Abstract
The Hsp90 molecular chaperone has been implicated as a contributor to evolution in several organisms by revealing cryptic variation that can yield dramatic phenotypes when the chaperone is diverted from its normal functions by environmental stress. In addition, as a cancer drug target, Hsp90 inhibition has been documented to sensitize cells to DNA-damaging agents, suggesting a function for Hsp90 in DNA repair. Here we explore the potential role of Hsp90 in modulating the stability of nucleotide repeats, which in a number of species, including humans, exert subtle and quantitative consequences for protein function, morphological and behavioral traits, and disease. We report that impairment of Hsp90 in human cells induces contractions of CAG repeat tracks by tenfold. Inhibition of the recombinase Rad51, a downstream target of Hsp90, induces a comparable increase in repeat instability, suggesting that Hsp90-enabled homologous recombination normally functions to stabilize CAG repeat tracts. By contrast, Hsp90 inhibition does not increase the rate of gene-inactivating point mutations. The capacity of Hsp90 to modulate repeat-tract lengths suggests that the chaperone, in addition to exposing cryptic variation, might facilitate the expression of new phenotypes through induction of novel genetic variation.
Keywords: Hsp90 chaperone, Stress-induced mutation, Repeat instability, Homologous recombination
Introduction
The Hsp90 molecular chaperone supports a diverse clientele of proteins, primarily kinases involved in signal transduction, cell cycle, and development (Citri et al. 2006; Neckers and Ivy 2003). Deviating from the classical description of chaperone function, Hsp90, under normal cellular conditions, appears to play a limited role in the indiscriminate de novo folding of proteins and instead assists in the maturation and stabilization of specific clients to facilitate their activation (Nathan et al. 1997) and assembly into larger complexes (Freeman and Yamamoto 2002). Pioneering work from Rutherford and Lindquist (1998) identified Hsp90 as a buffer of morphological variation (Queitsch et al. 2002; Sangster et al. 2008). The authors identified a molecular basis for the canalization of developmental traits, where Hsp90 suppresses cryptic genetic variation by stabilizing client proteins in spite of subtle changes in protein sequence. Under severe environmental stress, Hsp90 is recruited to repair damaged proteins, diverting the chaperone from its normal cellular functions. Under these conditions, the otherwise “silent” genetic variation is expressed, leading to an increase in morphological phenotypes. The Hsp90 chaperone therefore can facilitate rapid adaptation to environmental changes by releasing a broad range of “hidden” phenotypes (Palotai et al. 2008; Rutherford et al. 2007), of which a subset can be selected to accommodate environmental pressure (Cowen and Lindquist 2005).
Hsp90 is highly expressed in tumor cells, suggesting that the buffering and stabilizing functions of Hsp90 may also be vital to the survival and proliferation of malignant cells (Whitesell and Lindquist 2005). Pharmacological inhibitors of Hsp90, many of which are in clinical trials, have been effective in preferentially triggering cell death in several cancers (Banerji 2009). An unexpected consequence of treatment with Hsp90 inhibitors is the sensitization of tumors to DNA-damaging agents, prompting the use of these inhibitors in conjunction with conventional chemotherapies and radiotherapies. Impairment of Hsp90 has been implicated in the disruption of multiple aspects of DNA repair, most notably in double-strand break (DSB) repair (Camphausen and Tofilon 2007; Dote et al. 2006; Noguchi et al. 2006).
The observation that Hsp90 inhibition leads to improper resolution of DSBs, combined with the chaperone's known sensitivity to cellular stress, suggests a connection between genome instability and the environment. Microsatellite repeat mutation has been used as a biomarker or indicator of genome stability and cancer (Barber et al. 2006; Healy et al. 2006; Reuschenbach et al. 2009). In both yeast and mammalian cells, CAG repeat tracts have been shown to be naturally prone to DSBs (Jankowski et al. 2000; Meservy et al. 2003). We previously developed CAG-specific zinc-finger nucleases that introduce DSBs within CAG repeats and showed that they destabilize CAG repeats in a length-dependent manner in human and rodent cells, triggering a substantial induction of large repeat contractions (Mittelman et al. 2009). The susceptibility of CAG repeats to DSBs and the ability of Hsp90 inhibition to interfere with DSB repair led us to evaluate the possible role of Hsp90 in nucleotide repeat instability.
Using our previously described selection assay for measuring modifiers of CAG repeat contraction frequency (Lin et al. 2006), we find that impairment of Hsp90 significantly destabilizes CAG repeat tracts in human cells. Repeat destabilization appears to be mediated via effects on the recombinase Rad51, which is important for homology-dependent repair of DSBs. We show that inhibition of Hsp90 leads to a decrease in Rad51 protein, and that small interfering RNA (siRNA) knockdown of Rad51, in the absence of Hsp90 inhibition, leads to the same repeat instability as interference with Hsp90. These results suggest the intriguing possibility that Hsp90, apart from buffering and exposing cryptic variation, possesses the capacity to modulate the lengths of repeat tracts in the genome and, as a result, to induce a novel genetic variation.
Materials and methods
Human cell culture
The construction and growth conditions of the HT1080-derived HPRT− FLAH25 cell line have been described in detail elsewhere (Lin et al. 2006). Doxycycline (2 µg/ml) was added to the growth medium to induce the transcription of the HPRT reporter gene, which is controlled by the pTRE-CMVmini promoter. We performed selection for HPRT+ cells by plating 500,000 cells on 10-cm plates, supplemented with 0.1 mM hypoxanthine, 0.4 µM aminopterin, and 16 µM thymine, for 2 weeks. Surviving colonies were stained with 1% Coomassie blue for enumeration. HPRT+ frequencies, which were computed as the number of HPRT+ colonies divided by the number of viable cells, are the averages of results from at least three independent experiments. To select for HPRT− cells in HPRT+ HT1080 cells, we plated 500,000 cells on 10-cm plates, supplemented with 5 µg/ml 6-thioguanine, for 2 weeks. HPRT− frequencies were computed as described previously for the HPRT+ selection.
siRNA and drug treatments
We distributed 100,000 FLAH25 cells to each plate and transfected them 24 h later using Oligofectamine (Invitrogen) with siRNA duplexes for Hsp90α, Rad51, or Rad51c (Dharmacon). The sequences for the siRNA duplexes are as follows: Hsp90α siRNA-1, 5′-GUUUGAGAACCUCUGCAAA (Compton et al. 2006); Hsp90α siRNA-2, 5′-GGAAAGAGCUGCAUAUUAA (Chatterjee et al. 2007); Rad51 siRNA-1, 5′-GAGCUUGACAAACUACUUC (Ito et al. 2005); Rad51 siRNA-2, 5′-UGUAGCAUAUGCUCGAGCG (Ko et al. 2008); and Rad51C siRNA-1, 5′-CACCUUCUGUUCAGCACUAGA (Rodrigue et al. 2006). Each transfection included a specific siRNA at a concentration of 100 nM, along with a control siRNA at 100 nM targeted to vimentin but which has no discernable effect on cells (Lin et al. 2006). Each experiment also included a set of control cells transfected with vimentin siRNA alone at 200 nM. At 4 h posttreatment with siRNA, the cells were supplemented with a medium containing serum. The cells were grown for 72 h, and then retransfected with siRNA and cultured in the presence of doxycycline (2 µg/ml). Selection for HPRT+ cells was initiated 72 h after the second transfection. For drug treatments, the Hsp90 inhibitor 17-(allylamino)-17-demethoxygeldanamycin (17-AAG) (Sigma) was reconstituted in dimethyl sulfoxide (DMSO) at a concentration of 100 mM and added daily for 3 days to the cell medium, which contained doxycycline (2 µg/ml). Selection for HPRT+ cells was initiated 24 h after the final drug treatment. We used Student's t test to evaluate the significance of the differences between the means of specific siRNA treatments and the means of the control siRNA treatment.
Immunoblotting
We harvested cells at 24, 48, and 72 h posttreatment and used Western blot analysis to analyze the level of each target protein. For each analysis, 2–4 million cells were lysed in 200 μl of cell lysis buffer [300 mM Tris (pH 6.8), 2% sodium dodecyl sulfate, and 10% glycerol] on ice. Proteins (30 μg/sample, as quantified by a BCA protein assay) were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (pH 8.8) with a 4% stacking gel (pH 6.8) and transferred to nitrocellulose membranes at 30 mA overnight. Membranes were blocked with 5% dry milk in TBST [0.05 M Tris–HCl (pH 8.0), 0.15 M NaCl, and 0.05% Tween 20] at room temperature for 30 minutes and incubated with primary antibody (GAPDH, SC-32233, Santa Cruz Biotechnology; Hsp90α, SPA-840, Assay Designs; Rad51, PC130, EMD; Rad51C, MAB3696, Millipore), according to the manufacturer's instructions, in 3% dry milk in TBST for 2 h at room temperature. The membranes were incubated with 1:5000 secondary antibody IgG–horseradish peroxidase conjugate (Santa Cruz Biotechnology) in 3% dry milk in TBST for 1 hour at room temperature. We detected bands after exposure to ECL reagent (GE) and quantified the intensity of the bands using ImageJ software (National Institutes of Health).
Quantification of mRNA levels
To quantify HPRT messenger RNA (mRNA) levels, we harvested cells 24 h posttreatment, homogenized them in TRIzol reagent by four passes through a 24-gauge needle, added chloroform (20% of the total volume), vortexed the sample, and then centrifuged it at 10,000 × g for 15 min at 4°C. RNA was extracted from the aqueous layer using the RNeasy minikit (Qiagen) according to the manufacturer's recommendations. We used 50 ng of total RNA in each real-time RT-PCR using a SYBR Green RT-PCR kit (Qiagen) and normalized HPRT mRNA to β-actin mRNA. The HPRT primers were 5′-CGGCTACAAGGACGACTCTAG and 5′-TTGATGTAATCCAGCAGGTCAGC; the β-actin primers were 5′-AGAGAGGCATCCTCACCCTG and 5′-CATGAGGTAGTCAGTCAGGT. Conditions for RT-PCR were as follows: 50°C for 30 min, 95°C for 15 min, followed by 45 repeated cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 30 s. The relative levels of HPRT and β-actin mRNA were calculated by comparing the number of cycles (generally between 15 and 25 cycles) at which the PCR products became detectable above the basal threshold. The percentages of HPRT mRNA in siRNA-treated samples relative to vimentin controls were calculated as: % = 100 × 2exp[(HPRTcontrol − actincontrol) − (HPRTsample − actinsample)]. With the HPRT mRNA levels in vimentin-siRNA-treated cells defined as 100%, the value for Hsp90-siRNA-treated cells was 90 ± 25%, that for Rad51-siRNA-treated cells was 95 ± 1%, and that for Rad51C-siRNA-treated cells was 105 ± 6%.
Results and discussion
Drug and genetic inhibition of Hsp90
To test for a possible connection between Hsp90 function and the stability of repeat tracts, we employed a selection assay in human FLAH25 cells, which we have previously used to identify modifiers of CAG repeat stability (Gorbunova et al. 2003, 2004; Lin et al. 2006, 2010; Lin and Wilson 2007; Mittelman et al. 2009). FLAH25 cells carry an integrated HPRT minigene that contains a single intron interrupted by 95 copies of a CAG repeat (Fig. 1). This long CAG repeat is spliced into the HPRT mRNA, rendering the reporter gene inactive. If the repeat contracts to less than 39 copies, however, the CAG repeat is not efficiently spliced into the transcript, and a sufficient quantity of normal HPRT mRNA is produced to restore the activity of the reporter gene. Cells that experience adequate contractions in the CAG repeat survive selection for HPRT+ cells. The magnitude of an effect on contraction frequency can be assessed from the analysis of surviving HPRT+ colonies.
Fig. 1.
Selection assay for CAG repeat contraction in human cells. The long CAG tract in the intron of the HPRT gene prevents proper splicing and disrupts the production of functional HPRT protein. Contraction events that reduce the size of the CAG tract to 38 or fewer units allow the transcript to be efficiently spliced and sufficient normal HPRT protein to be produced, allowing the cell to survive HPRT+ selection. Selection for HPRT+ colonies allows for recovery of cells that have undergone a contraction in the CAG tract
We measured the mutagenic consequences of blocking Hsp90 function in FLAH25 cells by treatment with 17-AAG, a specific inhibitor of Hsp90. 17-AAG is a geldanamycin derivative that binds the N-terminal ATPase domain of Hsp90 and blocks all ATP-dependent chaperone activities (Kamal et al. 2003). We treated cells with 17-AAG dissolved in DMSO, or an equivalent concentration of DMSO alone, for 72 h and then assayed for contractions by selecting for HPRT+ cells. Pharmacological inhibition of Hsp90 with 17-AAG increased the frequency of HPRT+ colonies tenfold above the control values (P < 0.0001) (Fig. 2a). To identify alterations to the repeat tract, we isolated 15 HPRT+ colonies and characterized their CAG repeats by PCR amplification and DNA sequencing. We found that the CAG repeat tracts in 13 of the HPRT+ colonies were contractions to fewer than 39 repeats; the other two events were deletions that extended into the sequences flanking the repeat tract. This analysis is consistent with our previous characterizations of the selection assay (Gorbunova et al. 2004; Lin et al. 2006). Thus, inhibiting Hsp90 with 17-AAG destabilizes CAG repeat tracts.
Fig. 2.
Drug-induced inhibition and siRNA-mediated knockdowns. a Effects of 17-AAG and siRNA knockdowns on the frequency of HPRT+ colonies. The frequencies of HPRT+ colonies were calculated as the number of HPRT+ colonies divided by the number of viable cells; values were averaged over at least three independent experiments per treatment. The values (× 10− 6) for each condition are as follows: vimentin siRNA (8.1 ± 2.7; N =.7), 17-AAG (80 ± 20; N = 5), Hsp90α siRNA-1 (95 ± 24; N = 5), Hsp90α siRNA-2 (93 ± 18; N = 3), Rad51 siRNA-1 (76 ± 10; N = 6), Rad51 siRNA-2 (98 ± 21; N = 5), and Rad51C siRNA (52 ± 17; N = 3). An asterisk indicates that P < 0.0001 for a given treatment, in comparison to treatment with vimentin siRNA. b Representative Western blots demonstrating that siRNA treatments decreased the amounts of the target proteins, relative to GAPDH, after 24 h (D1), 48 h (D2), and 72 h (D3). As shown in the first column of each Western blot, 72 h of treatment with vimentin siRNA (VIM) had no effect on any of the target proteins. c Representative Western blot demonstrating that treatment with 17-AAG decreased the amount of Rad51 in a dose-dependent manner relative to GAPDH
To confirm that the observed effect on repeat contraction was the result of Hsp90 inhibition rather than a nonspecific effect of 17-AAG, we directly inhibited the expression of Hsp90 by siRNA knockdown. There are two isoforms of cytosolic Hsp90 in mammalian cells—Hsp90α and Hsp90β, and 17-AAG functionally inhibits both. Hsp90α is the inducible and more abundant form of mammalian Hsp90, while Hsp90β is fairly insensitive to cellular stress and has functions that are distinct from those of Hsp90α (Sreedhar et al. 2004). We therefore chose to use Hsp90α-specific siRNAs to confirm the effect on repeat instability. As shown in Fig. 2b, treatment with Hsp90α siRNA substantially reduced the level of cellular Hsp90α. siRNA knockdown of Hsp90α increased the frequency of HPRT+ colonies more than 11-fold above the control values (P < 0.0001) (Fig. 2a), which is about the same increase we obtained by inhibiting Hsp90 with 17-AAG. Because treatments with 17-AAG and Hsp90α-specific siRNAs destabilize CAG repeat tracts, we conclude that Hsp90 normally functions to maintain CAG repeat stability.
We previously showed that transcription through the CAG repeat tract in FLAH25 cells increases the frequency of repeat contractions (Lin et al. 2006). In our Hsp90 experiments, transcription was fully induced by addition of doxycycline; however, it was possible that inhibition or knockdown of Hsp90 could increase repeat instability by further stimulating transcription through the HPRT gene. To investigate this possibility, we measured the levels of HPRT mRNA induced by doxycycline when Hsp90 functioned normally and when it was knocked down by siRNA treatment. Knocking down Hsp90 function gave about 90% as much HPRT expression as occurred when the cells were treated with control siRNA. These results rule out the trivial possibility that Hsp90 inhibition destabilizes CAG repeats by stimulating transcription through the HPRT gene.
Molecular basis for Hsp90-mediated effects on CAG repeat instability
It is unlikely that Hsp90 modulates CAG repeat instability through a direct interaction with DNA. Rather, Hsp90 likely influences instability via an indirect effect on DNA metabolism. It is known, for example, that inhibition of Hsp90 sensitizes tumor cells to genotoxic agents through its effects on client proteins in one or more DNA repair pathways (Arlander et al. 2003; Dote et al. 2006; Noguchi et al. 2006; Yao et al. 2007). These effects are best characterized for proteins involved in DSB repair. Impairment of Hsp90 prevents radiation-induced activation of ATM, which is necessary for heterochromatic DSB repair (Dote et al. 2006; Goodarzi et al. 2008). Hsp90 is required to stabilize FancA, and pharmacological inhibition of Hsp90 blocks damage-mediated FancD2 activation (Yamashita et al. 2007). Hsp90 is also required for the persistence and proper function of Chk1, which is essential for the proper repair of DSBs by homologous recombination (HR) in mammalian systems (Arlander et al. 2003; Sorensen et al. 2005). A common downstream component of these pathways is the Rad51 recombinase, which is also depleted upon treatment with Hsp90 inhibitors (Noguchi et al. 2006; Yao et al. 2007). If inhibition of Hsp90 modulated CAG repeat instability via its effects on these pathways, then we should be able to reproduce its effects by inhibiting Rad51.
To test the possibility that impairment of Rad51 may contribute to CAG repeat instability, we used two different siRNAs to knock down the expression of Rad51 (Fig. 2b). We found that siRNA knockdown increased the frequency of HPRT+ colonies about tenfold above control levels (P < 0.0001) (Fig. 2a), similar to the effect that we observed with Hsp90 inhibition by 17-AAG and siRNA. Additionally, we established that the level of Rad51 decreases with increasing concentrations of 17-AAG in a dose-dependent manner (Fig. 2c). Moreover, the decrease in Rad51 with 100 nM 17-AAG (Fig. 2c) is comparable to the knockdown obtained with Rad51 siRNA (Fig. 2b). Since inhibition of Hsp90 by 17-AAG and siRNA knockdown of Rad51 have equivalent effects on Rad51 levels and CAG repeat instability, it seems likely that Hsp90-mediated effects on CAG repeat instability are the result of its effects on Rad51.
The principal cellular role of Rad51 is to promote the strand invasion step of HR. If Rad51 impairment destabilizes repeats by inhibiting HR, other components in the HR pathways should also affect repeat instability. To test this possibility, we knocked down Rad51C, a Rad51 paralog that is required in combination with Rad51 to promote strand invasion (Sigurdsson et al. 2001). We found that Rad51C knockdown increased the frequency of HPRT+ colonies about sixfold above control levels (P < 0.0001) (Fig. 2a), which is only slightly less than what we observed with Rad51 knockdown. Thus, we conclude that knockdown of Rad51 increases CAG repeat instability by impairing HR. Deciphering the complete pathway by which Hsp90 induces Rad51-mediated effects on CAG repeat instability will be a challenge, as there are multiple proteins, such as FancD2 and Chk1, that regulate Rad51 but are also in turn regulated by Hsp90.
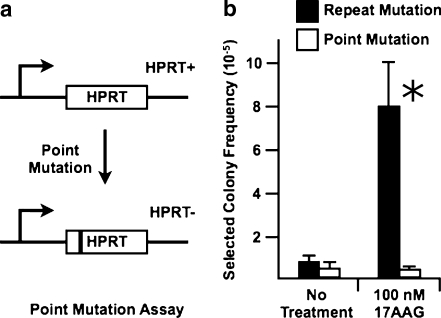
We have focused here on the role of Hsp90 in CAG repeat instability; however, it is possible that Hsp90 inhibition, in addition to inducing repeat instability, might also increase the frequency of other types of mutational event such as point mutation. To examine this possibility, we applied the same 17-AAG treatment to the HT1080 cells from which the FLAH25 cell line was derived. These cells harbor a wild-type copy of HPRT at the endogenous locus. HPRT-inactivating mutations, which can be detected by selection for loss of HPRT function, are dominated by point mutations (Sculley et al. 1992). Although treatment with 17-AAG induces a dramatic increase in repeat instability in FLAH25 cells, it did not detectably increase the frequency of HPRT− colonies in HT1080 cells (Fig. 3). These results suggest that the role of Hsp90 in CAG repeat instability is not part of a broader function in generalized genomic mutation.
Fig. 3.
Effects of 17-AAG on HPRT-inactivating mutations. a Assay for point mutations in the endogenous HPRT gene in HT1080 cells, from which FLAH25 cells were derived. Selection for the loss of HPRT activity recovers cells that have undergone an HPRT-inactivating mutation, most of which are point mutations (Sculley et al. 1992). b Comparison of the effects of 17-AAG on point mutations in HT1080 cells with its effects on repeat mutations in FLAH25 cells. For HT1080 cells, the values represent the frequencies of HPRT− colonies; for FLAH25 cells, the values are the frequencies of HPRT+ colonies. The values (× 10− 6) for each condition in the point mutation assay are an average of three independent experiments: no treatment (5.6 ± 0.6; N = 3) and 17-AAG treatment (5.0 ± 2.6; N = 3). An asterisk indicates that P < 0.0001 for 17-AAG treatment in comparison to no treatment. The values for the FLAH25 cells were taken from Fig. 2a
In summary, our results indicate that Hsp90 normally functions to promote the stability of CAG repeats, apparently through its effects on Rad51-mediated HR. Interference with Hsp90 decreases the amount of Rad51, which is the primary recombinase involved in the homology-dependent repair of DSBs. In mammalian cells, DSBs are repaired by two well-defined homology-dependent repair pathways: strand invasion, which depends on Rad51, and single-strand annealing (SSA), which does not (Paques and Haber 1999). The ability of Hsp90 to modulate Rad51 suggests that it controls the relative activity of these two pathways. Under stress, when Hsp90 is diverted from its normal function, the amount of Rad51 will decrease, thereby enhancing the SSA pathway. Because the SSA pathway fixes DSBs by pairing homologous sequences on either side of the break, it would naturally lead to changes in the lengths of the repeat tract (Richard et al. 1999). Consistent with our results, a previous study of stress-induced mutations in cultured murine cells revealed increases in the instability of minisatellites, although these studies did not evaluate the role of Hsp90 (Li et al. 2001). A more recent study found that Hsp90 inhibition activates transposon-mediated mutagenesis in the Drosophila genome (Specchia et al. 2010). Our observation that Hsp90 inhibition destabilizes microsatellite repeats, along with these other recent studies, suggests an emerging role for Hsp90 in the maintenance of genome integrity.
It will be interesting to see if Hsp90 inhibition can instantiate CAG repeat expansion, to which our selection assay is blind. Also, despite having specifically studied CAG repeats, we expect that repair by SSA would have similar destabilizing effects on other microsatellite repeats. Therefore, the effects of Hsp90 inhibition on Rad51 may represent a generalized way to manipulate the length of repeats in the genome, of which several have been implicated in the evolution of gene transcription (Vinces et al. 2009) and protein function (Fondon and Garner 2004). However, to make this connection, it will be important to establish whether this modulation of genome repeats occurs in the germline. The observation that the inducible form of Hsp90 is predominately expressed in the testis and brain (Vamvakopoulos 1993) supports a role for Hsp90 in germline repeat stability and additionally suggests a role for stress-induced microsatellite instability in the progression of neurological disorders caused by the instability of triplet repeats.
Acknowledgments
We would like to thank Dr. John W. Fondon III and Dr. Steve W. Lockless for insightful discussions and manuscript suggestions. We also would like to thank Drs. Jason Shohet and Zaowen Chen for technical assistance with real-time RT-PCR measurements. In addition, we thank the members of the Wilson laboratory for helpful suggestions and critical comments. This work was supported by a T-32 grant from the National Institutes of Health (EY07001) to D.M., an F-32 grant from the National Institutes of Health (NS064762) to D.M., a National Institute of Diabetes and Digestive and Kidney Diseases training grant (DK007696) to K.S., and an R01 grant from the National Institutes of Health (GM38219) to J.H.W.
References
- Arlander SJ, Eapen AK, Vroman BT, McDonald RJ, Toft DO, Karnitz LM. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278:52572–52577. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- Banerji U. Heat shock protein 90 as a drug target: some like it hot. Clin Cancer Res. 2009;15:9–14. doi: 10.1158/1078-0432.CCR-08-0132. [DOI] [PubMed] [Google Scholar]
- Barber RC, Hickenbotham P, Hatch T, Kelly D, Topchiy N, Almeida GM, Jones GD, Johnson GE, Parry JM, Rothkamm K, Dubrova YE. Radiation-induced transgenerational alterations in genome stability and DNA damage. Oncogene. 2006;25:7336–7342. doi: 10.1038/sj.onc.1209723. [DOI] [PubMed] [Google Scholar]
- Camphausen K, Tofilon PJ. Inhibition of Hsp90: a multitarget approach to radiosensitization. Clin Cancer Res. 2007;13:4326–4330. doi: 10.1158/1078-0432.CCR-07-0632. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Jain S, Stuhmer T, Andrulis M, Ungethum U, Kuban RJ, Lorentz H, Bommert K, Topp M, Kramer D, Muller-Hermelink HK, Einsele H, Greiner A, Bargou RC. STAT3 and MAPK signaling maintain overexpression of heat shock proteins 90alpha and beta in multiple myeloma cells, which critically contribute to tumor-cell survival. Blood. 2007;109:720–728. doi: 10.1182/blood-2006-05-024372. [DOI] [PubMed] [Google Scholar]
- Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. Hsp90 recognizes a common surface on client kinases. J Biol Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- Compton SA, Elmore LW, Haydu K, Jackson-Cook CK, Holt SE. Induction of nitric oxide synthase-dependent telomere shortening after functional inhibition of Hsp90 in human tumor cells. Mol Cell Biol. 2006;26:1452–1462. doi: 10.1128/MCB.26.4.1452-1462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- Fondon JW, III, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci U S A. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Dion V, Sandor Z, Meservy JL, Wilson JH. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol Cell Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mittelman D, Wilson JH. Genome-wide demethylation destabilizes CTG⋅CAG trinucleotide repeats in mammalian cells. Hum Mol Genet. 2004;13:2979–2989. doi: 10.1093/hmg/ddh317. [DOI] [PubMed] [Google Scholar]
- Healy C, Wade M, McMahon A, Williams A, Johnson DA, Parfett C. Flow cytometric detection of tandem repeat mutations induced by various chemical classes. Mutat Res. 2006;598:85–102. doi: 10.1016/j.mrfmmm.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Ito M, Yamamoto S, Nimura K, Hiraoka K, Tamai K, Kaneda Y. Rad51 siRNA delivered by HVJ envelope vector enhances the anti-cancer effect of cisplatin. J Gene Med. 2005;7:1044–1052. doi: 10.1002/jgm.753. [DOI] [PubMed] [Google Scholar]
- Jankowski C, Nasar F, Nag DK. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc Natl Acad Sci U S A. 2000;97:2134–2139. doi: 10.1073/pnas.040460297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- Ko JC, Hong JH, Wang LH, Lin YW. The role of repair protein Rad51 in synergistic cytotoxicity and mutagenicity induced by epidermal growth factor receptor inhibitor (Gefitinib, IressaR) and benzo[a]pyrene in human lung cancer. Exp Cell Res. 2008;314:1881–1891. doi: 10.1016/j.yexcr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Li CY, Little JB, Hu K, Zhang W, Zhang L, Dewhirst MW, Huang Q. Persistent genetic instability in cancer cells induced by non-DNA-damaging stress exposures. Cancer Res. 2001;61:428–432. [PubMed] [Google Scholar]
- Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG⋅CAG repeats. Proc Natl Acad Sci U S A. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meservy JL, Sargent RG, Iyer RR, Chan F, McKenzie GJ, Wells RD, Wilson JH. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol Cell Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelman D, Moye C, Morton J, Sykoudis K, Lin Y, Carroll D, Wilson JH. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci U S A. 2009;106:9607–9612. doi: 10.1073/pnas.0902420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci U S A. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Yu D, Hirayama R, Ninomiya Y, Sekine E, Kubota N, Ando K, Okayasu R. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351:658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- Palotai R, Szalay MS, Csermely P. Chaperones as integrators of cellular networks: changes of cellular integrity in stress and diseases. IUBMB Life. 2008;60:10–18. doi: 10.1002/iub.8. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Reuschenbach M, Kloor M, Morak M, Wentzensen N, Germann A, Garbe Y, Tariverdian M, Findeisen P, Neumaier M, Holinski-Feder E, von Knebel Doeberitz M (2009) Serum antibodies against frameshift peptides in microsatellite unstable colorectal cancer patients with Lynch syndrome. Fam Cancer Epub ahead of print [DOI] [PMC free article] [PubMed]
- Richard GF, Dujon B, Haber JE. Double-strand break repair can lead to high frequencies of deletions within short CAG/CTG trinucleotide repeats. Mol Gen Genet. 1999;261:871–882. doi: 10.1007/s004380050031. [DOI] [PubMed] [Google Scholar]
- Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Rutherford S, Knapp JR, Csermely P. Hsp90 and developmental networks. Adv Exp Med Biol. 2007;594:190–197. doi: 10.1007/978-0-387-39975-1_16. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Undurraga S, Milo R, Schellenberg K, Lindquist S, Queitsch C. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci U S A. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sculley DG, Dawson PA, Emmerson BT, Gordon RB. A review of the molecular basis of hypoxanthine–guanine phosphoribosyltransferase (HPRT) deficiency. Hum Genet. 1992;90:195–207. doi: 10.1007/BF00220062. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S, Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B–Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D'Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Sreedhar AS, Kalmar E, Csermely P, Shen YF. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/S0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- Vamvakopoulos NO. Tissue-specific expression of heat shock proteins 70 and 90: potential implication for differential sensitivity of tissues to glucocorticoids. Mol Cell Endocrinol. 1993;98:49–54. doi: 10.1016/0303-7207(93)90235-C. [DOI] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Oda T, Sekimoto T. Hsp90 and the Fanconi anemia pathway: a molecular link between protein quality control and the DNA damage response. Cell Cycle. 2007;6:2232–2235. doi: 10.4161/cc.6.18.4653. [DOI] [PubMed] [Google Scholar]
- Yao Q, Weigel B, Kersey J. Synergism between etoposide and 17-AAG in leukemia cells: critical roles for Hsp90, FLT3, topoisomerase II, Chk1, and Rad51. Clin Cancer Res. 2007;13:1591–1600. doi: 10.1158/1078-0432.CCR-06-1750. [DOI] [PubMed] [Google Scholar]