Summary
Bacteriophage T4 provides an important model system for studying the mechanism of homologous recombination. We have determined the crystal structure of the T4 UvsX recombinase, and the overall architecture and fold closely resembles that of RecA, including a highly conserved ATP binding site. Based on this new structure, we reanalyzed EM reconstructions of UvsX-DNA filaments and docked the UvsX crystal structure into two different filament forms, a compressed filament generated in the presence of ADP and an elongated filament generated in the presence of ATP and aluminum fluoride. In these reconstructions, the ATP binding site sits at the protomer interface, as in the RecA filament crystal structure. However, the environment of the ATP binding site is altered in the two filament reconstructions, suggesting that nucleotide cannot be as easily accommodated at the protomer interface of the compressed filament. Finally, we show that the phage helicase UvsW completes the UvsX-promoted strand-exchange reaction, allowing the generation of simple nicked circular product rather than complex networks of partially exchanged substrates.
Keywords: DNA repair, recombination, RecA, electron microscopy, X-ray crystallography
Introduction
Homologous recombination is a key process in DNA metabolism.1,2 Simple cut-and-paste mechanisms for recombination involve the alignment of homologous regions of two DNA molecules, strand exchange, formation of a Holliday Junction (HJ), HJ branch migration, and HJ resolution. In addition, homologous recombination pathways often involve DNA replication events, either localized or extensive, where DNA polymerase and associated proteins gain access to the invading 3’ end within a recombination intermediate. Although homologous recombination is typically associated with the generation of genetic diversity, it has critical roles in DNA repair3 and the rescue of stalled and collapsed replication forks.4
The central protein in strand exchange is the recombinase (e.g. bacterial RecA) that cooperatively and non-specifically polymerizes on ssDNA, and forms helical filaments that bind double-stranded DNA (dsDNA). Subsequent events include homology search, alignment of homologous regions and strand exchange using an ATP-dependent mechanism.1,5 Although relatively well understood biochemically, the structural basis for homologous strand pairing and exchange has yet to be fully elucidated. Electron microscopy (EM) and single molecule studies have provided important insights,6–8 and crystallographic studies have now revealed how RecA molecules interact with individual and paired ssDNA molecules within the filament.9
Bacteriophage T4 is an attractive system to study recombination and DNA metabolism in general. The majority of T4 DNA replication occurs by a recombination-dependent mechanism, allowing detailed study of the coupling of replication and recombination.10 Invasive single-strand DNA ends are prepared for recombination by the exonuclease complex encoded by phage genes 46 and 47,11 and the subsequent recombination process is apparently mediated by just four proteins: the recombinase UvsX, the recombination-mediator protein (RMP) UvsY and the single-stranded DNA-binding protein gp32,12 and the branch-specific helicase UvsW (see below).
UvsX is a 44 kDa RecA-like protein that forms similar helical assemblies on ssDNA13,14 and promotes joint molecule formation between homologous ds- and ssDNA molecules.15–18 UvsX utilizes an accessory protein, UvsY, to form stable filaments. UvsY is classified as an RMP,19 and it stimulates the ssDNA-dependent ATPase activity of UvsX, and lowers the critical concentration of UvsX required for activity.19–21 The interaction between UvsX and UvsY displaces the T4 ssDNA-binding protein gp32 from ssDNA prior to filament formation.22–24 UvsX and UvsY are functionally homologous to the eukaryotic Rad51 and Rad52 proteins, respectively.19
UvsW is a 58 kDa SF2 helicase that unwinds branched DNA molecules including static-X structures that resemble HJs.25,26 The UvsW helicase has multiple roles in T4 DNA replication, recombination and repair.25,27–29 The biochemical properties of UvsW have been extensively characterized,30,31 and we have determined its crystal structure.32,33
Here we report the crystal structure of a large fragment of UvsX and demonstrate its close structural and functional similarities to RecA. We also demonstrate that the structure can be fitted into image reconstructions of the active UvsX filament obtained from EM, in a manner consistent with the published RecA active filament structure.9 Finally, we present direct evidence for a central role of UvsW helicase in promoting the strand-exchange reaction.
Results
Fragment characterization and structure determination of UvsX
To initiate crystallization trials, full-length UvsX (UvsX1–391) was purified as a 6xHis tagged fusion protein, with four additional residues remaining at the N-terminus after cleavage with thrombin. It was necessary to add reducing agents to concentrate the protein higher than 5 mg/ml. Two UvsX species of lower molecular weight were consistently present when the concentrated protein was analyzed using SDS PAGE, and mass spectrometry revealed that one species was missing 15 residues at the N-terminus and the other was lacking an additional 32 residues at the C-terminus. Based on further fragment analysis using limited trypsin digestion and mass spectrometry, together with sequence comparison with RecA, three additional constructs were prepared for crystallization trials, residues 1–358 (UvsX1–358), 30–391(UvsX30–391) and 30–358 (UvsX30–358). Only UvsX30–358 yielded crystals and many conditions were eventually identified between pH 7.0 and 9.5. However, all crystals were morphologically similar and in space group P61 with cell dimensions a = b =96.0 Å, c=131.3 Å.
The initial structure was determined using SAD techniques at 2.8 Å resolution from selenomethionine-labeled crystals, and the electron density map allowed a complete tracing of the molecule which revealed two monomers in the asymmetric unit. The final structure was refined at 2.4 Å using native data, and the data collection and refinement statistics are shown in Table 1. In Chain A, residues 152–165, 197–211, 228–237 and 341–358 were not visible in the electron density map and were omitted from the structure. In addition, the side chains of residues 35, 151, 226–227, 238, 261–263, 286, 297, 301 and 307 were not defined and built as alanines. In Chain B, residues 152–166, 198–211, 228–238 and 343–358 were omitted, and residues 31, 35, 151, 226 –227, 261–263, 286 and 307 were built as alanines.
Table 1.
Crystallographic data collection and refinement statistics of UvsX30–358.
| Data Collection | Native | SAD |
|---|---|---|
| Wavelength (Å) | 1.0 | 0.97931 |
| Space group | P61 | P61 |
| Unit Cell (Å) | a=b=95.97 c=131.27 | a=b=95.48 c=132.30 |
| Resolution (Å) | 83.04 - 2.35 | 50 - 2.8 |
| Mosaicity (°) | 0.549 | 1.157 |
| Unique Reflections | 28005 | 16391 |
| Redundancy | 7.9 | 14.4 |
| Completeness (%)* | 99.3 (92.8) | 97.2 (81.2) |
| I/σ*† | 29.5 (3.2) | 27.4 (2.1) |
| Rsym (%)*‡ | 5.9 (34.5) | 11.6 (44.9) |
| Refinement | ||
| Rcrystal / Rfree(%)*§ | 21.0/24.8 (31.1/36.3) | |
| Average B-Factors (Å2) | ||
| Chain A (Protein) | 23.08 | |
| Chain B (Protein) | 23.13 | |
| Chain C (Water) | 28.40 | |
| Chain D (PO4) | 42.43 | |
| Ramachandran (%) | ||
| Favored | 92.2 | |
| Allowed | 7.8 | |
| Outliers | 0 | |
| Rmsd | ||
| Bond Length(Å) | 0.018 | |
| Bond Angles(°) | 1.759 | |
| Chirality | 0.127 |
Values in parentheses are from the highest resolution shell.
I/σ is the mean reflection intensity divided by the estimated error.
Rsym = (Σ|Ihkl − <I>|)/(ΣIhkl), where the average intensity <I> is taken over all symmetry equivalent measurements and Ihkl is the measured intensity for any given reflection.
Rcrystal = ||Fo| − |Fc||/|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree is equivalent to Rcrystal, but calculated for 5% of the reflections chosen at random and omitted from the refinement process.
Crystal structure of dimeric UvsX30–358
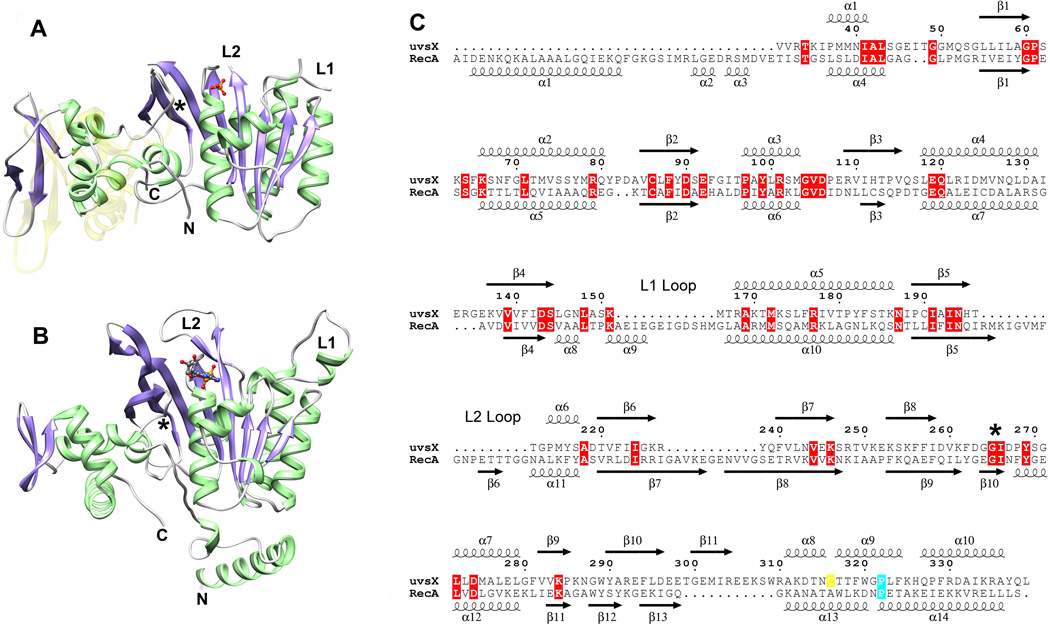
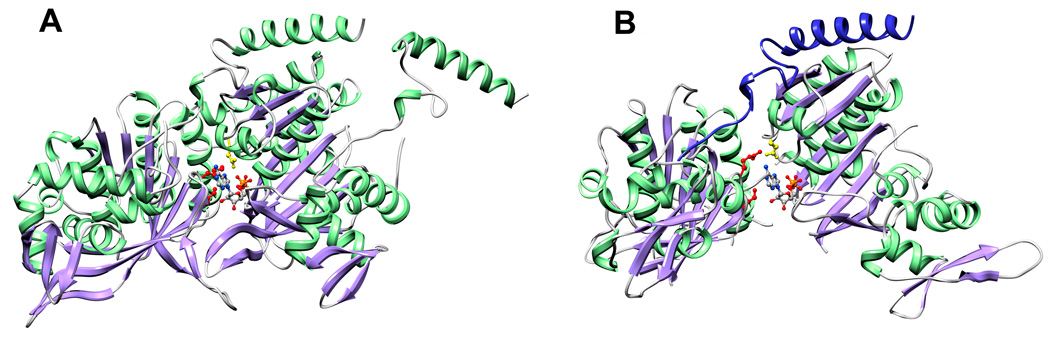
The two molecules in the asymmetric unit are virtually identical apart from loop regions, and superimpose with an RMSD on C alpha carbons of 0.44 Å. The molecule contains a larger N-terminal (‘core’) αβ domain and smaller C-terminal domain with mainly α-helical structure (Figure 1A). The core domain contains a mixed 8-stranded β-sheet in which strands β3 (residues 109–115), β2 (83–91), β4 (137–144), β5 (189–195), β1 (55–60) and β6 (220–226) are parallel, and strands β7 (239–246) and β8 (253–261) form a β-hairpin that associates with strand β6 and leads into the C-terminal domain. The β-sheet is flanked by two long α-helices on one side (α4, 118–131 and α5, 168–186) and two more on the opposite side (α1, 37–44 and α2, 66–80) together with the C-terminal domain. In common with other RecA-like structures that do not contain DNA, residues within loops L1 (152–165) and L2 (198–211) are unstructured. The C-terminal domain contains a small three-stranded β-sheet (β9, 281–285; β10, 290–297; β11, 300–306) in which strands β10 and β11 create an extended β-hairpin. This β-sheet is exposed on the surface of UvsX, and three α-helices in the domain (α8, 272–279; α9, 317–325; α10, 326–337) pack against the core domain. Residues 342–358 at the C-terminus of our construct are not visible in the electron density map. Despite the relatively poor sequence conservation,34 the overall fold of UvsX30–358 closely resembles that of the equivalent fragment of RecA (residues 39–333) (Figure 1B and Figure 1C).
Fig. 1.
Comparison of the UvsX30–358 monomer with RecA. (A) The structure of the UvsX30–358 monomer. α-helices are shown in green, β-strands in purple and loops in grey. The N-terminal ATP-binding αβ core domain is oriented towards the right and the helical C-terminal domain is on the left. ATP is not present in the structure, but the phosphate group (orange/red ball-and-stick) occupies the pocket that accommodates the ATP β-phosphate. Loops L1 and L2 are not visible in the structure, but their locations are labeled together with the N- and C-termini. (B) The RecA monomer as determined by Chen and coworkers9 (PDB ID: 3CMX) shown in the same orientation as UvsX in (A) based on an alignment of the core domain secondary structures. The RecA structure includes loops L1 and L2 (labeled) and a bound ADP (shown in ball-and stick). Note that the N-terminal oligomerization α-helix that was removed in UvsX to facilitate crystallization is present in RecA. (C) Structure-based sequence alignment of UvsX and RecA derived from (A) and (B). Sequence numbering refers to UvsX. Cys316 in UvsX is highlighted in yellow; RecA Pro313 and UvsX Pro322 are marked cyan; other identical residues are marked with red boxes. The figure was prepared with ESPript.64 In (A), the semi-transparent yellow structure shows the predicted location of the C-terminal domain based on its location in RecA. The ~15° rotation between the observed and predicted positions occurs around a conserved Gly-Ile motif indicated by an asterisk in (A), (B) and (C).
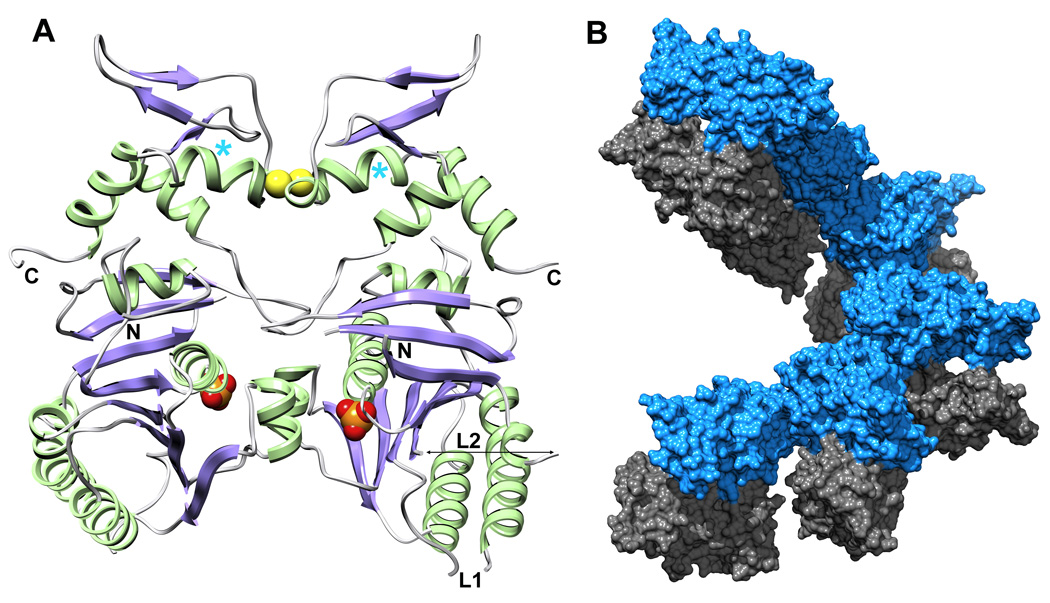
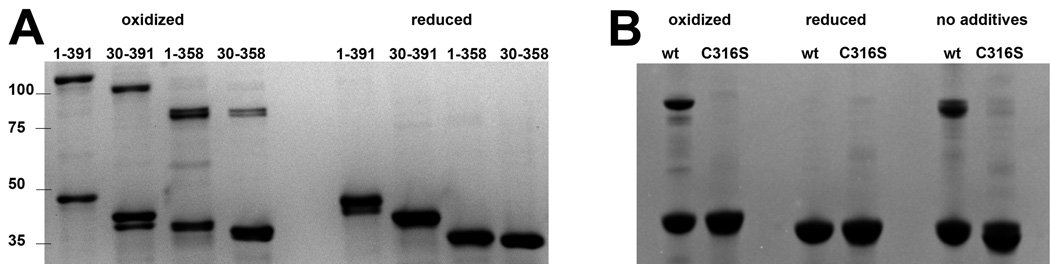
The two monomers in the asymmetric unit are related by a local two-fold axis and interact via two surfaces, one on each domain (Figure 2A). The larger surface on the core domain (1334 Å2) is centered on residues Phe65, Arg79, Tyr99 and Met103, and the smaller interaction surface on the C-terminal domain (357 Å2) is centered on Cys316 and Thr317. The electron density of the paired Cys316 sidechains suggests that a fraction of the dimers in the crystal are connected by a disulfide bond. To test whether the disulfide bond can exist in solution, full-length UvsX and the three truncated versions were analyzed by non-reducing SDS PAGE (Figure 3A). Samples were extensively dialyzed to remove all reducing agents and then fully oxidized using 1% H2O2. In all four constructs, a significant fraction of the protein appeared as a band of twice the molecular weight that was confirmed as UvsX by mass spectrometry, and this higher molecular weight band disappeared after addition of 5 mM TCEP (tris (2-carboxyethyl) phosphine) as reducing agent. UvsX1–391, UvsX30–391 and UvsX1–358 all showed a similar ratio of dimers and monomers under oxidizing solution conditions (approximately one half) whereas the dimeric species was less prominent in UvsX30–358 (Figure 3A). A C316S mutation in the context of full-length UvsX eliminated the dimeric species under oxidizing conditions (Figure 3B). Note that the full-length dimer is present in the absence of oxidizing agent and eliminated in the C316S mutant (Figure 3B).
Fig. 2.
The dimeric arrangement of UvsX30–358 in the crystal asymmetric unit. (A) UvsX30–358 is colored as in Figure 1A. The ATP-binding sites are indicated by the phosphate groups (orange/red CPK), and are occluded at the dimer interface. The paired Cys316 residues at the dimer interface appear to form a partially occupied disulfide bridge (yellow CPK) in the crystal and in solution (see Figure 3). The positions of the N- and C-termini and Loops L1 and L2 are indicated. The positions of Pro322 highlighted in Figure 1C are marked with cyan asterisks. (B) Six dimers create one turn of a helical array in the P61 unit cell. Interactions between successive monomer A molecules (blue) mediate the helical filament with no contributions from monomer B (grey).
Fig. 3.
SDS PAGE analysis of oxidized and reduced UvsX. (A) Wild type full length and truncated UvsX proteins show partial dimerization when oxidized and a single band when reduced. The N-terminal deletion mutant UvsX30–391 shows degradation when oxidized, but the reduced sample runs as a single species. The C-terminal deletion mutant UvsX1–358 and the core domain UvsX30–358 migrate similarly on the gel. The band positions of protein standards are marked with their molecular weights (kDa) on the left. Note that the dimeric fractions of both samples which lack the disordered acidic C-terminal region run at a molecular weight more consistent with the calculated molecular weight. The calculated molecular weight values for each dimer are: 93 kDa for UvsX1–391, 86 kDa for UvsX30–391, 84 kDa for UvsX1–358 and 80 kDa for UvsX30–358. (B) The C316S point mutant of full-length UvsX fails to dimerize in oxidizing conditions. Wild type (wt) UvsX exhibits dimerization in the loading buffer without addition of oxidizing agents (right hand side). Reduced UvsX protein was supplemented with 5 mM TCEP, and oxidized protein with 1% H2O2.
With some exceptions,35,36 RecA-like proteins crystallize in space group P61, which reflects their natural tendency to form filaments with approximate six-fold screw axes. Based on other recombinase structures, we assumed that UvsX30–358 is missing the protein-protein interaction region at the N-terminus and therefore unable to form active filaments. This proved to be correct. In the UvsX30–358 structure, the P61 crystal packing does not recapitulate the natural filament and the P61 crystal filament is constructed from two UvsX monomers. Chain A actually creates the filament and chain B decorates the filament with no interactions with its own symmetry mates (Figure 2B). Inter-filament crystal contacts are mediated by a third UvsX30–358-UvsX30–358 interaction surface (1019.5Å2) between β10, β11 and α10 in chain A and α4, α5, α7 and the loop connecting β7 and β8 in chain B.
The UvsX ATPase domain
Superposition of the UvsX and RecA9 structures reveals that the ATP-binding site of UvsX is conserved both in terms of location (Figure 1A and Figure 1B) and residue composition (Figure 1C). The Walker A or ‘P loop’ of RecA encompasses residues 65–73 and is equivalent to residues 59–67 of UvsX. The Walker B motif of RecA (residues 139–144) is also present in UvsX (residues 138–143) and contains the highly conserved aspartic acid (Asp144 in RecA and Asp143 in UvsX) that coordinates the Mg2+ ion in the ATP complex. In RecA, the ATP adenine ring π-stacks with Tyr103, and Tyr99 of UvsX is suitably located to perform the equivalent function. Finally, the catalytic glutamic acid in RecA, Glu96, corresponds to Glu92 in UvsX. In RecA, the loop encompassing residues 261–269 encloses the adenine and ribose moieties of ATP, but the equivalent loop in UvsX (residues 259–267) is not suitably positioned in our structure to perform this role. This suggests that ATP is required to fix its position, although we note that this loop is poorly defined in our structure and located at the dimer interface where its conformation is probably distorted. Most notably, Tyr264 in RecA interacts closely with the adenine ring, but its probable functional counterpart in UvsX, Phe262, is not visible. In our dimer structure of UvsX, the ATP binding site is occluded at the dimer interface, which likely explains why we have not been able to introduce nucleotide into our crystals. However, each Walker A motif contains strong electron density consistent with a bound phosphate group that occupies the β phosphate binding site of ATP (Figure 1A and Figure 1B).
EM reconstruction of UvsX filaments
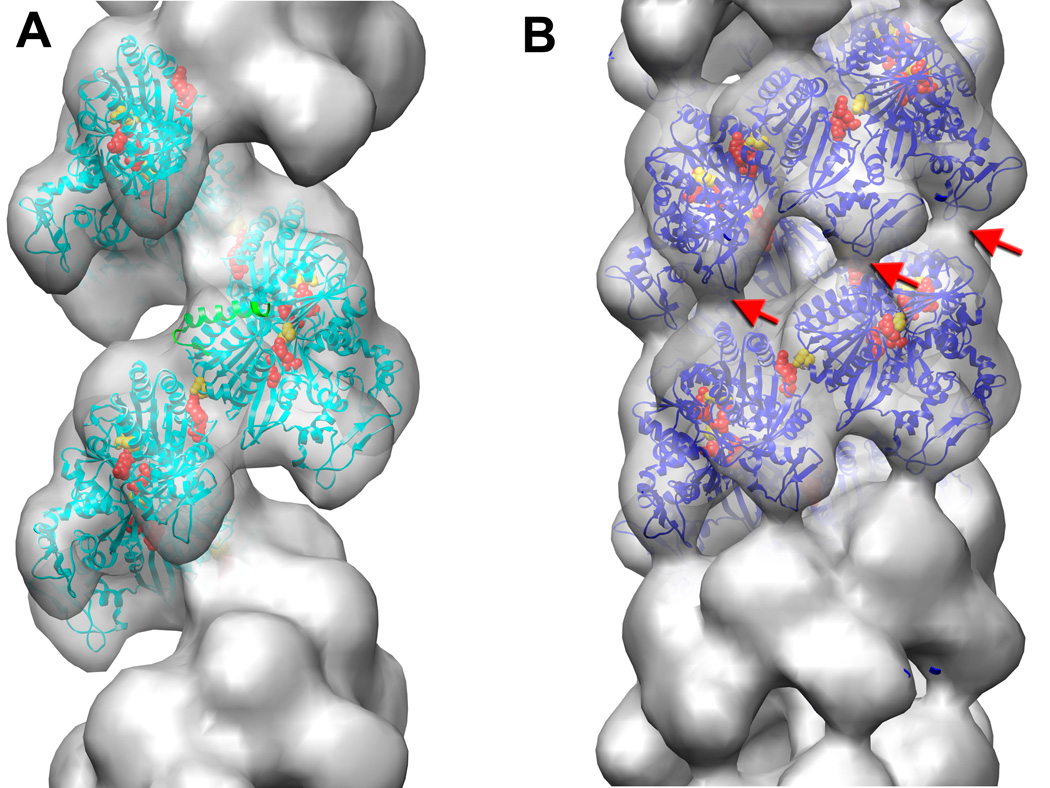
Reconstructions of UvsX-DNA filaments from EM were analyzed several years ago,37 and these images were reanalyzed using the UvsX crystal structure. UvsX filaments formed on dsDNA in the presence of ATP and aluminum fluoride possess structural heterogeneity. A dataset of 3,833 images analyzed using the iterative helical real space reconstruction (IHRSR) procedure38 did not converge to one unique solution. After sorting the images by twist based on the pitch of the one start helix, two classes emerged with twists of 6.2 and 7.0 units/turn. After 60 iterations, the most populated class (2,154 images) converged to a filament with a rotation per subunit of 58.5° (6.15 units/turn) and an axial rise per subunit of 16.1 Å (pitch of 99 Å). UvsX filaments formed on dsDNA in the presence of ADP are structurally more homogeneous and compressed than those formed in the presence of ATP and aluminum fluoride. Sorting by pitch revealed very small deviations from the mean value of 70Å, and a dataset of 3,898 images was used in the IHRSR procedure that yielded a filament with a rotation per subunit of 55.7° (6.46 units/turn) and axial rise per subunit of 10.8 Å (pitch of 69.8 Å). The 3D-reconstructions of the extended and compressed filaments are shown in Figure 4A and Figure 4B, respectively.
Fig. 4.
Electron microscopy of UvsX recombination filaments. (A) Reconstruction of the extended ‘active’ filament (grey) formed in the presence of dsDNA and ATP into which the UvsX crystal structure has been fitted (cyan). The C-terminal helical domain is pointing down towards the large groove. The filament has a rotation per subunit of 58.5° and axial rise per subunit of 16.1 Å. The 28 N-terminal residues of RecA were used to model the missing N-terminal UvsX residues (green ribbons). The positions of three residues in UvsX at the monomer-monomer interface that correspond to those in RecA involved in the ATP hydrolysis are shown as red (Lys246, Arg248), and yellow (Glu92) spheres. (B) The compressed ‘inactive’ filament formed in the presence of dsDNA and ADP in which the fitted UvsX structure is shown in dark blue. The filament has a rotation per subunit of 55.7° and axial rise per subunit of 10.8 Å. A bridge of density across the groove, corresponding to an interaction between residues 130–132 of one monomer and residues 285–288 of the other monomer, is indicated by red arrows.
The crystal structure of UvsX was docked into the ATP/aluminum fluoride filament without any perturbations, which yielded the atomic model of the UvsX filament in the extended state (Figure 4A, cyan ribbons). The UvsX structure is lacking the first 30 residues, and there was a corresponding empty region of the envelope on the top of each protomer. We aligned a subunit from the crystal structure of the RecA active filament9 with the UvsX protomer, and the first 28 residues of RecA (shown as green ribbons in Figure 4A) nicely filled the empty density in our reconstruction. The crystal structure of the RecA filament revealed that the ATP binding site sits at the monomer interface with residues 248’–254’ from the adjacent monomer providing a platform that encloses the exposed surface of the adenine and ribose rings. Also, Lys248’ and Lys250’ reach across the interface to form salt bridge interactions with the ATP phosphate groups and the active site Glu96 (Figure 5A). In the context of the UvsX EM reconstruction, these interactions are all present and mediated by residues 246’–252’, Lys246’, Arg248’ and Glu92, respectively (Figure.5B). Thus, the EM reconstruction of the UvsX active filament is entirely consistent with the active RecA filament crystal structure.
Fig. 5.
Monomer-monomer interactions in the RecA and UvsX active filaments. (A) RecA-RecA interaction derived from the structure of Chen and coworkers.9 ADP is bound at the interface and the residues involved in ATP hydrolysis, Lys248 (red), Lys250 (red) and Glu96 (yellow), are indicated. (B) The UvsX-UvsX interaction derived from the EM reconstruction. The ADP at the interface and the missing N-terminal α-helix (dark blue) are modeled from the RecA superposition shown in Figure 1B. By comparison with the RecA structure, residues Lys246 (red), Arg248 (red) and Glu92 (yellow), mediate ATP hydrolysis at the interface.
The same docking procedure was used to build up an atomic model of the UvsX filament in the compressed state (Figure 4B, blue ribbons). The protomers are located at a higher radius that those in the extended state, but the overall relative orientation of adjacent protomers is very similar to that in the extended filament with the missing N-terminus able to mediate their interaction. The reconstruction shows density linking the ATPase core and C-terminal lobe of adjacent protomers in successive turns of the helix (Figure 4B, red arrows) which may stabilize the compressed filament, and the atomic model reveals that this is mediated by residues 130–132 and residues 285–288, respectively. One difference between the compressed and elongated filaments is the environment of the ATP binding site at the protomer interface. The 248’–254’ platform that encloses the adenine and ribose moieties has moved by 4 Å and is replaced by the C-terminus of helix α6. This suggests that nucleotide is less easily accommodated at the protomer interface in the compressed filament.
The role of UvsW in the UvsX-promoted strand exchange reaction
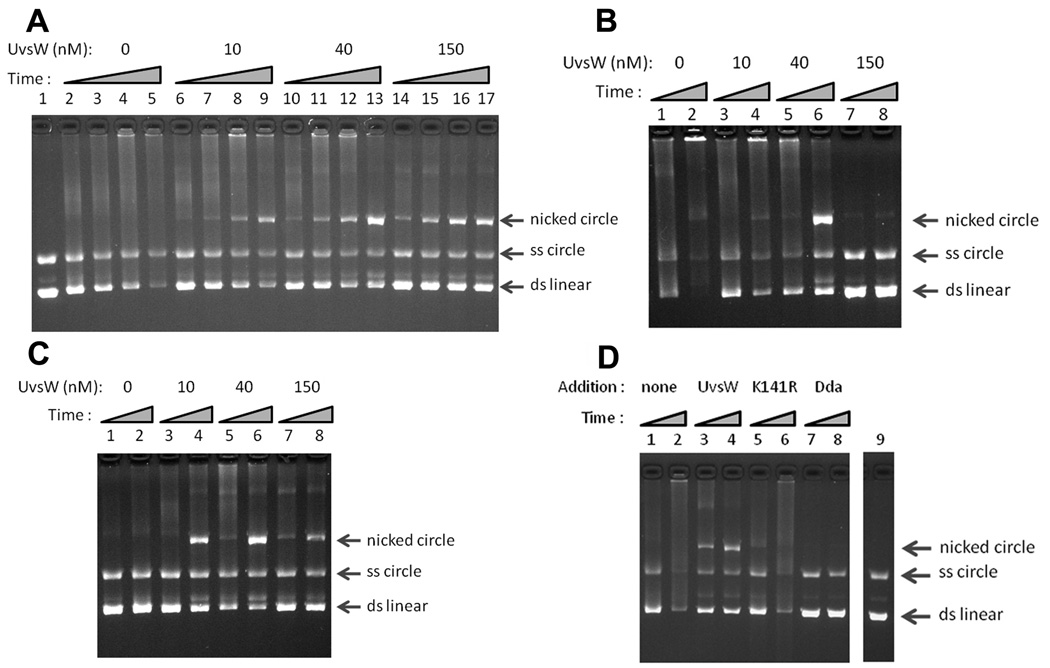
We next analyzed the UvsX-promoted strand exchange reaction, using 5.4-kb duplex linear DNA and homologous single-strand circles (Figure 6A). Similar to results from past studies,20,21,39,40 reactions containing UvsX, UvsY and gp32 generated a smear of slowly migrating forms and DNA networks that do not enter the gel. These have been previously characterized as partial strand-exchange products containing multiple exchanges.41 Strikingly, the addition of UvsW protein at nanomolar concentrations allowed the robust generation of the complete strand-exchange product: nicked circular DNA (Figure 6A, lanes 6–17). This result argues that UvsX is limited in its ability to promote branch migration during strand exchange, and that UvsW fulfills this role. UvsW promoted nicked circular product formation under a variety of concentrations of UvsX, UvsY and gp32 (Figure 6 and data not shown). UvsW could promote nicked circular product formation by UvsX and gp32 in the absence of UvsY (Figure 6B), showing that UvsY is not required for the stimulatory effect of UvsW. High concentrations of UvsY can also inhibit the reaction,40 and under these conditions, the strand-exchange reaction was strikingly rescued by the presence of UvsW. Once again the reaction proceeded to complete nicked circular product (Figure 6C).
Fig. 6.
UvsW allows completion of UvsX-promoted strand exchange. The migrations of nicked circular, single-strand circular and double-strand linear markers are indicated by arrows at the right (top to bottom, respectively). (A) Strand exchange reactions contained UvsX (1 µM), UvsY (0.5 µM), gp32 (2.5 µM), and the indicated concentration of UvsW, along with the DNA substrates (linear duplex DNA and homologous single-strand circles). Samples were removed at the following times: 2.5 min (lanes 2, 6, 10 and 14), 5 min (lanes 3, 7, 11 and 15), 10 min (lanes 4, 8, 12 and 16), and 22.5 min (lanes 5, 9, 13 and 17). The reaction in lane 1 contained UvsX, UvsY, and gp32 but no ATP. (B) UvsW allows completion of UvsX-promoted strand exchange in the absence of UvsY. Strand exchange reactions contained UvsX and gp32, each at 2.5 µM, the indicated concentration of UvsW, and no UvsY protein. Samples were removed at 5 min (lanes 1, 3, 5 and 7) and 22.5 min (lanes 2, 4, 6 and 8). (C) UvsW promotes strand exchange under conditions where UvsY is inhibitory. Strand exchange reactions contained UvsX, UvsY, and gp32, each at 2.5 µM, and the indicated concentration of UvsW. Samples were removed at 5 min (lanes 1, 3, 5 and 7) and 22.5 min (lanes 2, 4, 6 and 8). (D) Dda helicase and UvsW-K141R do not facilitate completion of UvsX-promoted strand exchange. Strand exchange reactions contained UvsX (2.5 µM), UvsY (0.5 µM), gp32 (2.5 µM), and, where indicated, UvsW (50 nM), UvsW-K141R (50 nM) or Dda (75 nM). Samples were removed at 5 min (lanes 1, 3, 5 and 7) and 25 min (lanes 2, 4, 6 and 8). The reaction in lane 9 was a control that contained UvsX, UvsY, and gp32, but no ATP.
We conclude that UvsW can stimulate the strand-invasion reaction and plays a major role in heteroduplex extension (3-strand branch migration) after the initial strand invasion. As expected, the inactive mutant UvsW-K141R did not promote nicked circular product formation (Figure 6D). Similar to a previous report,42 the T4 Dda helicase inhibited the strand invasion reaction and did not promote formation of nicked circular product (Figure 6D). Finally, we found that high concentrations (e.g., 150 nM) of UvsW can inhibit the strand exchange reaction (Figure 6A, Figure 6B and Figure 6C). Thus, as with the other three components of the T4 strand exchange system, the efficiency of the reaction depends exquisitely on the balance of protein concentrations20,39,40
Discussion
In this study, we report the crystal structure of the T4 recombinase UvsX and show that it closely resembles the structure of RecA. Moreover, we demonstrate using EM reconstructions that the monomer-monomer interactions that mediate the ‘active’ recombination filament in T4 are very similar to those that mediate the RecA active filament. In terms of its primary structure, this close similarity to RecA was not particularly apparent34 although the structural homology was predicted.43 The structural similarities justify use of the T4 system to study the molecular basis of homologous recombination. Our structures of the T4 filament in both the extended ‘active’ and compressed ‘inactive’ conformations, together with the in vitro assay described in this report that incorporates the required T4 accessory proteins, will facilitate future studies to probe this key event.
The binding of ATP and ssDNA appears to stabilize an extended conformation of the filament which facilitates the strand exchange process, and subsequent ATP hydrolysis generates an ADP-bound compressed filament that more readily disassembles. Insights into the transition between the two conformations, and the roles of ATP hydrolysis and DNA tension in controlling this transition have recently been provided by single-molecule studies on Rad5144–46 and RecA.47 Crystallographic analyses of RecA filaments have also revealed how ATP binding at the protomer-protomer interface stabilizes the extended filament in the presence of ssDNA.9 Although the molecular details of how these filament dynamics mediate the actual recombination reaction remain unclear, our structural studies on UvsX support a common fundamental mechanism for the DNA recombinase family. Compared to higher organisms, T4 recombination is relatively unencumbered by control mechanisms and accessory proteins, and is thus well suited for mechanistic analyses.
Does the crystallographic dimer have biological relevance? Our UvsX30–358 construct that yielded crystals lacks the N-terminal multimerization segment and the dimer may therefore be an artifact. Comparison with the RecA structure supports this. In RecA, Pro313 is at the N-terminus of an α-helix (α14) that forms part of a 3-helix bundle within the C-terminal domain (Figure 1C). In UvsX, this proline appears to be conserved as Pro322, but is located within helix α9 (Figure 1C and Figure 2A) and the 3-helix bundle is not present. The Cys316-mediated disulfide bond may therefore distort the 3-helix bundle and place Pro322 within α9. The formation of the crystal dimer also requires that the C-terminal domain be rotated by some 15° relative to that of RecA to avoid steric clash (Figure 1A), but this rotation may reflect a functionally required flexibility between the two domains. The rotation occurs around a conserved Gly-Ile motif located at residues 266–267 (267–268 in RecA) located in the loop connecting β8 in the core domain with α7 in the C-terminal domain (Figure 1). There is evidence that the C-terminal domain of RecA engages dsDNA during recombination,48 and a flexible connecting loop would facilitate the docking of dsDNA within the filament in the manner suggested by Shin and coworkers.35
However, the possibility remains that the dimer may resemble a functional state of UvsX. We consistently detect a UvsX dimer in solution (Figure 3), and the non-conserved Cys316 at the dimer interface may serendipitously stabilize it and allow its detection. We also note that the missing N-terminal multimerization α-helix would be located on the outer surface of the crystal dimer and would not prevent its formation (Figure 2A). The dimer may represent a storage form of UvsX in solution, as proposed for the heptameric and octameric double rings of the Pyrococcus furiosus RadA35 and human Dmc1,49 that requires interaction with gp32 and ATP to form filaments on DNA. RecA dimers have been invoked in filament assembly,50,51 although other studies suggest that assembly occurs by the addition of monomers.6
We have demonstrated that UvsW helicase promotes the strand-exchange reaction catalyzed by UvsX in the presence or absence of UvsY. UvsW can both stimulate the overall level of strand invasion and promote the generation of the final product, nicked circular DNA. Past biochemical studies of UvsX revealed mostly complex DNA networks resulting from multiple, incomplete strand-exchange reactions.1 Thus, UvsW fills an important role in the T4 recombination reaction, allowing efficient branch migration during both the early 3-strand phase of the reaction (Figure 6) and after generation of a HJ.26 Because UvsW unwinding activity is specific to branched DNA,25,26,30 we presume that it functions directly at the branch point during these reactions.
We recently demonstrated a structural similarity between UvsW and eukaryotic Rad54 protein,33 suggesting that the core recombinatorial triad of Rad51/Rad52/Rad54 in eukaryotic cells is functionally equivalent to the UvsX/UvsY/UvsW triad in T4. Our current studies extend this apparent functional parallel between the two triads. Thus, Rad54, similar to UvsW, has been shown to both stimulate Rad51-dependent strand exchange and increase the levels of strand invasion.52–55 Also, in addition to promoting strand exchange between a double and a single-stranded substrate, Rad54 and UvsW have both been shown to catalyze HJ branch migration.26,55 With the crystal structures of UvsW, UvsX and gp3256 now available, and a robust in vitro assay to monitor T4 recombination, we are now positioned to investigate at the molecular level how these three proteins interact to catalyze homologous recombination with the assistance of the RMP protein UvsY.
Materials and Methods
Cloning, expression and purification of UvsX
The full-length uvsX gene was PCR amplified from phage T4 genomic DNA and subcloned into a pET28b expression vector (Novagen) with an N-terminal 6xHis tag and a thrombin cleavage site. BL21 DE3 cells transformed with this vector were grown in LB media to OD600 of 0.6 and induced with 1 mM IPTG overnight at 16° C. The cells were harvested by 20 min. centrifugation at 4,000×g, resuspended in lysis buffer (0.5 M NaCl, 50 mM NaPO4 pH 7.8, 40 mM imidazole, 10 mM MgCl2, 10 mM β-mercaptoethanol) and lysed in a microfluidizer (Microfluidics). The lysate DNA was then digested for 30 min with DNAse I (0.1 U/ml) followed by 30 min centrifugation at 30,000×g. The supernatant was loaded on a 5 ml Ni2+ chelation sepharose column, and bound impurities were removed with 50 ml equilibration buffer (0.5 M NaCl, 20 mM Tris-HCl, pH 8.0, 40 mM imidazole, 5 mM β-mercaptoethanol). This was followed by a linear gradient from 40 to 500 mM imidazole (20 column volumes) in the same buffer, during which essentially pure protein was eluted. The affinity tag was removed with thrombin (0.1 U/ml) digestion and, following dialysis against equilibration buffer, the protein was passed over a 1 ml Ni2+ chelation sepharose column to remove residual His-tagged UvsX. The protein was dialyzed against 175 mM NaCl, 20 mM Tris-HCl, pH 8.0, and further purified on a 25 ml Q-sepharose column using a linear elution gradient from 0.1 M to 1 M NaCl in 20 mM Tris-HCl pH 8.0, 5 mM β-mercaptoethanol (10 column volumes). The pooled fractions were estimated to be >98% pure by SDS PAGE with Coomassie blue staining. For crystallization trials, the sample was dialyzed into 200 mM NaCl, 20 mM Tris-HCl pH 8.5, 2 mM DTT, concentrated to 15 mg/ml, flash frozen in liquid nitrogen and stored at −80° C.
Deletion mutants UvsX1–358, UvsX30–391 and UvsX30–358 were generated by PCR methods directly from the full-length expression construct. Using appropriate primers flanking the region to be deleted, the entire expression plasmid was copied minus the deletion, and then resealed via an engineered restriction site. The same technique was used to create the C316S point mutant within full-length UvsX. UvsX30–358 labeled with selenomethionine (SeMet) was expressed as described elsewhere.57 All the deletion mutants and the SeMet-labeled protein were purified using the protocol described above.
SDS PAGE analysis of oxidized and reduced UvsX
UvsX proteins purified as described above were dialyzed exhaustively at 1–2 mg/ml concentration in a modified storage buffer (200 mM NaCl, 20 mM Tris-HCl pH 8.5, 0.5 mM EDTA) without reducing agents. Proteins were oxidized by adjusting a final concentration of 1% H2O2 and reduced by a final concentration of 5 mM TECP prior to 5 min denaturation at 95°C in 2% SDS, 20% glycerol, 50 mM Tris-HCl pH 6.8. Samples were run on 4–20% acrylamide gels and stained with Coomassie blue.
Crystallization, data collection and structure solution of UvsX
Purified full-length UvsX, and truncated versions UvsX1–358, UvsX30–391 and UvsX30–358 were all subjected to crystallization screens. Native UvsX30–358 crystals grew to 0.2×0.2×0.6 mm3 within 3 weeks at 18° C in 1.8 M ammonium sulfate, 0.2 M ammonium phosphate, 0.1 M HEPES, pH 8.0, 5 mM DTT. None of the other protein variants yielded crystals, either in the apo form or in presence of ADP-Mg2+. Virtually identical SeMet-labeled UvsX30–358 crystals grew within 4 days in 0.8 M succinate pH 7.0, 0.1 M HEPES-Na pH 8.0, 5 mM DTT. Crystals were cryoprotected in their mother liquor supplemented with serially increasing concentrations of 10%, 25% and 35% ethylene glycol prior to flash freezing in liquid nitrogen. Diffraction data were collected at the SERCAT ID-22 beamline (APS, Argonne). Native crystals diffracted to 2.4 Ǻ resolution, and SeMet anomalous scattering data sets were collected to 2.8 Ǻ resolution. The initial structure solution was obtained from the selenomethionine peak signal using the SAD method with the Phenix software.58 The model building was performed using Coot,59 and the structure was refined at 2.4 Å against the native structure factors with a combination of CNS60 and Refmac61 using NCS restraints and TLS refinement.
Image analysis of UvsX filaments
EM images of UvsX-DNA complexes obtained by Yang et al.37 were used for the analysis. Negatives were scanned with a Nikon Coolscan 8000 as 16-bit images using a raster of 4.2 Å/pixel. The EMAN package was used to extract filament images from micrographs62 and overlapping segments (60 pixels long) of the filaments were collected from the images and used in the Iterative Helical Real Space Reconstruction (IHRSR) procedure.38 UCSF Chimera software63 was used to fit crystal structures into the experimental maps. Atomic coordinates from crystal structures were converted to density maps, and these were filtered to the resolution of the experimental map and docked manually.
Strand-exchange reactions
Full-length UvsX protein was purified in one step with Ni2+ sepharose affinity chromatography as described above, omitting thrombin cleavage and Q-sepharose chromatography. UvsY was also expressed as N-terminal 6xHis tagged fusion protein from a pET28b construct and purified on a Ni2+ chelating sepharose column using the same buffers as described for UvsX. UvsX and UvsY were dialyzed into a storage buffer containing 500 mM NaCl, 20 mM Tris-HCl pH 7.8, 1 mM DTT, 0.5 mM EDTA and 10% (w/v) glycerol and stored at −20° C. UvsW was purified as previously described,26 and gp32 was obtained from New England Biolabs. Strand exchange reactions contained ϕX174 single-strand circular DNA (15 µM in nucleotide), linearized ϕX174 double-strand circular DNA (10 µM in nucleotide), the indicated T4 proteins, and an ATP regeneration system (2 mM ATP, 15 mM creatine phosphate, and creatine phosphokinase at 25 µg/ml), in a buffer consisting of 30 mM Tris-HCl, pH 7.8, 90 mM sodium acetate, 10 mM magnesium acetate, 1 mM DTT and BSA at 0.1 mg/ml. Reactions were incubated at 37° C for 2 min without ATP, and then initiated by the addition of ATP. At the indicated times, an aliquot was removed and treated with one-tenth volume of 10X stop solution (100 mM EDTA, 5% SDS, 20% Ficoll, proteinase K at 5 mg/ml), followed by 10 min incubation at 37° C. The reaction products were separated by agarose gel electrophoresis and stained with ethidium bromide.
Acknowledgements
This work was supported by NIH grants GM066934 (to SWW and KNK) and GM035269 (to EHE), Cancer Center core grant CA21765 and the American Lebanese Syrian Associated Charities (ALSAC). Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We thank Dr. Xiaoping He for providing Dda protein. The structure factors and refined coordinates have been deposited in the PDB (accession code 3IO5).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 2.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 5.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 6.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 7.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 8.Prasad TK, Yeykal CC, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 10.Kreuzer KN, Morrical SW. Initiation of DNA replication. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington, DC: ASM Press; 1994. pp. 28–42. [Google Scholar]
- 11.Bleuit JS, Xu H, Ma Y, Wang T, Liu J, Morrical SW. Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc Natl Acad Sci U S A. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosig G. Homologous recombination. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington, DC: ASM Press; 1994. pp. 54–82. [Google Scholar]
- 13.Yu X, Egelman EH. DNA conformation induced by the bacteriophage T4 UvsX protein appears identical to the conformation induced by the Escherichia coli RecA protein. J Mol Biol. 1993;232:1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- 14.Ando RA, Morrical SW. Single-stranded DNA binding properties of the UvsX recombinase of bacteriophage T4: binding parameters and effects of nucleotides. J Mol Biol. 1998;283:785–796. doi: 10.1006/jmbi.1998.2124. [DOI] [PubMed] [Google Scholar]
- 15.Yonesaki T, Minagawa T. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 1985;4:3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 17.Hinton DM, Nossal NG. Cloning of the bacteriophage T4 uvsX gene and purification and characterization of the T4 uvsX recombination protein. J Biol Chem. 1986;261:5663–5673. [PubMed] [Google Scholar]
- 18.Harris LD, Griffith J. Visualization of the homologous pairing of DNA catalyzed by the bacteriophage T4 UvsX protein. J Biol Chem. 1987;262:9285–9292. [PubMed] [Google Scholar]
- 19.Beernink HT, Morrical SW. RMPs: recombination/replication mediator proteins. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 20.Yonesaki T, Minagawa T. Synergistic action of three recombination gene products of bacteriophage T4, uvsX, uvsY, and gene 32 proteins. J Biol Chem. 1989;264:7814–7820. [PubMed] [Google Scholar]
- 21.Kodadek T, Gan DC, Stemke-Hale K. The phage T4 uvsY recombination protein stabilizes presynaptic filaments. J Biol Chem. 1989;264:16451–16457. [PubMed] [Google Scholar]
- 22.Bleuit JS, Ma Y, Munro J, Morrical SW. Mutations in a conserved motif inhibit single-stranded DNA binding and recombination mediator activities of bacteriophage T4 UvsY protein. J Biol Chem. 2004;279:6077–6086. doi: 10.1074/jbc.M311557200. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Bond JP, Morrical SW. Mechanism of presynaptic filament stabilization by the bacteriophage T4 UvsY recombination mediator protein. Biochemistry. 2006;45:5493–5502. doi: 10.1021/bi0525167. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Qian N, Morrical SW. Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions. J Biol Chem. 2006;281:26308–26319. doi: 10.1074/jbc.M604349200. [DOI] [PubMed] [Google Scholar]
- 25.Carles-Kinch K, George JW, Kreuzer KN. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb MR, Plank JL, Long DT, Hsieh TS, Kreuzer KN. The phage T4 protein UvsW drives Holliday junction branch migration. J Biol Chem. 2007;282:34401–34411. doi: 10.1074/jbc.M705913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamlett NV, Berger H. Mutations altering genetic recombination and repair of DNA in bacteriophage T4. Virology. 1975;63:539–567. doi: 10.1016/0042-6822(75)90326-8. [DOI] [PubMed] [Google Scholar]
- 28.Derr LK, Drake JW. Isolation and genetic characterization of new uvsW alleles of bacteriophage T4. Mol Gen Genet. 1990;222:257–264. doi: 10.1007/BF00633826. [DOI] [PubMed] [Google Scholar]
- 29.Derr LK, Kreuzer KN. Expression and function of the uvsW gene of bacteriophage T4. J Mol Biol. 1990;214:643–656. doi: 10.1016/0022-2836(90)90283-R. [DOI] [PubMed] [Google Scholar]
- 30.Nelson SW, Benkovic SJ. The T4 phage UvsW protein contains both DNA unwinding and strand annealing activities. J Biol Chem. 2007;282:407–416. doi: 10.1074/jbc.M608153200. [DOI] [PubMed] [Google Scholar]
- 31.Nelson SW, Perumal SK, Benkovic SJ. Processive and unidirectional translocation of monomeric UvsW helicase on single-stranded DNA. Biochemistry. 2009;48:1036–1046. doi: 10.1021/bi801792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sickmier EA, Kreuzer KN, White SW. The crystal structure of the UvsW helicase from bacteriophage T4. Structure. 2004;12:583–592. doi: 10.1016/j.str.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Kerr ID, Sivakolundu S, Li Z, Buchsbaum JC, Knox LA, Kriwacki R, White SW. Crystallographic and NMR analyses of UvsW and UvsW.1 from bacteriophage T4. J Biol Chem. 2007;282:34392–34400. doi: 10.1074/jbc.M705900200. [DOI] [PubMed] [Google Scholar]
- 34.Fujisawa H, Yonesaki T, Minagawa T. Sequence of the T4 recombination gene, uvsX, and its comparison with that of the recA gene of Escherichia coli. Nucleic Acids Res. 1985;13:7473–7481. doi: 10.1093/nar/13.20.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, Volkmann N, Tsuruta H, Blundell TL, Venkitaraman AR, Tainer JA. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 37.Yang S, VanLoock MS, Yu X, Egelman EH. Comparison of bacteriophage T4 UvsX and human Rad51 filaments suggests that RecA-like polymers may have evolved independently. J Mol Biol. 2001;312:999–1009. doi: 10.1006/jmbi.2001.5025. [DOI] [PubMed] [Google Scholar]
- 38.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 39.Harris LD, Griffith JD. UvsY protein of bacteriophage T4 is an accessory protein for in vitro catalysis of strand exchange. J Mol Biol. 1989;206:19–27. doi: 10.1016/0022-2836(89)90520-2. [DOI] [PubMed] [Google Scholar]
- 40.Morrical SW, Alberts BM. The UvsY protein of bacteriophage T4 modulates recombination-dependent DNA synthesis in vitro. J Biol Chem. 1990;265:15096–15103. [PubMed] [Google Scholar]
- 41.Kodadek T, Wong ML, Alberts BM. The mechanism of homologous DNA strand exchange catalyzed by the bacteriophage T4 uvsX and gene 32 proteins. J Biol Chem. 1988;263:9427–9436. [PubMed] [Google Scholar]
- 42.Kodadek T. Inhibition of protein-mediated homologous pairing by a DNA helicase. J Biol Chem. 1991;266:9712–9718. [PubMed] [Google Scholar]
- 43.Story RM, Bishop DK, Kleckner N, Steitz TA. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- 44.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci U S A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson RB, Moses DN, Kwon Y, Chan P, Chi P, Klein H, Sung P, Greene EC. Structural transitions within human Rad51 nucleoprotein filaments. Proc Natl Acad Sci U S A. 2009;106:12688–12693. doi: 10.1073/pnas.0811465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishinaka T, Doi Y, Hashimoto M, Hara R, Shibata T, Harada Y, Kinosita K, Jr, Noji H, Yashima E. Visualization of RecA filaments and DNA by fluorescence microscopy. J Biochem. 2007;141:147–156. doi: 10.1093/jb/mvm033. [DOI] [PubMed] [Google Scholar]
- 48.Aihara H, Ito Y, Kurumizaka H, Terada T, Yokoyama S, Shibata T. An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing. J Mol Biol. 1997;274:213–221. doi: 10.1006/jmbi.1997.1403. [DOI] [PubMed] [Google Scholar]
- 49.Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, Shibata T, Kurumizaka H, Yokoyama S. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 50.Lauder SD, Kowalczykowski SC. Asymmetry in the recA protein-DNA filament. J Biol Chem. 1991;266:5450–5458. [PubMed] [Google Scholar]
- 51.Forget AL, Kudron MM, McGrew DA, Calmann MA, Schiffer CA, Knight KL. RecA dimers serve as a functional unit for assembly of active nucleoprotein filaments. Biochemistry. 2006;45:13537–13542. doi: 10.1021/bi060938q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 53.Solinger JA, Heyer WD. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc Natl Acad Sci U S A. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J Mol Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- 55.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 56.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 57.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods in Enzymology. 1997;276:523–530. [PubMed] [Google Scholar]
- 58.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 63.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 64.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]