Abstract
Heat shock protein 90 (HSP90) targets a broad spectrum of client proteins with divergent modes of interaction and consequences. The homologous epidermal growth factor receptor (EGFR) and ERBB2 receptors as well as kinase-deficient mutants thereof differ in their requirement for HSP90 in the nascent versus mature state of the receptor. Specific features of the kinase domain have been implicated for the selective association of HSP90 with mature ERBB2. We evaluated the role of HSP90 for the homologous ERBB3 receptor. ERBB3 is naturally kinase deficient, a central mediator in cell survival and stress response and the primary dimerization partner for ERBB2 in signaling. Cellular studies indicate that, similar to EGFR, the geldanamycin (GA) sensitivity of ERBB3 and HSP90 binding resides in the nascent state and is dependent on the presence of the kinase domain of ERBB3. Furthermore, despite its intrinsic lack of kinase activity and in contrast to the reported GA sensitivity of mature and kinase-deficient EGFR, the GA sensitivity of the nascent state of ERBB3 appears to be exclusive. Geldanamycin disrupts the interaction of ERBB3 and HSP90 and inhibits ERBB3 maturation at an early stage of synthesis, prior to export from the ER. Studies with a photo-convertible fusion protein of ERBB3 suggest geldanamycin sensitivity at a later stage in maturation, possibly through the putative role of HSP90 in structural proofreading.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-009-0166-1) contains supplementary material, which is available to authorized users.
Keywords: HER3, ERBB3, HSP90, Nascent kinase domain, Geldanamycin
Introduction
The human family of ERBB receptor tyrosine kinases includes epidermal growth factor receptor (EGFR; ERBB1), ERBB2 (HER2), ERBB3 (HER3), and ERBB4 (HER4). While single-receptor systems exist for example in Caenorhabditis elegans (Let23) and Drosophila (DER), the four human ERBB receptors are structurally highly homologous but have diversified in many aspects, including ligand binding, catalytic activity, and dimerization preferences. In addition, ERBB receptors have diverged in their dependency on HSP90 which can stabilize the receptors in both the nascent and the mature state.
Since the overexpression or deregulation of both EGFR and ERBB2 is a hallmark of a wide range of cancers, both have been targets of considerable efforts in drug development. While the ERBB3 receptor is structurally highly homologous to other ERBB receptors, it is distinct in that its kinase domain is catalytically impaired (Guy et al. 1994; Sierke et al. 1997), and overexpression of ERBB3 in isolation does not appear to be a significant event in human cancers. However, ERBB3 is a potent partner in heterodimerization events with other ERBB family members. In such heterodimers, the kinase domain of ERBB3 contributes a conserved interface needed for the allosteric cross-activation of heterodimers in trans (Zhang et al. 2006), and its extracellular domains bind activating ligands of the neuregulin (heregulin) family of epidermal-growth-factor-like ligands. Furthermore, the cytoplasmic tail region of ERBB3 carries a unique set of adaptor sites, most notably multiple copies of binding sites for the regulatory subunit p85 of phosphoinositide 3 kinase (PI3K). This makes phosphorylated ERBB3 one of the strongest known activators of PI3K/Akt signaling, thereby providing a strong cellular prosurvival signal. ERBB3-mediated signaling is an important component in the cellular response induced by stress and radiation (Dent et al. 2003), and ERBB3 confers and predicts resistance to the radiosensitization induced by HSP90 inhibitors (Dote et al. 2005). In addition, it has recently been proposed that the emergence of resistance to kinase inhibitor therapy aimed at ERBB2 or EGFR correlates with a rebound of the levels of phosphorylated ERBB3 in the face of strong sustained but incomplete inhibition of the kinase activity of its heterodimerization partners (Sergina et al. 2007). Despite its lack of intrinsic kinase activity, ERBB3 has therefore emerged as an important drug target in its own right.
HSP90 is a very abundant protein, estimated to represent 1–2% of total cytosolic protein (Wegele et al. 2004). Further elevated levels of HSP90 occur in some cancers such as breast cancers where HSP90 overexpression correlates with a lower survival rate in breast carcinoma (Pick et al. 2007). Indeed, the inhibition of HSP90 with the quinone ansamycin antibiotics geldanamycin (GA), herbimycin A, or 17-allylaminogeldanamycin (17-AAG) has been shown early on to reduce the steady-state levels of its client proteins including the protein kinases Src (Whitesell et al. 1994), EGFR (Murakami et al. 1994), platelet-derived growth factor receptor (Sakagami et al. 1999), and BCR/ABL (Okabe et al. 1994). The reversal of Src-induced cellular transformation by herbimycin A was an early example of the efficacy of ansamycin antibiotics, and this approach has since been expanded to a broad range of targets.
Although HSP90 can act as a classic chaperon that aids in early protein folding events and recognizes hydrophobic patches exposed in unfolded or partially unfolded proteins (Smith 1998), this function may be more associated with HSP70 than HSP90 in a cellular setting (Nathan et al. 1997). In vivo, HSP90 may predominantly act at a later stage of protein maturation with a broad but nevertheless specific subset of clients (Pratt et al. 1999). A system-wide analysis of HSP90 clients in yeast identified nearly 10% of the yeast proteome as putative clients, but the pool of clients is heavily biased towards proteins associated with functions in signal transduction (Millson et al. 2005; Zhao et al. 2005). Additional specificity in HSP90–client interactions may be provided by cochaperones. This is exemplified by CDC37, which confers at least partial and phosphorylation-regulated selectivity towards protein kinase clients (Roe et al. 2004; Vaughan et al. 2008).
In the case of c-Jun N-terminal kinases (JNKs), the interaction of the nascent kinase with HSP90/CDC37 complexes has been localized to the kinase domain of JNKs itself (Prince and Matts 2004). Subsequent studies have indicated that during maturation HSP90/CDC37 may temporarily take the place of an additional N-terminal structural component in JNKs and thereby stabilize the N-terminal lobe of the kinase. In mature JNKs, this additional structural element shields the single β-sheet of the N-terminal lobe and renders the JNKs independent of HSP90 binding (Prince and Matts 2005). However, mutant JNKs that lack this additional N-terminal structural motif retain binding of HSP90 but not CDC37, even in the presence of geldanamycin, albeit at greatly reduced affinity (Prince and Matts 2005). This reduced affinity binding mode of HSP90 has also been reported for wild-type Akt and in a manner that is sensitive to its phosphorylation state (Yun and Matts 2005). Hence, the ability of HSP90 to interact with either nascent or mature kinase clients may not be categorized in a binary fashion but may instead represent quantitative shifts in the relative affinity of HSP90 for client surfaces that are presented in both states of the kinase domain.
HSP90 interactions play a critical role in ERBB receptor signaling. The molecular nature of the interaction with HSP90 has been studied in significant detail for the mature forms of EGFR and ERBB2. Both nascent and mature ERBB2 are sensitive to GA while the sensitivity of wild-type and catalytically competent EGFR is limited to the nascent state (Xu et al. 2002b). For ERBB2, the rapid destabilization of mature receptors by ansamycins involves the dissociation of HSP90 from ERBB2 and the interaction of ERBB2 with the ubiquitin ligase CHIP followed by ubiquitination-dependent receptor degradation (Citri et al. 2002; Mimnaugh et al. 1996; Xu et al. 2002a; Zhou et al. 2003). This degradation of ERBB2 has been shown to coincide with either an overall increase in internalization and degradation (Lerdrup et al. 2007, 2006) or a change in sorting that directs more ERBB2 towards degradation instead of recycling (Austin et al. 2004).
The region conferring the very pronounced sensitivity of mature ERBB2 to GA has been mapped to the kinase domain of ERBB2 (Xu et al. 2001). Several studies (Citri et al. 2006; Tikhomirov and Carpenter 2003; Xu et al. 2005) further narrowed down the region of interest to the αC-β4 loop of the N-terminal lobe of the kinase domain using mutagenesis. Previous homology modeling of the kinase domains of EGFR and ERBB2 as well as a comparison with a broad range of human client kinases for HSP90 demonstrated that the absence of a negative surface charge in the surface area around the αC-β4 loop is critical for the ability of mature ERBB2 to interact with HSP90. Conversely, the presence of negative surface charges in this three dimensionally but to a lesser extent primary sequence defined region accounts for the inability of mature and catalytically competent EGFR to interact with HSP90. HSP90 binding and GA sensitivity of the mature state can be reversed for EGFR and ERBB2 by the corresponding charge-altering mutations. Notably, while truncations of the kinase domain of ERBB2 result in a loss of GA sensitivity in the nascent and mature state, mutations in ERBB2 that eliminate the sensitivity of the mature receptor to HSP90 retain sensitivity of the nascent protein to GA (Xu et al. 2005). These data suggested that the interaction of HSP90 with the cytoplasmic kinase domain is required in the nascent and mature state, but while interactions are clearly distinct in nature, the unique aspects of the interaction in the nascent state are at present not known.
Binding of HSP90 to mature kinases appears to be sensitive to their activation state as it is defined by the conformation of the kinase domain, apparently regardless of catalysis. The binding of HSP90/CDC37 to the Src kinase Lck is stabilized by the Src family specific kinase inhibitor PP2 (Giannini and Bijlmakers 2004). For ERBB2, HSP90 binding to the mature receptor is sensitive to the irreversible and ERBB-specific kinase inhibitor CI-1033 (Citri et al. 2002), and the inhibition of the ERBB2/HSP90 interaction by 17-AAG can suppress the escape from tyrosine kinase inhibition in ERBB2-overexpressing SKBR3 cells (Pashtan et al. 2008). Interestingly, kinase-deficient mutants of both EGFR and ERBB2 display enhanced sensitivity to geldanamycin (Citri et al. 2002), and this enhanced sensitivity to geldanamycin by kinase-deficient EGFR resides largely with the mature and cell-surface-localized species. This is in contrast to wild-type EGFR, which displays a pattern of reduced and delayed sensitivity to geldanamycin, thought to reflect sensitivity primarily at the nascent stage.
Our surface charge modeling for the ERBB3 kinase domain (Fig. 1) would predict EGFR-like behavior for ERBB3, specifically a lack of sensitivity in the mature state. On the other hand, ERBB3 is unique among ERBB receptors in that it naturally represents a catalytically deficient kinase. Since kinase-deficient mutants of EGFR had exhibited a reversal in GA sensitivity in that they created sensitivity in the mature state, this would give rise to a prediction for ERBB3 that contradicts surface charge features. The question whether or not ERBB3 interacts with HSP90 is also relevant with respect to the mechanism of signal transduction. For ERBB2, its primary heterodimerization partner, binding of the mature receptor to HSP90 reduces its ability to form heterodimers (Citri et al. 2004), and HSP90 dissociation enhances autoactivation followed by Src-mediated tyrosine phosphorylation at tyrosine 877 and subsequent degradation of ERBB2 (Xu et al. 2007). A dependency or alternatively an uncoupling of mature ERBB3 from HSP90 is also relevant in light of the central role of ERBB3 in stress and radiation responses as well as its reported ability to counteract the efficacy of HSP90 inhibitors in therapeutic applications. Ultimately, the answer to this question is of importance not only in the context of ongoing efforts to target the interaction of HSP90 and ERBB3 receptors clinically but also with respect to our mechanistic understanding of how HSP90 may carry out functionally distinct interactions at different stages of the receptors’ lifetime.
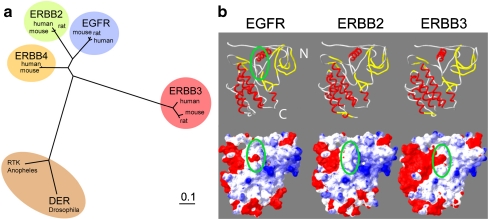
Fig. 1.
The kinase domain of EGFR resembles ERBB3 with respect to the charge distribution in the HSP90 interface but is closer to ERBB2 in phylogeny. a Phylogenetic nearest neighbor analysis of ERBB kinase domains and their orthologs. Drosophila DER is added as a reference point. Phylogenetically, ERBB3 is the most divergent of the mammalian ERBB kinase domains. b Ribbon diagram and surface charge representation of the kinase domains of EGFR (1 M17) and homology models of the kinase domains of ERBB2 and ERBB3 with highlighted HSP90 interface (green oval). The absence of negative charge (red) in the HSP90 interface of ERBB2 has been shown previously to correlate with the binding of mature ERBB2 to HSP90
Our studies on ERBB3 place a special emphasis on excluding an impact of GA on the mature receptor, and we demonstrate that decreases in the level of nascent receptor are likewise not the result of lower levels of ERBB3 message. Furthermore, inhibition studies with Brefeldin A (BFA) and studies with fusion proteins of ERBB3 with photo-convertible fluorescent protein indicate that the point of GA sensitivity resides before the export from the ER but at a point past the initial stages of synthesis and early folding. Consistent with data obtained for ERBB2, the GA sensitivity and HSP90 interaction of ERBB3 are dependent on the presence of the receptor kinase domain. Receptors lacking the kinase domain are insensitive to GA, show no physical interaction between ERBB3 and HSP90, yet proceed efficiently to the cell surface. This suggests an exclusive involvement of HSP90 in the late stages of ERBB3 maturation, possibly in the structural proofreading of its kinase domain.
Materials and methods
Homology modeling of the ERBB3 kinase domain
A homology model for the kinase domain of ERBB3 and ERBB2 was based on the available crystal structure for the kinase domain of EGFR in the active conformation (1M17, Stamos et al. 2002). While the inactive conformation of EGFR (1XKK, Wood et al. 2004) may arguably be a better representation of the resting state of ERBB3, large segments of the kinase domain, most notably the activation loop, are not fully resolved in the structure representing the inactive state of EGFR, yet the charge distribution in the putative HSP90 interface for mature receptors remains intact when both crystal structures of EGFR are compared (Supplementary Fig. 1). For model construction, primary sequences of all three receptor kinase domains were aligned using clustalW, and models were created using Swiss Model builder (Schwede et al. 2003). The surface charges for all three kinase domains were calculated in Swiss viewer using the default parameters.
Cell culture and reagents
MCF7 cells were obtained from the American Type Culture Collection and grown in Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% fetal bovine serum (FBS). Cells were transfected with either Lipofectamine Plus (Invitrogen) for the coimmunoprecipitation or ExGen (Fermentas) for microscopy. The following antibodies were used: cytoplasmic ERBB2 (C18) and ERBB3 (C-17) from Santa Cruz Biotechnology, extracellular ERBB2 (Ab-5) and ERBB3 (Ab-4) from EMD, and HSP90 (SPA-835) from Stressgen. Geldanamycin was obtained from A.G. Scientific; Brefeldin A was from Fluka Biochemika; and cycloheximide (CHX) was from Sigma. Endonuclease H was obtained from Roche.
Steady-state measurements
MCF7 cells were treated with 3 µM GA or equivalent volume of dimethyl sulfoxide (DMSO) as a vehicle control in RPMI supplemented with 10% FBS for time points ranging from 0 to 15 h. Cells were lysed in mild lysis buffer (MLB) [20 mM Tris base pH 8.0, 137 mM NaCl, 1% triton X-100, 10% glycerol, 5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, protease inhibitor cocktail (Sigma)] supplemented with 1% sodium dodecyl sulfate (SDS). Lysates for all assays were normalized for protein content (BCA assay, Pierce Biotechnology). Samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted using antibodies to the C terminus of ERBB3 (C-17) or ERBB2 (C-18).
Cell surface biotinylation
MCF7 cells were cell-surface-biotinylated for 40 min with 1 mg/ml NHS-biotin (Pierce Biotechnology) in ice-cold phosphate-buffered saline (PBS) supplemented with MgCl2 (1 mM) and CaCl2 (0.1 mM; pH 7.5), followed by quenching of the reaction with PBS supplemented with 100 mM glycine and 5 mM EDTA (pH 8.0). Cells were then washed twice with 37°C RPMI/10%FBS prior to treatment with 3 µM GA or equivalent volume of DMSO for 6 h. Cells were then lysed in MLB supplemented with 1% SDS. Lysates were diluted 1:10 in MLB, incubated at 25°C for 30 min and run through an Ultrafree-MC 0.22-μm spin filter (Millipore) to remove any precipitated cell debris. Biotinylated proteins were pulled down with streptavidin-conjugated agarose beads (Pierce Biotechnology), resolved by SDS-PAGE, and analyzed by Western blot using antibodies against ERBB3 (C-17) or ERBB2 (C-18). Densitometry measurements were taken from the Western blot, and the percent change in protein levels for GA-treated samples was normalized relative to levels in DMSO samples after 6 h.
Chemical cross-linking of surface receptors
Chinese hamster ovary (CHO) cells were transfected with pFlag ERBB2 or pFlag ERBB3 using Lipofectamine Plus (Invitrogen). Twenty-four hours after transfection, cells were treated with 50 μM MG115 for 3 h and with 50 mM phenylarsine oxide (PAO) for the last 30 min of MG115 treatment. Cells were then treated with both MG115 and PAO and either 3 μM GA or equivalent volume of DMSO for 1 h. All treatments were done at 37°C followed by cross-linking on ice for 30 min with 1 mM BS3 (Pierce Biotechnology) with all drugs present. Cells were then washed once in PBS and lysed and analyzed by SDS-PAGE analysis and Western blotting using antibodies against the C terminus of ERBB2 (C-18) and ERBB3 (C-17).
35S isotope labeling
MCF7 cells were pulse-labeled with 35S cysteine and methionine (Pro-mix l-[35S], Amersham) in RPMI lacking cysteine and methionine for 16 h, followed by two washes with RPMI containing no label. Cells were allowed to recover from washing at 37°C, and 1.5 h after the 35S label was removed, lysates were collected at increasing time points from 0 to 16 h. Cells were lysed in MLB supplemented with 1% SDS, followed by 1:10 dilution in MLB and run through spin filters. Samples were immunoprecipitated (IP) with anti-ERBB3 antibody (C-17) and resolved by SDS-PAGE, and radiolabeled ERBB3 was visualized by scanning on a Typhoon 9410 Laser densitometer (GE Healthcare-Amersham). Control (C) samples contained an ERBB3 peptide (SC-285P, Santa Cruz) incubated with the ERBB3 antibody and cell lysates and were used to indicate the amount of nonspecific binding in the IP. Samples were analyzed in duplicate, and a representative set is shown.
Cycloheximide treatment
MCF7 cells were treated with 20 μM CHX, 3 μM GA, or CHX and GA together for time points ranging from 0 to 16 h. Cells were harvested; samples were resolved by SDS-PAGE and immunoblotted with ERBB2 or ERBB3 antibodies.
Quantitative PCR
MCF7 cells were treated with 3 μM GA or equivalent volume of DMSO for 2 h, followed by collection of mRNA using RNA Stat 60 ( Iso-Tex Diagnostics, Inc., TX, USA) mRNA was subjected to a reverse transcriptase reaction, and the resulting cDNA was quantified using quantitative polymerase chain reaction (PCR). Measurements were normalized against β-actin cDNA levels. Primer sequences used were: ERBB3 fwd: GTGGCACTCAGGGAGCATTTA, ERBB3 rev: TCTGGGACTGGGGAAAAGG; human β-actin fwd: GATGAGATTGGCATGGCTT, human β-actin rev: CACCTTCACCGTTCCAGTTT. Analysis and PCR were done using the iCycler (Bio-Rad).
Brefeldin A and endonuclease H treatment
MCF7 cells were treated with 3 µM GA and/or 10 mg/ml BFA in RPMI supplemented with 10% FBS for the times indicated, followed by lysis in MLB supplemented with 1% SDS. Equivalent protein amounts were loaded. For the additional identification of the nascent band following BFA treatment, lysates in MLB were treated directly with EndoH (o.1 U/20 μl, Roche) for 1 h at 37°C prior to analysis by SDS-PAGE.
Dendra2 fusion constructs and fluorescence microscopy
A fusion protein of the photo-convertible fluorescent protein Dendra2 and ERBB3 was created by in frame insertion of full-length and PCR-amplified Dendra2 (Dendra2, Evrogen) into the C-terminal Xba1/Cla1 cassette of the previously published pFLAG-ERBB3 expression vectors (Park et al. 2008). MCF7 cells were transfected with pFlag-ERBB3-Dendra2 (Dendra2, Evrogen) and grown for 24 h posttransfection on fibronectin-coated (Roche) glass chambers. Cells were imaged and photoconverted by illumination for 40 s using a hydroxycoumarin filter set (405/20 excitation, Chroma Technology, VT, USA). Photoconverted cells were incubated for 2 h in the presence or absence of either 20 μM CHX or 3 μM GA cycloheximide as indicated and imaged again. Images were acquired on a Zeiss Axiovert 200 M fluorescence microscope equipped with an ORCA-ER digital camera. For the detection of green Dendra2, a filter cube with HQ480/40 excitation, HQ535/50 emission, and Q505lp beam splitter configuration was used. Photoconverted Dendra2 was visualized using a Texas red filter set, using a HQ560/55 excitation, HQ645/75 emission, and Q595lp beam splitter configuration (both Chroma Technology, VT, USA). Due to multiple media changes during data acquisition, images were normalized for uniform background intensity. All imaging and drug treatment for imaging experiments were done in Phenol red-free RPMI 1640 (Invitrogen) supplemented with 10% FBS.
A Dendra2 fusion of a variant of ERBB3 lacking the kinase domain was generated by deletion of 331 amino acids starting with serine in position 669 (residue numbering based on alignment of kinase domains of EGFR, ERBB2, and ERBB4 in Qiu et al. 2008) and including aspartic acid in position 1000 (SIEPLDP … DLEAEED). This removes juxtamembrane section B together with the entire kinase domain. As a consequence the basic juxtamembrane, segment A is fused directly to the C-terminal tail of ERBB3. Following selection of transfected, neomycin-resistant MCF7 cells that carried either soluble Dendra2, ERBB3-Dendra2, or kinase domain lacking ERBB3(ΔKD)-Dendra2, cells were sorted by fluorescence-activated cell sorting to obtain uniform populations. Based on the presence of the shared C-terminal epitope for an anti-ERBB3 antibody (C17, Santa Cruz), both Dendra2 fusions of ERBB3 expressed stably at approximately 40–100% of endogenous levels of ERBB3 in MCF7 (i.e., approximately 4,000–10,000 recombinant receptors per cell). Expression levels of Dendra2 fusions can be evaluated selectively by direct Western blot analysis with anti-Dendra2 antibodies (Evrogen).
HSP90 coimmunoprecipitation
The coimmunoprecipitation of ERBB receptors and HSP90 was performed in both directions. For immune precipitations of ERBB3 followed by detection of HSP90, we adapted a previously published protocol (Citri et al. 2002). Equal numbers of MCF7 cells were seeded and transfected with pFlag-ERBB2 or pFlag-ERBB3, a constitutive expression vector carrying an N-terminal FLAG epitope tag. After 24 h, cells were treated with 5 μM GA in RMPI/FBS at 37°C for the samples and times indicated. All subsequent procedures were done on ice or at 4°C. Cells were washed in PBS supplemented with 1 mM phenylmethylsulfonyl fluoride and lysed in TMISV buffer (150 mM Tris–HCl, pH 7.5, 150 mM NaCl, 20 mM NaMoO4, 0.09% Igepal, 1 mM NaVO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and protease inhibitor cocktail (Sigma)). Cells were scraped of the cell culture dishes, and lysates were passed repeatedly through a 26-gauge needle, followed by centrifugation at 15,000×g for 15 min. The cleared lysate was incubated for 2 h on a rocker with or without antibodies against the N terminus of either ERBB2 (Ab-5, EMD Biosciences) or ERBB3 (Ab-4, EMD Biosciences), followed by incubation with protein A/G beads (Santa Cruz Biotech, CA, USA) for an additional 2 h. Immunoprecipitated samples were analyzed by SDS-PAGE and Western blotting against HSP90 (SPA-835, Assay Designs), ERBB2 (C18, Santa Cruz), or ERBB3 (C17, Santa Cruz) as indicated. For the inverse immune precipitation, lysates of transfected or untransfected MCF7 cells were prepared as described above. HSP90 was immunoprecipitated with the rat monoclonal antibody 9D2 (Stressgen). Endogenous ERBB3 (C17, Santa Cruz) in untransfected MCF7 cells or Dendra2 fusion proteins in stably transfected MCF7 cells were detected by Western blotting after binding and processing at room temperature as indicated above.
Results
A structural homology model of the ERBB3 kinase domain indicates that the putative HSP90 interface in ERBB3 resembles EGFR more than ERBB2
ERBB3 is considered to be a client of HSP90 (Citri et al. 2006; Zheng et al. 2000) based on an observed sensitivity of steady-state levels of ERBB3 to ansamycins, but the direct interaction of ERBB3 and HSP90 has not been demonstrated. The extent to which this sensitivity reflects dependency in both the nascent and mature state or exclusively the nascent state has likewise not been experimentally tested. Compared to ERBB2, steady-state levels of ERBB3 display only a slow and incomplete decay in response to treatment with ansamycin antibiotics (Citri et al. 2002; Zheng et al. 2000), and in this respect ERBB3 resembles wild-type EGFR more than ERBB2. However, ERBB2 and ERBB3 are also known to exhibit pronounced differences in the nature and rate of protein turnover. ERBB3 is fundamentally distinct from both EGFR and ERBB2 in that its kinase domain is catalytically impaired. ERBB3 resembles ERBB2 in its strict dependence on heterodimerization for signaling but is similar to EGFR in its ability to bind ligand through its extracellular domains. Lastly, based on the sequence homology of their kinase domains, EGFR and ERBB2 are more closely related to each other than either one is to ERBB3 (Fig. 1a).
Previous studies had already demonstrated that the absence of a negative surface charge in the area around the αC-β4 loop is critical for the ability of mature ERBB2 to interact with HSP90. The importance of the spatial context and overall surface charge is reflected in the fact that there is no strong consensus sequence that discriminates clients that bind only in the nascent state versus clients that require HSP90 in the mature state. Since previous studies did not include a homology model and surface charge analysis of the kinase domain of ERBB3, we made this the starting point of our analysis (Fig. 1b).
Based on the high sequence conservation between ERBB receptors, we used the existing crystal structure of the EGFR kinase domain (1M17, Stamos et al. 2002) to create a homology model of the kinase domain of ERBB2 and ERBB3. For the lapatinib-stabilized kinase domain of EGFR in the inactive conformation (1XKK, Wood et al. 2004), the negative surface charge in the putative HSP90 interface remains prominent although significant portions of the kinase domain are disordered in the inactive conformation (Suppl. Fig. 1). We therefore modeled the surrounding of the αC-β4 loop of ERBB3 using the better-defined above structure of the active state of EGFR (Fig. 1b). This makes the ERBB3 homology model also more comparable to previous comparisons between EGFR and ERBB2 (Citri et al. 2006; Xu et al. 2005). Whether the catalytic deficiency of ERBB3 favors the inactive over the active conformation is not known. The putative HSP90 interface is circled in green and includes the αC-β4 loop of the N-terminal lobe of the kinase domain. In our homology model of the kinase domain of ERBB3, the characteristic negative charge of EGFR, the absence of which correlates with the binding of mature ERBB2 to HSP90, is clearly prominent in ERBB3. For ERBB3, this analysis suggests that the putative HSP90 interface in ERBB3 resembles EGFR more than ERBB2 despite the fact that the kinase domains of ERBB2 and EGFR are more similar to each other in their overall surface charge distribution and indeed their sequence homology.
Geldanamycin treatment reduces the steady-state levels of ERBB3
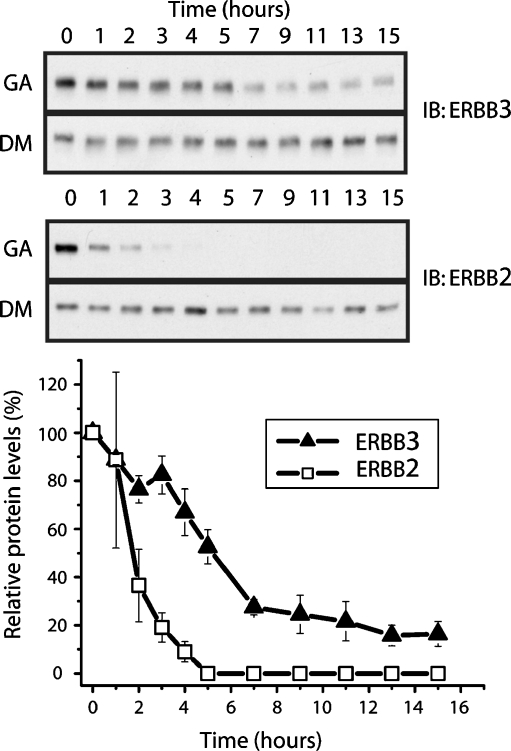
Previous studies had indicated that ERBB3 and wild-type EGFR are relatively insensitive to GA when compared to the rapid response seen for ERBB2. However, long-term exposure to GA does result in decreases of both EGFR and ERBB3 receptor levels (Citri et al. 2006; Zheng et al. 2000). However, the underlying mechanisms or the extent to which this decrease is a direct or off-target effect was not clear for ERBB3. We focused our initial efforts on a model system with low and endogenous levels of both ERBB2 and ERBB3 to avoid potential changes in the stability of receptors as consequence of exogenous overexpression. MCF7 cells are a widely used breast-cancer-derived cell line that carries both modest levels of ERBB3 as well as approximately matching levels of ERBB2 (Aguilar et al. 1999). When MCF7 cells are treated with GA or vehicle control (DMSO) over a course of 15 h, both receptors show a different response to treatment with geldanamycin (Fig. 2). While steady-state levels of ERBB2 drop rapidly after the addition of geldanamycin, ERBB3 levels show a pronounced decrease, but at a significantly slower rate. In addition, ERBB3 steady-state levels plateau after 8 h of treatment at approximately 20% of their starting value while ERBB2 levels are effectively undetectable at this point. Hence, the low, endogenous levels of ERBB3 in MCF7 exhibit a pattern of sensitivity to geldanamycin that resemble published reports for EGFR and that is distinct from the response of its primary signaling partner, ERBB2.
Fig. 2.
Geldanamycin (GA) treatment leads to a slow decrease in the steady-state levels of ERBB3 in MCF7 cells. Cells were treated with 3 µM GA or DMSO as a vehicle control for 0–15 h and lysed at indicated time points. Samples were resolved by SDS-PAGE and immunoblotted using antibodies to the C terminus of ERBB3 (C17) or ERBB2 (C18)
Cell surface ERBB3 is not destabilized by GA
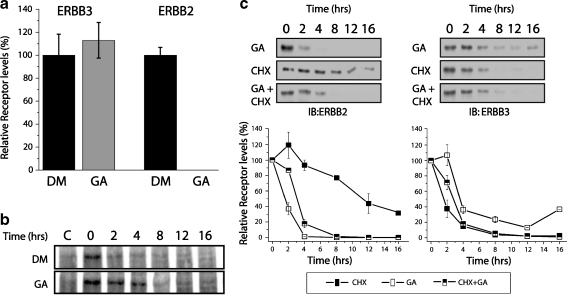
In order to evaluate whether the slow decrease in ERBB3 levels is a reflection of a reduced destabilization of the mature receptor, we measured the decay of cell surface biotinylated ERBB3 and ERBB2. Relative to the receptor levels obtained by natural turnover in DMSO-treated controls, cell-surface-localized ERBB2 was completely degraded after 6 h of GA treatment. In contrast, the levels of cell surface ERBB3, when normalized for steady-state turnover, did not diminish relative to untreated controls (Fig. 3a). In fact, we consistently noticed a modest stabilization of cell surface ERBB3 by GA compared to the steady-state turnover in untreated samples (see also Fig. 3b). Hence, under the same conditions at which absolute steady-state level diminish by 60% (Fig. 2), cell surface ERBB3 shows no indication of being destabilized by GA.
Fig. 3.
Mature ERBB3 is not destabilized by GA. a Levels of cell surface ERBB2 and ERBB3 after GA treatment, relative to unchallenged turnover. MCF7 cells were cell-surface-biotinylated with 1 mg/ml NHS-biotin and subsequently treated with 3 µM GA or DMSO (vehicle control) for 6 h prior to lysis. Biotinylated proteins were enriched on streptavidin-conjugated agarose beads and analyzed by SDS gel electrophoresis and Western blotting using antibodies against ERBB3 (C17) or ERBB2 (C18). Densitometry measurements were taken from the Western blot, and the relative changes for GA-treated samples were normalized to levels of DMSO control samples. b Turnover of 35S-labeled, mature ERBB3 is not accelerated by GA. MCF7 cells were pulse-labeled with 35S cysteine and methionine for 16 h, followed by two washes with media containing no label. Cells were allowed to recover from washing. One and half hours after the 35S label was removed, lysates were collected at increasing time points from 0 to 16 h. Samples were immunoprecipitated with anti-ERBB3 antibody and resolved by SDS-PAGE, and radiolabeled ERBB3 was visualized by autoradiography. For control samples (c), an ERBB3-derived blocking peptide was added during immunoprecipitation to determine the amount of nonspecific binding. c Geldanamycin and cycloheximide have comparable effects on the steady-state levels of ERBB3. MCF7 cells were treated with cycloheximide (CHX), GA, or CHX and GA together for 0–16 h. Cells were harvested; samples were resolved by SDS-PAGE and immunoblotted against ERBB2 or ERBB3 antibodies
We confirmed the lack of GA sensitivity of cell surface ERBB3 by metabolic labeling. To this end, MCF7 cells were labeled overnight with [35S] (Fig. 3b). Without the use of recombinant overexpression in these initial studies, the signal obtained from the direct labeling of nascent receptor is insufficient for analysis due to the low total levels of endogenous receptors in MCF7 cells (approximately 15,000; Aguilar et al. 1999). In order to focus the analysis instead on the larger pool of mature receptor, cells were subjected to a 90-min chase prior to the addition of GA and onset of analysis. Despite low overall incorporation, our findings are consistent with the hypothesis that the turnover of metabolically labeled ERBB3 is not accelerated relative to DMSO control. Instead, we again observed a modest stabilization of labeled mature ERBB3, and our data indicate that cell surface ERBB3 is excluded from the GA-sensitive pool.
To confirm that the slow decrease in steady-state levels of ERBB3 is instead due to a lack of resupply, we compared the impact of GA with the protein synthesis inhibitor CHX (Fig. 3c). The treatment of MCF7 cells with cycloheximide alone provides an estimate for the half-life of the receptors and confirms previous observations that ERBB3 does have a significantly shorter half-life compared to ERBB2 (Warren et al. 2006). However, while the addition of GA significantly shortens the half-life of ERBB2, we again observed a modest increase in half-life for ERBB3. This is consistent with our earlier observation that GA slows down the turnover of mature ERBB3, thereby extending the pool of existing ERBB3 in the absence of resynthesis. However, CHX or the simultaneous presence of CHX and GA does eliminate the plateau of ERBB3 that can be observed following treatment with GA alone. This suggests that the plateau of residual ERBB3 reflects an incomplete sensitivity to GA or incomplete HSP90 dependency at an early stage of receptor maturation or synthesis, not a failure of the protein degradation machinery to remove preexisting and otherwise destabilized ERBB3 because of a prolonged exposure of cells to GA.
HSP90 does not associate with mature ERBB3
The above analysis indicates that mature ERBB3 is not sensitive to GA but does not exclude the possibility that the disruption of HSP90 interactions with ERBB3 may, in contrast to ERBB2, merely be without consequence on receptor stability. Given the low recovery in HSP90 coimmunoprecipitations, a failure to detect HSP90 after sequential ERBB3 and biotin-based pull downs of surface ERBB3 would not be conclusive in case of a negative finding. However, HSP90 binds its clients as a functional dimer, and even in the absence of any cochaperones, this would constitute a complex more than twice the size of the kinase domain of either ERBB2 or ERBB3. It is therefore not surprising that in a cellular setting the binding of HSP90 to ERBB2 interferes with receptor association events (Citri et al. 2004). We therefore relied on this steric interference of HSP90 with receptor interactions as an indirect readout under live cell conditions. Previous studies had demonstrated that, in contrast to ERBB2, ERBB3 shows a marked propensity to self-associate in the absence of ligand (Kani et al. 2005; Landgraf and Eisenberg 2000). This pronounced self-association can be visualized by chemical cross-linking with cell-surface-impermeable cross-linkers. We recently confirmed ERBB3 oligomerization at low endogenous receptor levels (approximately 10,000 receptors per cell) using live cell photo cross-linkable aptamers in MCF7 breast cancer cells (Park et al. 2008), and the identity of the cross-linked species as ERBB3 homodimers and oligomers had been demonstrated by dual-tagged pull-down experiments in both insect cells (Landgraf and Eisenberg 2000) and CHO cells (Park et al. 2008).
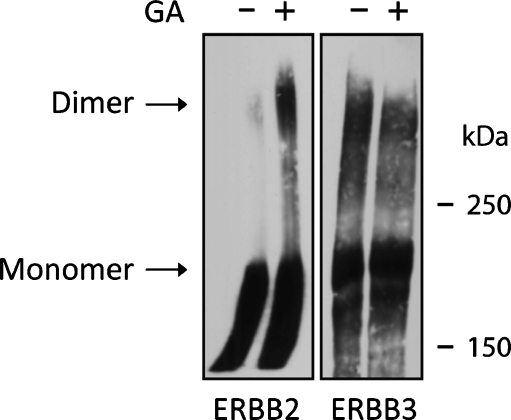
When ERBB2 or ERBB3 is individually overexpressed in CHO cells (Fig. 4), chemical cross-linking with the non-membrane-permeable and homobifunctional cross-linker BS3 indicates the enhanced formation of otherwise transient dimers of ERBB2 in the presence of GA. This is consistent with previous reports that GA induces enhanced constitutive activation of ERBB2 (Xu et al. 2007) and interferes with the heterodimerization of ERBB2 and ERBB3 (Citri et al. 2004). ERBB3 shows significantly elevated cross-linking, reflecting its enhanced constitutive self-association. However, in contrast to ERRBB2, the cross-linking properties of ERBB3 (including potential associations with other cell surface proteins that are subject to short-range cross-linkers) are not modulated in either direction by the addition of GA. This suggests, albeit indirectly, that the observed lack of sensitivity of ERBB3 to GA on the cell surface is likely to be a reflection of a lack of binding of HSP90 to the mature receptor which was further supported by subsequent immunoprecipitation studies (Fig. 5).
Fig. 4.
After geldanamycin treatment, receptor cross-linking increases substantially for cell surface ERBB2, but not for ERBB3. ERBB2 and ERBB3 were individually overexpressed in CHO cells. Following treatment with either DMSO or GA for 2 h at 37°C as indicated, receptors were cross-linked for 30 min on ice with the membrane impermeable cross-linker BS3 (50 mM ) and subsequently analyzed by SDS-PAGE and Western blotting. The location of the receptor monomer and dimer bands are indicated
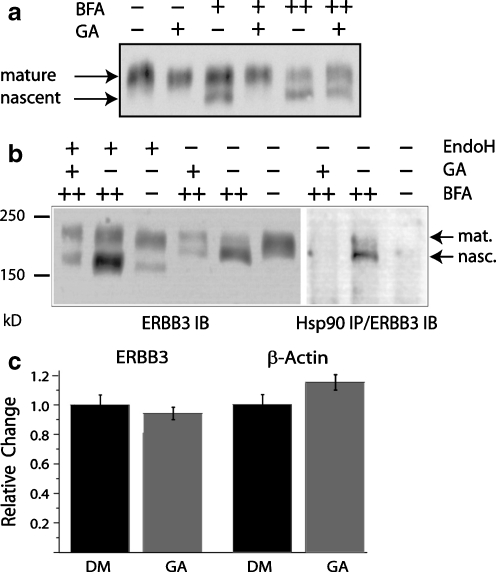
Fig. 5.
HSP90 interacts selectively with nascent ERBB3. a GA blocks the buildup of Brefeldin-A-enriched and nascent ERBB3. MCF7 cells were treated with 3 µM GA and 10 mg/ml Brefeldin A (BFA) as indicated. Nascent and mature ERBB3 appear as the lower and higher molecular weight bands, respectively. “+” indicates treatment for 2 h, and “++” indicates treatment for 4 h. During 4-h BFA treatments (++), GA was added during the last 2 h as indicated. b ERBB3 selectively binds to nascent ERBB3. Left: The low molecular weight and GA-sensitive species of ERBB3 is further reduced in molecular weight by treatment of lysates with endoglycosidase H (1 h, 0.1 U/20 μl), identifying it as nascent protein. Right: Nascent ERBB3 coimmunoprecipitates with HSP90. In the absence of BFA, the dominant mature ERBB3 species does not coimmunoprecipitate with HSP90. c The decrease in the levels of nascent ERBB3 protein levels to GA is not due to a decrease in mRNA levels. MCF7 cells were treated with GA or DMSO (DM) for 2 h, followed by collection of mRNA. The mRNA was reverse-transcribed, and the resulting cDNA was analyzed by quantitative PCR. The levels of β-actin cDNA are shown as a reference
Nascent ERBB3 is GA sensitive
At the low endogenous levels of ERBB3, found in most cell lines and in MCF7 cells in particular, the pool of nascent ERBB3 is expected to be very small. However, the progression of ERBB3 from nascent to matured and glycosylated receptors can be blocked with BFA. By inhibiting the activation of the small GTPase Arf1p, BFA interferes with the recruitment of coat proteins to transport vesicles and blocks anterograde transport from the endoplasmic reticulum (ER) to the Golgi (Jackson and Casanova 2000). As a consequence, BFA treatment results in the accumulation of ERBB receptors as an incompletely glycosylated and ER-resident species that can be distinguished initially based on its lower apparent molecular weight by SDS-PAGE. When MCF7 cells were treated with BFA for 2 h, ERBB3 was built up as a putative nascent form of lower molecular weight (Fig. 5a). For ERBB2, which exhibits a significantly longer half-life, this treatment did not result in a visible build up of nascent receptors within the time frame in which MCF7 cells tolerated BFA treatment. Prolonged BFA treatment beyond 4 h resulted in significant morphological changes and large-scale system-wide ubiquitination, thereby limiting the maximum duration of this assay (data not shown). When BFA treatment was combined with GA, we did not observe the faster migrating and putative nascent species of ERBB3 while the higher molecular weight and putative mature species remained. However, when MCF7 cells were pretreated with BFA for 2 h, followed by treatment with both BFA and GA for an additional 2 h, the depletion of the faster migrating species was incomplete. This may suggest that ERBB3, which accumulated during the first 2 h of BFA pretreatment, has progressed to a GA-insensitive but not fully glycosylated species. The relative fraction of the apparently GA-insensitive and nascent species varied between experiments, as is evident by comparison with Fig. 5b. The primary experimental variation however was consistently in the degree of buildup of the GA-sensitive nascent receptors, not the amount of GA-resistant nascent receptors that remained when compared to mature species. However, within the limitations of this assay, we also cannot exclude the possibility that 4 h of BFA treatment has merely compromised the degradation machinery involved in the GA-induced processing of ERBB3.
To further confirm that the nascent form of ERBB3 exhibits selective sensitivity to geldanamycin, we digested the lysate obtained from BFA and GA-treated MCF7 cells with endoglycosidase H (Fig. 5b). Endoglycosidase H cleaves carbohydrate moieties of nascent proteins while mature glycosylated protein is resistant to endoglycosidase H (Chavany et al. 1996). Following treatment with endoglycosidase H, the faster migrating band of ERBB3 is shifted to an even lower apparent molecular weight. This shift confirms the identity of the faster migrating and GA-sensitive band as nascent ERBB3.
In order to demonstrate that the GA sensitivity of the nascent species correlates with a physical interaction of ERBB3 and HSP90, we coimmunoprecipitated HSP90 and ERBB3 (Fig. 5b, right). The coimmunoprecipitation of HSP90 and ERBB3 following BFA treatment recovers a species of ERBB3 that correlates in its running behavior with the band confirmed by endoglycosidase H treatment to be nascent protein. By contrast, in the absence of BFA treatment, when mature protein represents the overwhelming majority of receptors, no mature receptor was coimmunoprecipitated. These findings support the indirect evidence obtained through receptor cross-linking (Fig. 4) that mature ERBB3 is not only insensitive to GA but does not bind HSP90. Likewise, no ERBB3 was recovered after 4 h of BFA treatment and 2 h of GA treatment. This is consistent with a model in which the residual nascent species that can be obtained when the addition of BFA precedes GA treatment by 2 h is resistant to GA and does not bind HSP90. However, the relative scarcity of the potentially GA-resistant species in the input material limits conclusions that are based on the absence of signal in the immunoprecipitation.
Previous studies have shown that GA does not significantly affect overall levels of protein synthesis (Fisher et al. 2000; Giannini and Bijlmakers 2004), but the destabilization of HSP90 clients could conceivably impact the regulation of small subsets of genes, including ERBB3, within the time course of our experiment. Such a selective suppression of ERBB3 expression would explain the observed sensitivity of ERBB3 to GA at an early stage of synthesis. Using quantitative PCR, we evaluated the levels of ERBB3 mRNA relative to a β-actin control in the presence or absence of GA (Fig. 5b). MCF7 cells were treated with GA or vehicle control for 2 h. Relative to a DMSO control, ERBB3 message levels decreased only 5%. When message levels were instead normalized for the modest increase in the levels for the β-actin control after GA treatment, ERBB3 mRNA levels decreased approximately 20% with GA treatment. This drop of 5% or 20% in mRNA levels, depending on the mode of normalization, is in contrast to the complete depletion of nascent ERBB3 over the same time period (Fig. 4a) and suggests that the inhibition of ERBB3 expression does not provide a sufficient explanation for the observed lack of nascent ERBB3 after 2 h of treatment with GA.
In order to further confirm the differential impact of GA on nascent ERBB3 by alternative and minimally invasive means, we made use of a C-terminal fusion protein of ERBB3 with the photo-convertible fluorophore Dendra2 (Fig. 6). Dendra2 is a mutated form of green fluorescent protein (GFP) that is expressed as a green fluorophore that can be photoconverted to a red fluorophore. Similar to GFP, protein folding occurs rapidly for Dendra2, but the subsequent spontaneous formation of its fluorophore through cyclization and oxidation of amino acid side chains is slow, requiring approximately 90 min (Zhang et al. 2007). In order to visualize the differential impact of GA on newly made and preexisting ERBB3, individual MCF7 cells expressing ERBB3–Dendra2 were imaged prior to photoconversion and the reemergence of green fluorescent ERBB3–Dendra2 after photoconversion was evaluated in the presence or absence of GA. Within 2 h, new green fluorescent ERBB3–Dendra2 emerged in untreated cells in the perinuclear region. Most of the reemerging green fluorescent protein could be observed in the presence of cycloheximide. This is a reflection of the 90-min time delay between synthesis and the emergence of fluorescence, imposed by the intramolecular formation of the Dendra2 fluorophore. However, in the presence of GA, we observed only a very modest increase in new green fluorescent protein.
Fig. 6.
a Live single-cell visualization of the destabilization of nascent ERBB3–Dendra2 fusion proteins in MCF7 cells. MCF7 cells were transiently transfected with ERBB3–Dendra2, and the expressed green fusion protein was photoconverted, giving rise to red ERBB3–Dendra2 (shown for untreated and geldanamycin-treated samples), which turns over and moves from perinuclear to diffuse cell surface localizations over the course of 2 h. Cells were treated for 2 h as indicated with GA, cycloheximide, or without drugs. Images were taken before and immediately after photo conversion, as well as 2 h later. The emergence of green fluorescent protein in the presence of cycloheximide reflects the time delay between synthesis and rapid folding and the relatively slow chromophore formation for Dendra2. However, most of this previously synthesized and folded ERBB3–Dendra2 fusion protein is sensitive to GA, indicating a point of GA sensitivity at a relatively late stage in maturation. b Stable expression of low levels of Dendra2, ERBB3–Dendra2 and ERBB3(ΔKD)–Dendra2 in MCF7 cells. Low-level expression significantly reduces perinuclear buildup of receptor fusion constructs, and cell surface localization is particularly visible at cell–cell contacts. Nuclei are stained with DAPI. c GA sensitivity of Dendra2 fusion proteins expressed stably at or below endogenous ERBB3 levels. MCF7 cells stably expressing the indicated Dendra2 constructs were treated with GA for the indicated time, and protein levels were evaluated by Western blotting for Dendra2. ERBB3–Dendra2 but not free Dendra2 exhibits GA sensitivity that mirrors endogenous ERBB3 (compare with Fig. 3). ERBB3(ΔKD)–Dendra2 exhibits only minimal GA sensitivity at prolonged exposure times, indicating a requirement for the kinase domain in GA sensitivity
Over the course of this assay, cell surface ERBB3–Dendra2 remained constant, consistent with an equilibrium of resupply and constitutive turnover. The recombinant overexpression of ERBB3–Dendra2 results in a larger proportion of receptors accumulating in perinuclear regions than was anticipated based on the small pool of nascent protein that was detected biochemically at low endogenous expression levels. However, the perinuclear pool of ERBB3–Dendra2 is stable over time and ultimately moves along the secretory pathway to replenish cell surface ERBB3–Dendra2. Although our experimental setup did not allow accurate quantitation of the flux of perinuclear-localized ERBB3–Dendra2, we did not see indications of an accelerated decrease of perinuclear-localized receptors in the presence of GA. In contrast to data obtained from BFA treatment, this fluorescence-based approach is expected to cause only minimal perturbation of cellular processes on a system-wide level. However, its findings are consistent with our biochemical findings of staggered BFA and GA treatment (Fig. 5a) and would suggest that perinuclear ERBB3 receptors that have progressed beyond a critical stage in their synthesis appear to be resistant to GA.
Additional insight into the time window of GA sensitivity is provided by the time delay between the synthesis and rapid folding of Dendra2 and the slow maturation of its fluorophore (Zhang et al. 2007). Due to this delay, most of the green fluorescent protein that reemerges after photoconversion was already synthesized prior to the addition of either GA or CHX. The sensitivity of this pool of ERBB3–Dendra2 to GA but not CHX indicates that GA sensitivity is not limited to the early stages of protein synthesis and early folding. In fact, the almost complete lack of reemergence of new green fluorescent ERBB3–Dendra2 places the point of sensitivity to GA at a relatively late stage within the 1.5-h period for fluorophore formation that follows the rapid folding of the fluorescent reporter.
The kinase domain of ERBB3 is required for GA sensitivity and the physical association with HSP90
The above fluorescent pulse chase experiment in the absence of BFA does require transient overexpression to significantly populate early states of maturation. In addition, Dendra2 may have a chaperon requirement on its own that could alter the processing of the ERBB3–Dendra2 fusion proteins. In order to quantitatively evaluate the GA sensitivity of ERBB3–Dendra2 fusions, we created stable cell lines in which the levels of fusion proteins were at or below the levels of endogenous ERBB3, as determined by Western blotting for the shared C-terminal ERBB3 epitope for the C17 antibody (Santa Cruz, data not shown). In addition, we created a fusion protein (ERBB3(ΔKD)–Dendra2) in which the kinase domain of ERBB3 has been removed, but all other cytoplasmic segments were retained. Cells stably expressing soluble, cytoplasmic Dendra2 were used as an additional control for the GA sensitivity of Dendra2. The cytoplasmic localization of Dendra2 and cell surface localization of the receptor fusions at low endogenous-like levels are shown in Fig. 6b. Upon treatment with GA and Western blot detection of Dendra2, ERBB3–Dendra2 shows a significant decline in receptor levels that is comparable to the rate of decrease seen for wild-type ERBB3 (Fig. 6c). By comparison, Dendra2 and ERBB3(ΔKD) show no significant decrease in protein levels. These findings confirm that Dendra2 does not alter the GA sensitivity of ERBB3 and indicate that the GA sensitivity seen for the full-length receptor is mediated by the presence of the kinase domain.
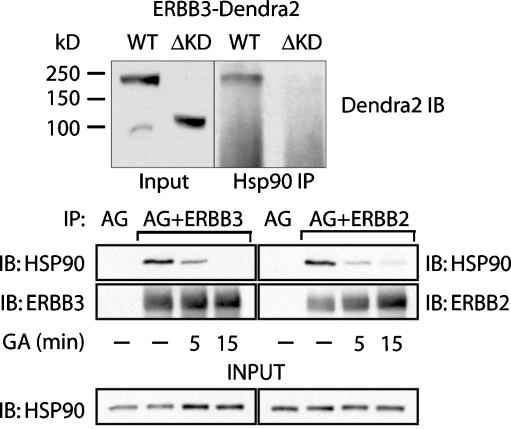
To test whether the loss in GA sensitivity for ERBB3(ΔKD) reflects a failure to physically interact with HSP90, we carried out a coimmunoprecipitation after a limited (2 h) treatment with BFA to enrich for nascent protein at low expression levels while minimizing potential artifacts that may arise from prolonged BFA treatment (Fig. 5b). At gel running conditions needed to visualize full-length as well as kinase domain deleted fusion proteins of Dendra2 and ERBB3, nascent and mature species did not resolve as distinct species. However, following immunoprecipitation of HSP90, a band matching the size of the ERBB3–Dendra2 fusion protein in the input control can be detected. By comparison, ERBB3(ΔKD)–Dendra2 did not coimmunoprecipitate with HSP90.
We also wanted to confirm the observed interaction of ERBB3 and HSP90 in the reverse coimmunoprecipitation and compare it to the already-established interaction of ERBB2 and HSP90. To this end, we overexpressed additional ERBB2 or ERBB3 in MCF7 cells, immunoprecipitated the receptors, and probed for coimmunoprecipitated HSP90. In contrast to the studies shown in Fig. 5, the addition of GA was limited to relatively short time periods prior to lysis, thus emphasizing the destabilization of existing complexes over the inhibition of new complex formation. Under those conditions, the direct interaction of ERBB3 and HSP90 could be confirmed in the reverse coimmunoprecipitation. Both ERBB3 and ERBB2 coimmunoprecipitations were sensitive to GA addition just prior to lysis, albeit with potentially different rates of complex dissociation.
Discussion
HSP90 interacts with a wide range of client proteins, yet proteins in signal transduction are heavily overrepresented as HSP90 clients. At the onset of this study, ERBB3 had been identified as a putative client based on the clear but delayed decrease of its steady-state levels upon treatment with GA, a mode of response similar to that of EGFR. Previous studies that focused on EGFR and ERBB2 had provided several pieces of evidence. On the one hand, the absence of a negative charge in a defined surface area of the kinase domain correlates with binding of mature ERBB2 receptors to HSP90 that can be abolished in the case of ERBB2 or created in the case of EGFR through targeted alterations of the surface charge features. However, these same features do apparently not determine sensitivity of nascent EGFR or ERBB2 to GA, yet the kinase domain is required for overall GA sensitivity, including that of the nascent state. These observations may be reconciled in different models. The means by which HSP90 “reads out” features of the kinase domain could involve distinct regions of the kinase domain in the nascent and mature state and hence represent truly distinct complexes, both in the nature of HSP90 interfaces and potential cochaperones. Alternatively, differences may reside mainly in the stability of complexes, and interactions of HSP90 with the nascent kinase domains may intrinsically involve weaker and more transient interactions. The strengthening and extension of these interactions into the mature state may differ between kinases and may depend on several factors. The nature of the previously identified “mature” kinase interface, or the activation state and correlating conformation of the kinase domain, or a combination of both factors could contribute to such a differential stability.
In contrast to EGFR or ERBB2, ERBB3 is constitutively deficient in its kinase activity, yet it displays overall GA sensitivity and a modeled surface charge distribution that would predict an HSP90 dependency similar to wild-type EGFR. We anticipated that a detailed analysis of the GA sensitivity of ERBB3 would shed light on two questions. First, does the lack of kinase activity in the absence of unnatural and potentially structure perturbing mutations within the kinase domain supersede the previously identified surface features with respect to GA sensitivity in the mature state? Second, does the anticipated lack of significant sensitivity in the mature state, especially in the context of a naturally kinase-deficient receptor, represent exclusive sensitivity in the nascent state or alternatively a GA sensitivity that is merely reduced in its intensity and impact in the mature state?
In terms of the specificity of the GA-induced depletion of ERBB3, our data indicate that steady-state levels as well as the levels of cell surface ERBB3 decrease at a rate equivalent to unchallenged steady-state turnover. This would suggest a sensitivity of nascent ERBB3 without a partial destabilization of mature cell surface receptors (Figs. 2 and 3). Note that this aspect of the analysis defines mature ERBB3 as cell-surface-localized ERBB3. In fact, instead of a destabilization of cell surface ERBB3, we saw a modest but consistent stabilization across several assays compared to the rapid destabilization of cell-surface-localized ERBB2. This may be the consequence of a GA-induced stabilization of factors that contribute to the steady-state turnover of ERBB3. However, the nature of this stabilization is not clear from our data.
Based on two independent observations, the complete lack of GA sensitivity of mature ERBB3 is likely a reflection of a lack of interaction with HSP90. For cell surface ERBB3, we evaluated this indirectly through the interference of HSP90 with receptor association events (Fig. 4). Consistent with the established binding of HSP90 to mature ERBB2, cross-linking of individually overexpressed cell surface ERBB2 but not ERBB3 was sensitive to GA. The lack of interaction of more broadly defined mature ERBB3 with HSP90 was also confirmed in coimmunoprecipitation studies after BFA treatment (Fig. 5b). This immunoprecipitation recovered nascent ERBB3 but not fully glycosylated, mature ERBB3, even under conditions when the mature band is by far the dominant species (ratio visible in Fig. 5a, b). Both the sensitivity to GA (Fig. 6) as well as the physical interaction with HSP90 (Fig. 7) was lost upon deletion of the kinase domain, consistent with previous studies on ERBB2 and EGFR. The observed decrease in steady-state levels of ERBB3 is also not due to a decrease in ERBB3 message (Fig. 5c). In combination, our findings indicate that despite its intrinsic deficiency in kinase activity, ERBB3 is selectively dependent on physical interactions with HSP90 in the nascent state of the receptor.
Fig. 7.
ERBB3 associates with HSP90 in a kinase-domain-dependent manner. a (top)MCF7 cells stably expressing ERBB3–Dendra2 or ERBB3(ΔKD)–Dendra2 were pretreated with BFA (2 h) to enrich nascent fusion protein. IPs were analyzed by Western blotting for Dendra2 to avoid cross-reactivity with endogenous ERBB3. ERBB3–Dendra2 but not ERBB3(ΔKD)–Dendra2 coimmunoprecipitates with HSP90. b (bottom) The direct immunoprecipitation of ERBB3 and ERBB2 with HSP90 was confirmed in the reverse direction by detecting HSP90 after immunoprecipitation of either ERBB2 or ERBB3. MCF7 cells were transiently transfected with either additional ERBB2 or ERBB3 without Dendra2 fusion. Prior to lysis, cells were treated with DMSO or 5 μM GA for 5 or 15 min as indicated. Cells were lysed and immunoprecipitations were performed with antibodies against the extracellular domains of ERBB2 or ERBB3. Protein A/G (AG)-conjugated beads were used in all samples, and beads without antibodies (AG) serve as a control for nonspecific binding to the resin
The definition of mature receptors as being cell-surface-localized reflects technical limitations. Studies with Brefeldin A and endoglycosidase H treatment more narrowly define the end point of the time window in which ERBB3 is sensitive to GA. The accumulation of nascent and incompletely glycosylated ERBB3 in the presence of Brefeldin A is blocked by GA within a 2-h window of simultaneous treatment with both drugs. On the other hand, ERBB3 that accumulates due to a prior BFA treatment is insensitive to GA (Fig. 5a). Similar results were obtained in the absence of BFA using overexpressed ERBB3–Dendra2 in fluorescent pulse chase studies. Preexisting (red converted) pools of perinuclear ERBB3–Dendra2 proceed gradually to the cell surface instead of being rapidly destabilized (Fig. 6a). This suggests a relatively early stage at which GA sensitivity is lost, possibly prior to the point of BFA-induced accumulation, which is the exit from the ER to the Golgi. Coimmunoprecipitation studies after staggered BFA and GA treatment (Fig. 5b) are in agreement with this model, although the overall low abundance of the potentially GA-resistant pool limits the conclusions that can be drawn from a lack of immunoprecipitation. Attempts to further exploit these observations and to narrow down the point of emerging GA insensitivity were limited on technical grounds. For studies using BFA treatment, we observed significant cellular toxicity and system-wide but also ERBB3-directed ubiquitination, even in the absence of GA treatment (data not shown) at periods of prolonged BFA treatment (4 h or longer). A prolonged BFA treatment would be needed to build up a significant pool of GA-insensitive receptors from the endogenous receptor pool.
Nevertheless, some additional insight into the time window of GA sensitivity is provided by the use of the ERBB3–Dendra2 fusion protein. The Dendra2 reporter construct requires approximately 90 min postfolding for fluorophore maturation. Consistent with the above BFA studies, previously accumulated and perinuclear receptors appear resistant to GA but the emergence of new green fluorescent ERBB3–Dendra2 is fully blocked by GA but not by cycloheximide. Hence, the point of GA sensitivity is substantially later than the time of initial synthesis and early folding events. This may indicate a role for HSP90 at a late stage in maturation or possibly structural proofreading. A relatively well-studied example for the role of HSP90 in structural proof reading is provided by the cystic fibrosis transmembrane conductance regulator (CFTR). For CFTR, HSP90 but not GRP94 or HSP70 is implicated in the control of endoplasmic-reticulum-associated degradation, although HSP70 does bind CFTR at an earlier stage in maturation (Loo et al. 1998). This suggested that for CFTR the role of HSP90 is not in early folding assistance. Furthermore, for CFTR, the transient nature of the interaction between HSP90 and CFTR is a critical requirement. Mutant CFTR, which fails to proceed to the cell surface, apparently suffers from an excessive stability and consequently enhanced duration of HSP90 binding (Wang et al. 2006). Combined with the short time window in which productive interactions would occur in the late ER just prior to export to the Golgi, this creates effectively a kinetic proofreading mechanism.
Our data indicate that mature ERRB3 is not only insensitive to GA but does not bind HSP90. The distinct lack of HSP90 binding to mature ERBB3, despite of its catalytically dead kinase domain, is expected to impact the initiation of signaling through ERBB3–ERBB2 heterodimers and may reflect the evolutionary optimization of this functionally specialized receptor pair. As a functional dimer, HSP90 far exceeds the size of the kinase domain of ERBB2 or ERBB3, even in the absence of additional cochaperones. The association of HSP90 with ERBB2 does block transient ERBB2 homodimerization (Fig. 4), suppresses ERBB2 autoactivation and subsequent phosphorylation of the receptor by activated Src (Xu et al. 2007), and interferes with heterodimerization (Citri et al. 2004). While HSP90 is not associated with activated ERBB2, it is not clear if activation results in the dissociation of HSP90 or whether HSP90 displacement has to precede catalytic activation. However, ligands initially bind to ERBB3 and in doing so destabilize the self-association of signaling incompetent ERBB3 receptors (Kani et al. 2005). Ligand-bound ERBB3, that is poised to allosterically activate ERBB2 through kinase domain interactions, is expected to be a more potent competitor with HSP90 for binding to the kinase domain of ERBB2. The absence of HSP90 association for mature ERBB3 may therefore aid in efficient ligand-mediated signal transmission while HSP90 association with ERBB2 imposes a barrier to stochastic activation events that result from transient interactions. The latter level of protection is also not needed for ERBB3.
The structural analysis of the features needed for the allosteric cross-activation of ERBB kinase domains has provided great insights into the selective conservation of structural kinase domain motifs (Zhang et al. 2006). Specifically, the variability of the most N-terminal amino acids of the ERBB3 kinase domain, compared to EGFR, ERBB2, and ERBB4, correlates nicely with the role of ERBB3 as a structurally conserved allosteric activator (through interactions mediated by its conserved C-terminal lobe) and inability to process incoming activating signal (mediated through the most N-terminal segments of the N-lobe in the case of EGFR, ERBB2, and ERBB4). However, the variability of the most N-terminal residues of ERBB3 that precede the first segment of the N-terminal β-sheet may also be relevant to the exclusion of HSP90 from interactions with mature ERBB3. This is apparently the case despite its intrinsic kinase deficiency, a catalytic state that otherwise can reintroduce GA sensitivity in the case of mature EGFR. In its divergent N terminus, the ERBB3 kinase domain may share some functional resemblance with JNKs. Instead of sequence variation, JNKs feature an additional 28-amino-acid structural motif that is thought to sterically cover elements in the underlying N-terminal lobe surrounding the αC-β4 loop, known to confer HSP90 binding determinants in the case of mature ERBB2. The exclusion of HSP90 from the mature state of select kinases may therefore depend on either blocked access to the HSP90 interfaces or alterations of adjacent motifs. In this model, binding of HSP90 to the mature state of a kinase would represent the default. Specific subsets of kinases may have evolved different but related mechanisms to curtail this dependency and to uncouple their mature state functionally from HSP90, possibly to facilitate their role in cellular stress or ligand-mediated responses.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Ribbon diagram and surface charge representation of the kinase domains of EGFR in the active conformation (1M17, Stamos et al. 2002) and lapatinib-stabilized inactive conformation (1XKK, Wood et al. 2004). Large segments (outlined in purple) are not resolved in the inactive conformation, making surface charge projections of this portion of the kinase domain inaccurate. However, the area around the αC-β4 loop is resolved and retains the characteristic negative surface charge projection, associated with a failure of mature EGFR to bind HSP90. (JPEG 4674 kb)
Acknowledgements
We thank Dr. Walter Scott for critical reading and suggestions during the writing of this manuscript and members of the Shuai lab at UCLA for help with qPCR analysis. This work was supported by funding from the National Institutes of Health (RL, CA098881-01A1, CA98881-05).
Footnotes
This work was supported by funding from the National Institutes of Health (RL, CA098881).
References
- Aguilar Z, Akita RW, Finn RS, et al. Biologic effects of heregulin/neu differentiation factor on normal and malignant human breast and ovarian epithelial cells. Oncogene. 1999;18:6050–6062. doi: 10.1038/sj.onc.1202993. [DOI] [PubMed] [Google Scholar]
- Austin CD, Maziere AM, Pisacane PI, et al. Endocytosis and sorting of ErbB2 and the site of action of cancer therapeutics trastuzumab and geldanamycin. Mol Biol Cell. 2004;15:5268–5282. doi: 10.1091/mbc.E04-07-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavany C, Mimnaugh E, Miller P, et al. p185erbB2 binds to GRP94 in vivo. Dissociation of the p185erbB2/GRP94 heterocomplex by benzoquinone ansamycins precedes depletion of p185erbB2. J Biol Chem. 1996;271:4974–4977. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- Citri A, Alroy I, Lavi S, et al. Drug-induced ubiquitylation and degradation of ErbB receptor tyrosine kinases: implications for cancer therapy. Embo J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Gan J, Mosesson Y, Vereb G, Szollosi J, Yarden Y. Hsp90 restrains ErbB-2/HER2 signalling by limiting heterodimer formation. EMBO Rep. 2004;5:1165–1170. doi: 10.1038/sj.embor.7400300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Harari D, Shohat G, et al. Hsp90 recognizes a common surface on client kinases. J Biol Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- Dent P, Yacoub A, Contessa J, et al. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res. 2003;159:283–300. doi: 10.1667/0033-7587(2003)159[0283:SARIAO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Dote H, Cerna D, Burgan WE, Camphausen K, Tofilon PJ. ErbB3 expression predicts tumor cell radiosensitization induced by Hsp90 inhibition. Cancer Res. 2005;65:6967–6975. doi: 10.1158/0008-5472.CAN-05-1304. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Mandart E, Doree M. Hsp90 is required for c-Mos activation and biphasic MAP kinase activation in Xenopus oocytes. Embo J. 2000;19:1516–1524. doi: 10.1093/emboj/19.7.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini A, Bijlmakers MJ. Regulation of the Src family kinase Lck by Hsp90 and ubiquitination. Mol Cell Biol. 2004;24:5667–5676. doi: 10.1128/MCB.24.13.5667-5676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy PM, Platko JV, Cantley LC, Cerione RA, KLr C. Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/S0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Kani K, Warren CM, Kaddis CS, Loo JA, Landgraf R. Oligomers of ERBB3 have two distinct interfaces that differ in their sensitivity to disruption by heregulin. J Biol Chem. 2005;280:8238–8247. doi: 10.1074/jbc.M410944200. [DOI] [PubMed] [Google Scholar]
- Landgraf R, Eisenberg D. Heregulin reverses the oligomerization of HER3. Biochemistry. 2000;39:8503–8511. doi: 10.1021/bi000953+. [DOI] [PubMed] [Google Scholar]
- Lerdrup M, Hommelgaard AM, Grandal M, Deurs B. Geldanamycin stimulates internalization of ErbB2 in a proteasome-dependent way. J Cell Sci. 2006;119:85–95. doi: 10.1242/jcs.02707. [DOI] [PubMed] [Google Scholar]
- Lerdrup M, Bruun S, Grandal MV, Roepstorff K, Kristensen MM, Hommelgaard AM, Deurs B. Endocytic down-regulation of ErbB2 is stimulated by cleavage of its C-terminus. Mol Biol Cell. 2007;18:3656–3666. doi: 10.1091/mbc.E07-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, Riordan JR. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. Embo J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryot Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.9.4974. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Mizuno S, Uehara Y. Accelerated degradation of 160 kDa epidermal growth factor (EGF) receptor precursor by the tyrosine kinase inhibitor herbimycin A in the endoplasmic reticulum of A431 human epidermoid carcinoma cells. Biochem J. 1994;301(Pt 1):63–68. doi: 10.1042/bj3010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Uehara Y, Noshima T, Itaya T, Kunieda Y, Kurosawa M. In vivo antitumor activity of herbimycin A, a tyrosine kinase inhibitor, targeted against BCR/ABL oncoprotein in mice bearing BCR/ABL-transfected cells. Leuk Res. 1994;18:867–873. doi: 10.1016/0145-2126(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Park E, Baron R, Landgraf R. Higher-order association states of cellular ERBB3 probed with photo-cross-linkable aptamers. Biochemistry. 2008;47:11992–12005. doi: 10.1021/bi8004208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashtan I, Tsutsumi S, Wang S, Xu W, Neckers L. Targeting Hsp90 prevents escape of breast cancer cells from tyrosine kinase inhibition. Cell Cycle. 2008;7:2936–2941. doi: 10.4161/cc.7.18.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, Kluger HM. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 2007;67:2932–2937. doi: 10.1158/0008-5472.CAN-06-4511. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Silverstein AM, Galigniana MD. A model for the cytoplasmic trafficking of signalling proteins involving the hsp90-binding immunophilins and p50cdc37. Cell Signal. 1999;11:839–851. doi: 10.1016/S0898-6568(99)00064-9. [DOI] [PubMed] [Google Scholar]
- Prince T, Matts RL. Definition of protein kinase sequence motifs that trigger high affinity binding of Hsp90 and Cdc37. J Biol Chem. 2004;279:39975–39981. doi: 10.1074/jbc.M406882200. [DOI] [PubMed] [Google Scholar]
- Prince T, Matts RL. Exposure of protein kinase motifs that trigger binding of Hsp90 and Cdc37. Biochem Biophys Res Commun. 2005;338:1447–1454. doi: 10.1016/j.bbrc.2005.10.100. [DOI] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Choi SH, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe SM, Ali MM, Meyer P, et al. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/S0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- Sakagami M, Morrison P, Welch WJ. Benzoquinoid ansamycins (herbimycin A and geldanamycin) interfere with the maturation of growth factor receptor tyrosine kinases. Cell Stress Chaperones. 1999;4:19–28. doi: 10.1379/1466-1268(1999)004<0019:BAHAAG>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J. 1997;322(Pt 3):757–763. doi: 10.1042/bj3220757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF. Sequence motifs shared between chaperone components participating in the assembly of progesterone receptor complexes. Biol Chem. 1998;379:283–288. doi: 10.1515/bchm.1998.379.4-5.505. [DOI] [PubMed] [Google Scholar]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Tikhomirov O, Carpenter G. Identification of ErbB-2 kinase domain motifs required for geldanamycin-induced degradation. Cancer Res. 2003;63:39–43. [PubMed] [Google Scholar]
- Vaughan CK, Mollapour M, Smith JR, et al. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell. 2008;31:886–895. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Venable J, LaPointe P, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Warren CM, Kani K, Landgraf R. The N-terminal domains of neuregulin 1 confer signal attenuation. J Biol Chem. 2006;281:27306–27316. doi: 10.1074/jbc.M512887200. [DOI] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J. Hsp70 and Hsp90—a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp 60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, Neckers L. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem. 2001;276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Mimnaugh EG, Kim JS, Trepel JB, Neckers LM. Hsp90, not Grp94, regulates the intracellular trafficking and stability of nascent ErbB2. Cell Stress Chaperones. 2002;7:91–96. doi: 10.1379/1466-1268(2002)007<0091:HNGRTI>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yuan X, Xiang Z, Mimnaugh E, Marcu M, Neckers L. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat Struct Mol Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- Xu W, Yuan X, Beebe K, Xiang Z, Neckers L. Loss of Hsp90 association up-regulates Src-dependent ErbB2 activity. Mol Cell Biol. 2007;27:220–228. doi: 10.1128/MCB.00899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BG, Matts RL. Hsp90 functions to balance the phosphorylation state of Akt during C2C12 myoblast differentiation. Cell Signal. 2005;17:1477–1485. doi: 10.1016/j.cellsig.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gurskaya NG, Merzlyak EM, et al. Method for real-time monitoring of protein degradation at the single cell level. Biotechniques. 2007;42:446. doi: 10.2144/000112453. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zheng FF, Kuduk SD, Chiosis G, Munster PN, Sepp-Lorenzino L, Danishefsky SJ, Rosen N. Identification of a geldanamycin dimer that induces the selective degradation of HER-family tyrosine kinases. Cancer Res. 2000;60:2090–2094. [PubMed] [Google Scholar]
- Zhou P, Fernandes N, Dodge IL, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Ribbon diagram and surface charge representation of the kinase domains of EGFR in the active conformation (1M17, Stamos et al. 2002) and lapatinib-stabilized inactive conformation (1XKK, Wood et al. 2004). Large segments (outlined in purple) are not resolved in the inactive conformation, making surface charge projections of this portion of the kinase domain inaccurate. However, the area around the αC-β4 loop is resolved and retains the characteristic negative surface charge projection, associated with a failure of mature EGFR to bind HSP90. (JPEG 4674 kb)