and is shared by cell types of distinct and divergent origin (Table 3). This includes the well-established cold response genes CIRBP and RBM3, as well as genes involved in cell proliferation and differentiation, metabolism, and signal transduction. It is therefore possible that some of the systemic effects of hypothermia might be mediated by widespread effects on the expression of genes that affect metabolism, cell cycle, and signal transduction, whereas others are likely to be the result of more cell-specific responses to hypothermia.
A significant criticism of the use of microarrays is their propensity to generate false–positive reports, given the large numbers of sequences examined simultaneously. In previous experiments comparing a single stress condition to a control, we used T-distribution-derived 95% confidence intervals on natural log-transformed expression ratios to identify statistically significant changes in expression, followed by aggressive post-hoc filtering to reduce the number of false–positive reports (Sonna et al. 2003, 2002b; Wood et al. 2004). While this approach is highly efficient at generating findings that can be reproduced by reverse transcription PCR, it cannot easily be applied to multi-factorial experimental designs such as the one reported here. In the present experiment, we first used two-way ANOVA to reduce the number of genes for further analysis by about 70% (from ∼22,400 to ∼6,600), then used additional, rigorous ANOVA-based techniques to identify the sources of difference across experimental conditions. The analytical approach used here produced findings that are consistent with the published literature, including the observation of increased expression of CIRBP and RBM3 after hypothermia, the pattern of HSP expression after heat shock (an initial increase in expression during thermal stress that broadens and intensifies during recovery) and the observation that rewarming after hypothermia leads to a response that has some overlap with the heat shock response. Additionally, we were able to confirm, by reverse transcription PCR, the responsiveness to moderate hypothermia of a number of the genes identified by the microarray. Finally, the observation that many “housekeeping” genes were unaffected adds to our confidence that the analytical approach used is in fact capable of reliably distinguishing changes in expression that occur as a result of thermal stress from those that occur by chance alone. Altogether, these considerations suggest that many of the changes in expression identified here reflect responses that will prove to be replicable. We have made available the analytical tools used in our studies of thermal intensity (Khatri et al. 2006).
There are several important limitations to the experimental results reported here. We used a hepatocellular tumor cell line to limit variance between experiments and thus reduce the number of arrays needed to identify significant findings. Although some of the changes detected are likely to be generalizable, it is likely that others will prove to be idiosyncratic to hepatocytes or to tumor cell lines. Additionally, the degree of hypothermia applied may not be bioequivalent to the intensity of the heat shock applied. Although both of the temperatures applied are within the survivable range for hepatocytes in culture, it is possible that a greater overlap between heat shock and moderate hypothermia responses would be observed at different intensities of thermal stress (with respect to temperature or duration of exposure). Furthermore, to minimize the confounding effects of unmeasured factors in serum, the experiments were performed in serum-free media. Although use of serum-free media did not prevent us from detecting expected changes in gene expression (such as HSPs after heat shock and increases in expression of CIRBP and RBM3 after hypothermia), this aspect of experimental design should be kept in mind when comparing the results of this study to data obtained in culture media containing serum or to results obtained in vivo. Temperature also alters cellular metabolic rate, and it is possible that some of the gene expression changes observed in the cells exposed to hypothermia for 16 h are the consequence of decreased rate of depletion of essential nutrients or altered rate of production of metabolites in the cell media, rather than a direct effect of hypothermia itself. Another potential source of experimental error involves the thermal and chemical stresses produced during the ∼10 min required to wash and trypsinize the cells prior to lysis for RNA extraction. This source of error would only affect genes capable of showing very rapid changes in expression and did not prevent us from detecting expected changes in expression of known heat- and cold-responsive genes. Yet another limitation of our results was the use of RT-PCR, which is qualitative in nature, rather than the use of quantitative real-time PCR. However, RT-PCR is adequate to detect very large changes in expression such as illustrated in Figs. 1 and 5. Finally, the post-hoc analyses applied cannot be expected to compensate entirely for the problem of simultaneous testing for significance of thousands of variables. We have dealt with this by reporting significance values (P values), to allow assessment of the likelihood that a given finding represents a false–positive response.
An important limitation of automated ontology tools, including Onto-Express, is that they rely on annotations of gene function in public databases that include categories that often appear to be overlapping. This can leave investigators with the difficult choice of presenting findings generated by the software as they are (which will reflect the fact that P values for each category were generated and used to include or exclude certain categories from the final report) or, alternatively, combining similar categories (and run the risk of statistical bias). In this manuscript, we chose to present the categories as they were generated by Onto-Express and have included their definitions in the supplemental materials to facilitate future analysis of the data.
In conclusion, the thermal exposure history of human HepG2 hepatocytes appears to be reflected in signature alterations in gene expression that occur as a function of time. Hypothermia to a degree similar to that used in therapeutic protocols results in distinct and significant changes in a large number of genes and includes a substantial inhibitory effect on gene expression, including well-studied families of genes such as HSPs. The genes affected differ substantially from those affected by heat shock, which suggests that the two thermal stresses produce changes in gene expression by different mechanisms during the period of thermal stress itself. Some of the gene expression changes induced by hypothermia, including induction of genes including ribonuclear binding proteins and the inhibitory effect of cold on HSP expression, have been reported in other human cell types, such as THP-1 cells (Sonna et al. 2006). By contrast, rewarming leads to a response that includes a heat shock gene expression signature, suggesting that there may be at least some common mechanistic elements involved in recovery from hypothermia and heat shock. Based on previous literature, heat shock factor-1 would appear worthy of investigation as a candidate mediator of these shared responses. Finally, our in vitro results suggest that future in vivo investigations of the effects of therapeutic hypothermia should consider the possibility that changes in gene expression might occur that could contribute to the observed effects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 502 kb)
Acknowledgments
The views, opinions, and findings contained in this publication are those of the authors and should not be construed as an official USUHS or US Department of the Army position, policy, or decision, unless so designated by other documentation.
Approved for public release; distribution unlimited.
Funding was provided by the US Army Medical Research and Materiel Command (USAMRMC). National Heart, Lung, and Blood Institute Grants HL-64104 and HL-072114 also supported this work.
The authors thank Dr. Michael Sawka of the US Army Research Institute of Environmental Medicine, Natick, MA, USA and Dr. Karen Fairchild of the University of Virginia, Charlottesville, VA, USA for critical review of this manuscript. The authors would also like to thank Dr. Jaques Reifman, whose input contributed to the development of the software used to analyze these data.
Footnotes
The figure thus does not include DNAJA3, whose largest significant change in expression was a decrease in expression of 0.75-fold relative to control at the T = 3 time point.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard SA, Gray TW, Buist MD, Jones BM, Silvester W, Gutteridge G, Smith K. Treatment of comatose survivors of out-of-hospital cardiac arrest with induced hypothermia. N Engl J Med. 2002;346:557–563. doi: 10.1056/NEJMoa003289. [DOI] [PubMed] [Google Scholar]
- Boss O, Samec S, Dulloo A, Seydoux J, Muzzin P, Giacobino JP. Tissue-dependent upregulation of rat uncoupling protein-2 expression in response to fasting or cold. FEBS Lett. 1997;412:111–114. doi: 10.1016/S0014-5793(97)00755-2. [DOI] [PubMed] [Google Scholar]
- Cotto JJ, Morimoto RI. Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem Soc Symp. 1999;64:105–118. [PubMed] [Google Scholar]
- Danno S, Nishiyama H, Higashitsuji H, Yokoi H, Xue JH, Itoh K, Matsuda T, Fujita J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- Denjean F, Lachuer J, Geloen A, Cohen-Adad F, Moulin C, Barre H, Duchamp C. Differential regulation of uncoupling protein-1, -2 and -3 gene expression by sympathetic innervation in brown adipose tissue of thermoneutral or cold-exposed rats. FEBS Lett. 1999;444:181–185. doi: 10.1016/S0014-5793(99)00056-3. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-tools, the toolkit of the modern biologist: onto-express, onto-compare, onto-design and onto-translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell RE., Jr . Determination of nucleic acid concentration and purity. RNA methodologies. San Diego: Academic; 1998. pp. 94–103. [Google Scholar]
- Fukui O, Kinugasa Y, Fukuda A, Fukuda H, Tskitishvili E, Hayashi S, Song M, Kanagawa T, Hosono T, Shimoya K, Murata Y. Post-ischemic hypothermia reduced IL-18 expression and suppressed microglial activation in the immature brain. Brain Res. 2006;1121:35–45. doi: 10.1016/j.brainres.2006.08.121. [DOI] [PubMed] [Google Scholar]
- Fuller BJ. Gene expression in response to low temperatures in mammalian cells: a review of current ideas. Cryo Lett. 2003;24:95–102. [PubMed] [Google Scholar]
- Ganta CK, Helwig BG, Blecha F, Ganta RR, Cober R, Parimi S, Musch TI, Fels RJ, Kenney MJ. Hypothermia-enhanced splenic cytokine gene expression is independent of the sympathetic nervous system. Am J Physiol Regul Integr Comp Physiol. 2006;291:R558–R565. doi: 10.1152/ajpregu.00846.2005. [DOI] [PubMed] [Google Scholar]
- Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- Khatri P, Chen D, Reifman J, Lilly CM, Sonna LA Software tool for analysis of variance of DNA microarray data. USARIEM Technical Report TR 07/05. 12-15-2006. Natick, MA, US Army Research Institute of Environmental Medicine
- Kobayashi MS, Asai S, Ishikawa K, Nishida Y, Nagata T, Takahashi Y. Global profiling of influence of intra-ischemic brain temperature on gene expression in rat brain. Brain Res Rev. 2008;58:171–191. doi: 10.1016/j.brainresrev.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Marion DW, Penrod LE, Kelsey SF, Obrist WD, Kochanek PM, Palmer AM, Wisniewski SR, DeKosky ST. Treatment of traumatic brain injury with moderate hypothermia. N Engl J Med. 1997;336(8):540–546. doi: 10.1056/NEJM199702203360803. [DOI] [PubMed] [Google Scholar]
- McIntyre LA, Fergusson DA, Hebert PC, Moher D, Hutchison JS. Prolonged therapeutic hypothermia after traumatic brain injury in adults: a systematic review. JAMA. 2003;289:2992–2999. doi: 10.1001/jama.289.22.2992. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Miura-Suzuki T, Yamashita H, Mori N. Distinct regulation of brain mitochondrial carrier protein-1 and uncoupling protein-2 genes in the rat brain during cold exposure and aging. Biochem Biophys Res Commun. 2000;278:691–697. doi: 10.1006/bbrc.2000.3859. [DOI] [PubMed] [Google Scholar]
- Niemann CU, Xu F, Choi S, Behrends M, Park Y, Hirose R, Maher JJ. Short passive cooling protects rats during hepatectomy by inducing heat shock proteins and limiting the induction of pro-inflammatory cytokines. J Surg Res. 2010;158:43–52. doi: 10.1016/j.jss.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama H, Higashitsuji H, Yokoi H, Itoh K, Danno S, Matsuda T, Fujita J. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997;204:115–120. doi: 10.1016/S0378-1119(97)00530-1. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Terao Y, Shintani Y, Kiyota Y. Therapeutic time window of post-ischemic mild hypothermia and the gene expression associated with the neuroprotection in rat focal cerebral ischemia. Neurosci Res. 2007;57:424–433. doi: 10.1016/j.neures.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Remick DG, Xioa H. Hypothermia and sepsis. Front Biosci. 2006;11:1006–1013. doi: 10.2741/1858. [DOI] [PubMed] [Google Scholar]
- Safar PJ, Kochanek PM. Therapeutic hypothermia after cardiac arrest. N Engl J Med. 2002;346:612–613. doi: 10.1056/NEJM200202213460811. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Fujita J, Gaffin SL, Lilly CM. Invited review: effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Gaffin SL, Pratt RE, Cullivan ML, Angel KC, Lilly CM. Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells. J Appl Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Cullivan ML, Sheldon HK, Pratt RE, Lilly CM. Effect of hypoxia on gene expression by human hepatocytes (HepG2) Physiol Genomics. 2003;12:195–207. doi: 10.1152/physiolgenomics.00104.2002. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Kuhlmeier MM, Carter HC, Hasday JD, Lilly CM, Fairchild KD. Effect of moderate hypothermia on gene expression by THP-1 cells: a DNA microarray study. Physiol Genomics. 2006;26:91–98. doi: 10.1152/physiolgenomics.00296.2005. [DOI] [PubMed] [Google Scholar]
- Wood RJ, Tchack L, Angelo G, Pratt RE, Sonna LA. DNA microarray analysis of vitamin D-induced gene expression in a human colon carcinoma cell line. Physiol Genomics. 2004;17:122–129. doi: 10.1152/physiolgenomics.00002.2003. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Sato Y, Mori N. Difference in induction of uncoupling protein genes in adipose tissues between young and old rats during cold exposure. FEBS Lett. 1999;458:157–161. doi: 10.1016/S0014-5793(99)01143-6. [DOI] [PubMed] [Google Scholar]
- Yenari MA, Han HS. Influence of hypothermia on post-ischemic inflammation: role of nuclear factor kappa B (NFkappaB) Neurochem Int. 2006;49:164–169. doi: 10.1016/j.neuint.2006.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(DOC 502 kb)