Abstract
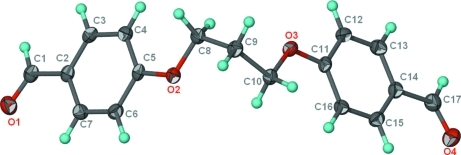
The title compound, C17H16O4, is a dialdehyde in which two formylphenoxy units are linked by a –CH2CH2CH2– chain; the molecule is V-shaped with the middle methylene C atom as the apex. The two benzene rings are aligned at 77.4 (1)°. In the crystal, molecules are linked into centrosymmetric dimers by pairs of non-classical C—H⋯O hydrogen bonds.
Related literature
For background to Schiff bases derived by condensing similar dialdehydes with primary amines, see: Zhang et al. (2008 ▶). For the crystal structure of the 2,2′-disubstituted analog, see: Hu et al. (2005 ▶).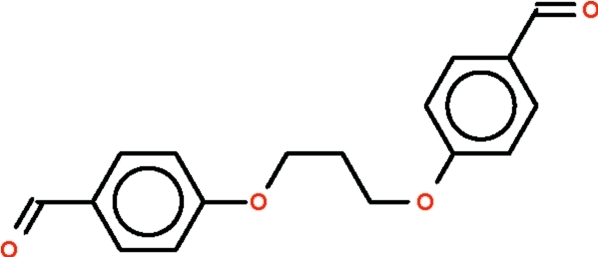
Experimental
Crystal data
C17H16O4
M r = 284.30
Monoclinic,
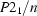
a = 15.3323 (15) Å
b = 4.6173 (5) Å
c = 20.2800 (19) Å
β = 104.783 (1)°
V = 1388.2 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.25 × 0.20 × 0.10 mm
Data collection
Bruker SMART APEXII diffractometer
8297 measured reflections
3113 independent reflections
2538 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.106
S = 1.03
3113 reflections
190 parameters
H-atom parameters constrained
Δρmax = 0.25 e Å−3
Δρmin = −0.22 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810021124/ci5094sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810021124/ci5094Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16⋯O1i | 0.95 | 2.41 | 3.287 (2) | 154 |
Symmetry code: (i) 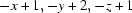 .
.
Acknowledgments
The authors thank the University of Karachi and the University of Malaya for supporting this study.
supplementary crystallographic information
Comment
The two-arm aldehyde is intended for condensation with primary amines to form Schiff bases, which, in a subsequent step, will be reacted with β-cyclodextrin to furnish inclusion compounds. The idea for this theme draws on a report on such compounds of poly(Schiff bases) (Zhang et al., 2008). The flexibilty of the Schiff base can be controlled by varying the position of the formyl group; the title compound has the the formyl groups in the 4,4'-positions. The crystal structure of the 2,2'-substituted compound has been reported (Hu et al., 2005). The molecule of C17H16O4 (Scheme I) is V-shaped with the middle methylene carbon as the apex (Fig. 1).
Experimental
4-Hydroxybenzaldehyde (1 g, 8.2 mmol) was dissolved in acetone (25 ml). To the solution was added potassium carbonate (2.3 g, 16.4 mmol). The mixture was heated for 1 h. 1,3-Dibromopropane (0.29 ml, 2.7 mmol) was added and the mixture heated for another hour. The mixture was set aside for 8 h. The solvent was removed and the solid material was extracted with ethyl acetate. The solvent was again removed and the product purified by column chromatography by using dichloromethane-hexane (1:4) as mobile phase. Single crystals were obtained by recrystallization from dichloromethane.
Refinement
H atoms were placed in calculated positions [C–H = 0.95–0.99 Å] and were included in the refinement in the riding model approximation, with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
Displacement ellipsoid plot (Barbour, 2001) of C17H16O4 at the 70% probability level; H atoms are drawn as spheres of arbitrary radius.
Crystal data
| C17H16O4 | F(000) = 600 |
| Mr = 284.30 | Dx = 1.360 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 2677 reflections |
| a = 15.3323 (15) Å | θ = 3.1–28.0° |
| b = 4.6173 (5) Å | µ = 0.10 mm−1 |
| c = 20.2800 (19) Å | T = 100 K |
| β = 104.783 (1)° | Plate, colourless |
| V = 1388.2 (2) Å3 | 0.25 × 0.20 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII diffractometer | 2538 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.027 |
| graphite | θmax = 27.5°, θmin = 1.5° |
| ω scans | h = −19→19 |
| 8297 measured reflections | k = −5→5 |
| 3113 independent reflections | l = −19→26 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.106 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0513P)2 + 0.4617P] where P = (Fo2 + 2Fc2)/3 |
| 3113 reflections | (Δ/σ)max = 0.001 |
| 190 parameters | Δρmax = 0.25 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.33791 (6) | 1.4272 (2) | 0.27971 (5) | 0.0247 (2) | |
| O2 | 0.37445 (6) | 0.5332 (2) | 0.52487 (5) | 0.0191 (2) | |
| O3 | 0.36450 (6) | 0.5695 (2) | 0.70403 (5) | 0.0191 (2) | |
| O4 | 0.58320 (6) | 1.2514 (2) | 0.97043 (5) | 0.0290 (3) | |
| C1 | 0.28543 (9) | 1.3387 (3) | 0.31097 (7) | 0.0193 (3) | |
| H1 | 0.2260 | 1.4154 | 0.2992 | 0.023* | |
| C2 | 0.30737 (8) | 1.1217 (3) | 0.36563 (7) | 0.0171 (3) | |
| C3 | 0.24107 (8) | 1.0286 (3) | 0.39613 (7) | 0.0183 (3) | |
| H3 | 0.1816 | 1.1028 | 0.3805 | 0.022* | |
| C4 | 0.25983 (8) | 0.8289 (3) | 0.44913 (7) | 0.0182 (3) | |
| H4 | 0.2137 | 0.7657 | 0.4694 | 0.022* | |
| C5 | 0.34744 (8) | 0.7227 (3) | 0.47213 (6) | 0.0166 (3) | |
| C6 | 0.41484 (8) | 0.8113 (3) | 0.44128 (7) | 0.0188 (3) | |
| H6 | 0.4742 | 0.7361 | 0.4566 | 0.023* | |
| C7 | 0.39463 (9) | 1.0082 (3) | 0.38860 (7) | 0.0192 (3) | |
| H7 | 0.4404 | 1.0678 | 0.3676 | 0.023* | |
| C8 | 0.30783 (8) | 0.4333 (3) | 0.55834 (7) | 0.0182 (3) | |
| H8A | 0.2590 | 0.3283 | 0.5257 | 0.022* | |
| H8B | 0.2810 | 0.5991 | 0.5771 | 0.022* | |
| C9 | 0.35588 (9) | 0.2331 (3) | 0.61526 (7) | 0.0190 (3) | |
| H9A | 0.3901 | 0.0869 | 0.5964 | 0.023* | |
| H9B | 0.3103 | 0.1290 | 0.6331 | 0.023* | |
| C10 | 0.42019 (8) | 0.3892 (3) | 0.67370 (7) | 0.0176 (3) | |
| H10A | 0.4544 | 0.2486 | 0.7074 | 0.021* | |
| H10B | 0.4635 | 0.5084 | 0.6567 | 0.021* | |
| C11 | 0.40426 (8) | 0.7236 (3) | 0.76107 (6) | 0.0164 (3) | |
| C12 | 0.34516 (8) | 0.8830 (3) | 0.78886 (7) | 0.0189 (3) | |
| H12 | 0.2821 | 0.8762 | 0.7685 | 0.023* | |
| C13 | 0.37858 (9) | 1.0507 (3) | 0.84602 (7) | 0.0202 (3) | |
| H13 | 0.3383 | 1.1611 | 0.8647 | 0.024* | |
| C14 | 0.47139 (8) | 1.0595 (3) | 0.87679 (7) | 0.0187 (3) | |
| C15 | 0.52930 (8) | 0.8986 (3) | 0.84844 (7) | 0.0189 (3) | |
| H15 | 0.5923 | 0.9038 | 0.8691 | 0.023* | |
| C16 | 0.49718 (8) | 0.7308 (3) | 0.79069 (7) | 0.0174 (3) | |
| H16 | 0.5375 | 0.6227 | 0.7716 | 0.021* | |
| C17 | 0.50495 (9) | 1.2379 (3) | 0.93787 (7) | 0.0235 (3) | |
| H17 | 0.4622 | 1.3516 | 0.9531 | 0.028* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0269 (5) | 0.0271 (5) | 0.0213 (5) | 0.0011 (4) | 0.0081 (4) | 0.0041 (4) |
| O2 | 0.0179 (4) | 0.0231 (5) | 0.0158 (5) | 0.0023 (4) | 0.0036 (4) | 0.0036 (4) |
| O3 | 0.0165 (4) | 0.0234 (5) | 0.0163 (5) | 0.0004 (4) | 0.0020 (3) | −0.0030 (4) |
| O4 | 0.0248 (5) | 0.0375 (6) | 0.0228 (6) | −0.0053 (4) | 0.0027 (4) | −0.0083 (5) |
| C1 | 0.0220 (6) | 0.0177 (6) | 0.0168 (7) | 0.0004 (5) | 0.0027 (5) | −0.0033 (5) |
| C2 | 0.0200 (6) | 0.0162 (6) | 0.0139 (6) | −0.0002 (5) | 0.0020 (5) | −0.0036 (5) |
| C3 | 0.0168 (6) | 0.0190 (6) | 0.0169 (7) | 0.0015 (5) | 0.0006 (5) | −0.0027 (5) |
| C4 | 0.0171 (6) | 0.0207 (6) | 0.0165 (7) | −0.0013 (5) | 0.0039 (5) | −0.0019 (5) |
| C5 | 0.0197 (6) | 0.0166 (6) | 0.0118 (6) | 0.0005 (5) | 0.0010 (5) | −0.0027 (5) |
| C6 | 0.0166 (6) | 0.0212 (7) | 0.0177 (7) | 0.0022 (5) | 0.0026 (5) | −0.0018 (5) |
| C7 | 0.0189 (6) | 0.0211 (7) | 0.0180 (7) | −0.0012 (5) | 0.0055 (5) | −0.0017 (5) |
| C8 | 0.0187 (6) | 0.0199 (7) | 0.0158 (7) | −0.0021 (5) | 0.0039 (5) | −0.0018 (5) |
| C9 | 0.0208 (6) | 0.0174 (6) | 0.0181 (7) | −0.0019 (5) | 0.0035 (5) | −0.0009 (5) |
| C10 | 0.0184 (6) | 0.0182 (6) | 0.0160 (7) | 0.0010 (5) | 0.0038 (5) | −0.0005 (5) |
| C11 | 0.0191 (6) | 0.0155 (6) | 0.0135 (6) | −0.0013 (5) | 0.0022 (5) | 0.0025 (5) |
| C12 | 0.0154 (6) | 0.0218 (7) | 0.0192 (7) | −0.0006 (5) | 0.0037 (5) | 0.0012 (5) |
| C13 | 0.0194 (6) | 0.0205 (7) | 0.0221 (7) | 0.0011 (5) | 0.0077 (5) | −0.0014 (6) |
| C14 | 0.0205 (6) | 0.0190 (6) | 0.0163 (7) | −0.0018 (5) | 0.0037 (5) | 0.0006 (5) |
| C15 | 0.0160 (6) | 0.0204 (7) | 0.0186 (7) | −0.0006 (5) | 0.0015 (5) | 0.0027 (5) |
| C16 | 0.0165 (6) | 0.0190 (6) | 0.0173 (7) | 0.0016 (5) | 0.0053 (5) | 0.0009 (5) |
| C17 | 0.0242 (7) | 0.0258 (7) | 0.0214 (7) | −0.0022 (6) | 0.0073 (5) | −0.0026 (6) |
Geometric parameters (Å, °)
| O1—C1 | 1.2152 (16) | C8—H8A | 0.99 |
| O2—C5 | 1.3620 (15) | C8—H8B | 0.99 |
| O2—C8 | 1.4389 (15) | C9—C10 | 1.5167 (17) |
| O3—C11 | 1.3623 (15) | C9—H9A | 0.99 |
| O3—C10 | 1.4382 (15) | C9—H9B | 0.99 |
| O4—C17 | 1.2145 (17) | C10—H10A | 0.99 |
| C1—C2 | 1.4682 (19) | C10—H10B | 0.99 |
| C1—H1 | 0.95 | C11—C12 | 1.3931 (18) |
| C2—C3 | 1.3863 (18) | C11—C16 | 1.3981 (17) |
| C2—C7 | 1.4014 (18) | C12—C13 | 1.3796 (19) |
| C3—C4 | 1.3894 (19) | C12—H12 | 0.95 |
| C3—H3 | 0.95 | C13—C14 | 1.4009 (17) |
| C4—C5 | 1.3938 (18) | C13—H13 | 0.95 |
| C4—H4 | 0.95 | C14—C15 | 1.3895 (19) |
| C5—C6 | 1.3989 (18) | C14—C17 | 1.4674 (19) |
| C6—C7 | 1.3765 (19) | C15—C16 | 1.3859 (18) |
| C6—H6 | 0.95 | C15—H15 | 0.95 |
| C7—H7 | 0.95 | C16—H16 | 0.95 |
| C8—C9 | 1.5153 (18) | C17—H17 | 0.95 |
| C5—O2—C8 | 117.78 (10) | C10—C9—H9A | 108.9 |
| C11—O3—C10 | 118.66 (9) | C8—C9—H9B | 108.9 |
| O1—C1—C2 | 124.65 (12) | C10—C9—H9B | 108.9 |
| O1—C1—H1 | 117.7 | H9A—C9—H9B | 107.7 |
| C2—C1—H1 | 117.7 | O3—C10—C9 | 105.71 (10) |
| C3—C2—C7 | 118.83 (12) | O3—C10—H10A | 110.6 |
| C3—C2—C1 | 119.72 (11) | C9—C10—H10A | 110.6 |
| C7—C2—C1 | 121.44 (12) | O3—C10—H10B | 110.6 |
| C2—C3—C4 | 121.36 (12) | C9—C10—H10B | 110.6 |
| C2—C3—H3 | 119.3 | H10A—C10—H10B | 108.7 |
| C4—C3—H3 | 119.3 | O3—C11—C12 | 115.02 (11) |
| C3—C4—C5 | 118.95 (12) | O3—C11—C16 | 124.28 (11) |
| C3—C4—H4 | 120.5 | C12—C11—C16 | 120.69 (12) |
| C5—C4—H4 | 120.5 | C13—C12—C11 | 119.74 (11) |
| O2—C5—C4 | 124.25 (12) | C13—C12—H12 | 120.1 |
| O2—C5—C6 | 115.34 (11) | C11—C12—H12 | 120.1 |
| C4—C5—C6 | 120.40 (12) | C12—C13—C14 | 120.51 (12) |
| C7—C6—C5 | 119.66 (12) | C12—C13—H13 | 119.7 |
| C7—C6—H6 | 120.2 | C14—C13—H13 | 119.7 |
| C5—C6—H6 | 120.2 | C15—C14—C13 | 118.95 (12) |
| C6—C7—C2 | 120.77 (12) | C15—C14—C17 | 121.73 (12) |
| C6—C7—H7 | 119.6 | C13—C14—C17 | 119.32 (12) |
| C2—C7—H7 | 119.6 | C16—C15—C14 | 121.44 (12) |
| O2—C8—C9 | 106.81 (10) | C16—C15—H15 | 119.3 |
| O2—C8—H8A | 110.4 | C14—C15—H15 | 119.3 |
| C9—C8—H8A | 110.4 | C15—C16—C11 | 118.67 (12) |
| O2—C8—H8B | 110.4 | C15—C16—H16 | 120.7 |
| C9—C8—H8B | 110.4 | C11—C16—H16 | 120.7 |
| H8A—C8—H8B | 108.6 | O4—C17—C14 | 124.94 (13) |
| C8—C9—C10 | 113.45 (11) | O4—C17—H17 | 117.5 |
| C8—C9—H9A | 108.9 | C14—C17—H17 | 117.5 |
| O1—C1—C2—C3 | −177.57 (13) | C11—O3—C10—C9 | 176.03 (10) |
| O1—C1—C2—C7 | 3.3 (2) | C8—C9—C10—O3 | 65.84 (13) |
| C7—C2—C3—C4 | 0.75 (19) | C10—O3—C11—C12 | −177.41 (11) |
| C1—C2—C3—C4 | −178.41 (12) | C10—O3—C11—C16 | 3.47 (18) |
| C2—C3—C4—C5 | 0.48 (19) | O3—C11—C12—C13 | −178.89 (12) |
| C8—O2—C5—C4 | 0.89 (18) | C16—C11—C12—C13 | 0.26 (19) |
| C8—O2—C5—C6 | −179.69 (11) | C11—C12—C13—C14 | −0.7 (2) |
| C3—C4—C5—O2 | 178.02 (12) | C12—C13—C14—C15 | 0.6 (2) |
| C3—C4—C5—C6 | −1.37 (19) | C12—C13—C14—C17 | −179.52 (13) |
| O2—C5—C6—C7 | −178.41 (11) | C13—C14—C15—C16 | 0.0 (2) |
| C4—C5—C6—C7 | 1.0 (2) | C17—C14—C15—C16 | −179.92 (12) |
| C5—C6—C7—C2 | 0.2 (2) | C14—C15—C16—C11 | −0.42 (19) |
| C3—C2—C7—C6 | −1.1 (2) | O3—C11—C16—C15 | 179.37 (12) |
| C1—C2—C7—C6 | 178.04 (12) | C12—C11—C16—C15 | 0.29 (19) |
| C5—O2—C8—C9 | −178.76 (10) | C15—C14—C17—O4 | −3.7 (2) |
| O2—C8—C9—C10 | 70.22 (13) | C13—C14—C17—O4 | 176.44 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6···O2i | 0.95 | 2.57 | 3.508 (2) | 168 |
| C16—H16···O1ii | 0.95 | 2.41 | 3.287 (2) | 154 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CI5094).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Hu, P.-Z., Ma, L.-F., Wang, J.-G., Zhao, B.-T. & Wang, L.-Y. (2005). Acta Cryst. E61, o2775–o2777.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst.43 Submitted.
- Zhang, Y., Deng, X., Wang, L. & Wei, T. (2008). J. Incl. Phenom. Macrocycl. Chem.60, 313–319.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810021124/ci5094sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810021124/ci5094Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report