Abstract
The asymmetric unit of the title compound, [Co2(C2O4)(C10H8N2)4][W6O19], consists of one half of the complex [Co2(C2O4)(C10H8N2)4]2+ cation and one half of the Lindqvist-type [W6O19]2− isopolyanion. Both constituents are completed by crystallographic inversion symmetry. In the dimeric cation, the CoII atom is surrounded in a distorted octahedral coordination by four N atoms from two chelating 2,2′-bipyridine ligands and by two O atoms from the chelating oxalate anion. The Lindqvist-type anion exhibits the characteristic W—O bond-length distribution, with the shortest bonds being the W—Oterminal bonds and the longest being those to the central O atom.
Related literature
For general background to polyoxidometalates, see: Pope & Müller (1991 ▶). For polyoxidometalates modified with amines, see: Zhang, Dou et al. (2009 ▶); Zhang, Wei et al. (2009 ▶); Zhang et al. (2010 ▶). For another structure comprising a Lindqvist-type isopolyanion, see: Meng et al. (2006 ▶). For a related structure, see: Li & Xu (2009 ▶).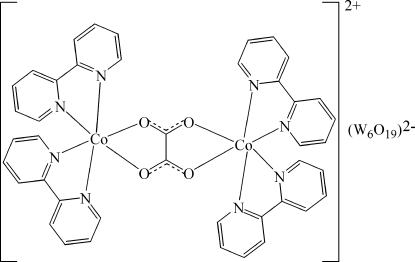
Experimental
Crystal data
[Co2(C2O4)(C10H8N2)4][W6O19]
M r = 2237.72
Triclinic,
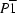
a = 9.4876 (15) Å
b = 9.8548 (15) Å
c = 14.174 (2) Å
α = 90.769 (2)°
β = 91.576 (2)°
γ = 91.113 (2)°
V = 1324.3 (4) Å3
Z = 1
Mo Kα radiation
μ = 13.67 mm−1
T = 293 K
0.12 × 0.10 × 0.08 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.291, T max = 0.408
9331 measured reflections
4613 independent reflections
3755 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.031
wR(F 2) = 0.076
S = 1.00
4613 reflections
368 parameters
H-atom parameters constrained
Δρmax = 2.41 e Å−3
Δρmin = −1.10 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT-Plus (Bruker, 2001 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810023007/wm2358sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810023007/wm2358Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Co1—O1 | 2.104 (6) |
| Co1—N1 | 2.101 (7) |
| Co1—N4 | 2.105 (6) |
| Co1—N2 | 2.114 (6) |
| Co1—N3 | 2.119 (7) |
| Co1—O2 | 2.134 (6) |
| O3—W2 | 1.690 (6) |
| O4—W2 | 1.919 (6) |
| O4—W3 | 1.926 (6) |
| O5—W3 | 1.904 (5) |
| O5—W2i | 1.935 (5) |
| O6—W3 | 1.698 (6) |
| O7—W3 | 1.915 (6) |
| O7—W1 | 1.931 (6) |
| O8—W3i | 2.3185 (4) |
| O8—W3 | 2.3185 (4) |
| O8—W1 | 2.3240 (4) |
| O8—W1i | 2.3240 (4) |
| O8—W2i | 2.3252 (5) |
| O8—W2 | 2.3252 (5) |
| O9—W2 | 1.912 (6) |
| O9—W1 | 1.915 (5) |
| O10—W1 | 1.914 (6) |
| O10—W3i | 1.914 (6) |
| O11—W1 | 1.696 (6) |
| O12—W1 | 1.922 (5) |
| O12—W2i | 1.920 (6) |
Symmetry code: (i) 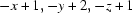 .
.
Acknowledgments
Financial support from the 973 Key Program of the MOST (2006CB932905 and 2007CB81532), the National Natural Science Foundation of China (20873160), the Chinese Academy of Sciences (KJCX2-YW—M02), Shandong Provincial Education Department and Shandong Institute of Education is gratefully acknowledged.
supplementary crystallographic information
Comment
There has been extensive interest in polyoxidometalates, owing to their fascinating properties and great potential applications in many fields such in catalysis, material science, medicine and magnetochemistry (Pope & Müller, 1991). Organic amines, such as 3-(2-pyridyl)pyrazole and pyrazine, are used to effectively modify polyoxidomolybdates or heteropolyoxidomolybdates under hydrothermal condictions (Zhang, Dou et al., 2009; Zhang, Wei et al., 2009; Zhang et al., 2010). Here, we describe the synthesis and structural characterization of the title compound.
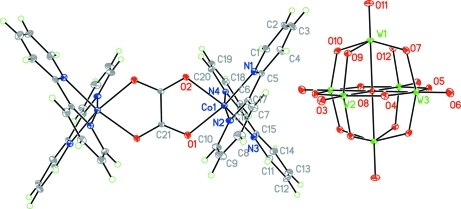
As shown in Figure 1, the title compound consists of two subunits, viz. of a binuclear complex [Co2(C2O4)(C10H8N2)4]2+ cation, and one Lindqvist-type [W6O19]2- isopolyanion. Both constituents exhibit 1 symmetry. The Co2+ cation is surrounded in a distorted octahedral coordination by four N atoms from two chelating 2,2'-bipyridine ligands and two O atoms from a chelating oxalate anion. The Co—N and Co—O bond lengths are in the range of 2.101 (7)—2.119 (7) and 2.104 (6)—2.134 (6) Å, respectively, and are in good agreement with the bond lenghts observed for catena-poly[[(2,2'-bipyridine-κN,N')cobalt(II)]-µ-oxalato- κ4O1,O2:O1',O2'] (Li & Xu, 2009).
The [W6O19]2- polyoxidoanion, possessing the well known Lindquist structure, is formed by six WO6 octahedra connected with each other through edge-sharing oxygen atoms. This anion approaches an approximate Oh symmetry, but actually has 1 symmetry. Three different kinds of oxygen atoms exist in the cluster, viz. terminal Oa, double-bridging Ob, and central Oc oxygen atoms. Therefore, W—O bond lengths can be grouped into three sets: W—Oa: 1.690 (6)—1.698 (6) Å; W—Ob: 1.904 (5)—1.935 (5) Å; W—Oc: 2.3185 (4)—2.3252 (5) Å; these bond lengths strictly follow the rule W—Oa < W—Ob < W—Oc, which is in agreement with the Lindqvist-type polyoxidotungstate reported by Meng et al. (2006).
Experimental
2,2'-bipyridine (0.5 mmoL 0.07 g) and p-carboxyphenylboronic acid were purchased from Jinan Henghua Science & Technology Co. Ltd. A mixture of 2,2'-bipyridine (0.5 mmol 0.07 g), tungstic acid (0.4 mmoL, 0.10 g), oxalic aicd (10 mmol, 0.09), p-carboxyphenylboronic acid (0.3 mmol, 0.05 g), and cobalt(II) sulfate heptahydrate (0.2 mmol, 0.05 g) in 14 ml distilled water was sealed in a 25 ml Teflon-lined stainless steel autoclave and was kept at 433 K for three days. Red crystals suitable for the X-ray experiment were obtained. Anal. Calc. for C42H32Co2N8O23W6: C, 22.53; H, 1.43; N, 5.01. Found: C, 22.26; H, 1.33; N, 4.85%.
Refinement
All hydrogen atoms bound to carbon were refined using a riding model with distance C—H = 0.93 Å, Uiso = 1.2Ueq(C). In the final difference Fourier map the highest peak is 2.60 Å from atom H1 and the deepest hole is 0.81 Å from atom W3. The highest peak is located in the voids of the crystal structure and may be associated with an additional water molecule. However, refinement of this position did not result in a reasonable model.
Figures
Fig. 1.
The cation and anion of the title compound with the atom-labelling scheme. Displacement ellipsoids are drawn at the 30% probability level; H atoms are given as spheres of arbitrary radius.
Crystal data
| [Co2(C2O4)(C10H8N2)4][W6O19] | Z = 1 |
| Mr = 2237.72 | F(000) = 1022 |
| Triclinic, P1 | Dx = 2.806 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.4876 (15) Å | Cell parameters from 3530 reflections |
| b = 9.8548 (15) Å | θ = 2.5–27.3° |
| c = 14.174 (2) Å | µ = 13.67 mm−1 |
| α = 90.769 (2)° | T = 293 K |
| β = 91.576 (2)° | Block, red |
| γ = 91.113 (2)° | 0.12 × 0.10 × 0.08 mm |
| V = 1324.3 (4) Å3 |
Data collection
| Bruker APEXII CCD diffractometer | 4613 independent reflections |
| Radiation source: fine-focus sealed tube | 3755 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| φ and ω scans | θmax = 25.0°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −11→11 |
| Tmin = 0.291, Tmax = 0.408 | k = −11→11 |
| 9331 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.031 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.076 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0375P)2 + 1.9139P] where P = (Fo2 + 2Fc2)/3 |
| 4613 reflections | (Δ/σ)max = 0.001 |
| 368 parameters | Δρmax = 2.41 e Å−3 |
| 0 restraints | Δρmin = −1.10 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.5864 (9) | 0.5558 (9) | 0.7481 (6) | 0.039 (2) | |
| H1 | 0.6557 | 0.5423 | 0.7040 | 0.046* | |
| C2 | 0.4503 (10) | 0.5210 (10) | 0.7228 (7) | 0.050 (3) | |
| H2 | 0.4275 | 0.4831 | 0.6638 | 0.060* | |
| C3 | 0.3498 (10) | 0.5440 (10) | 0.7869 (7) | 0.050 (3) | |
| H3 | 0.2562 | 0.5220 | 0.7714 | 0.060* | |
| C4 | 0.3841 (9) | 0.5996 (10) | 0.8753 (6) | 0.045 (2) | |
| H4 | 0.3150 | 0.6150 | 0.9192 | 0.054* | |
| C5 | 0.5239 (8) | 0.6314 (8) | 0.8957 (5) | 0.0262 (18) | |
| C6 | 0.5713 (9) | 0.6925 (8) | 0.9874 (5) | 0.031 (2) | |
| C7 | 0.4828 (10) | 0.7231 (11) | 1.0590 (7) | 0.050 (3) | |
| H7 | 0.3869 | 0.7028 | 1.0521 | 0.061* | |
| C8 | 0.5343 (12) | 0.7834 (12) | 1.1405 (7) | 0.062 (3) | |
| H8 | 0.4747 | 0.8067 | 1.1889 | 0.074* | |
| C9 | 0.6775 (12) | 0.8085 (12) | 1.1486 (7) | 0.064 (3) | |
| H9 | 0.7157 | 0.8488 | 1.2035 | 0.077* | |
| C10 | 0.7625 (11) | 0.7754 (10) | 1.0781 (6) | 0.046 (2) | |
| H10 | 0.8589 | 0.7931 | 1.0851 | 0.055* | |
| C11 | 0.8337 (10) | 0.9655 (10) | 0.8619 (7) | 0.046 (2) | |
| H11 | 0.7816 | 0.9641 | 0.9166 | 0.055* | |
| C12 | 0.8660 (11) | 1.0894 (10) | 0.8234 (7) | 0.052 (3) | |
| H12 | 0.8396 | 1.1705 | 0.8515 | 0.062* | |
| C13 | 0.9399 (12) | 1.0868 (11) | 0.7404 (8) | 0.062 (3) | |
| H13 | 0.9621 | 1.1676 | 0.7105 | 0.074* | |
| C14 | 0.9796 (10) | 0.9684 (10) | 0.7032 (7) | 0.047 (2) | |
| H14 | 1.0299 | 0.9669 | 0.6478 | 0.057* | |
| C15 | 0.9459 (8) | 0.8496 (9) | 0.7469 (6) | 0.0306 (19) | |
| C16 | 0.9845 (8) | 0.7135 (9) | 0.7095 (6) | 0.0294 (19) | |
| C17 | 1.0644 (9) | 0.6960 (10) | 0.6295 (6) | 0.040 (2) | |
| H17 | 1.1000 | 0.7709 | 0.5981 | 0.048* | |
| C18 | 1.0897 (9) | 0.5675 (10) | 0.5977 (6) | 0.043 (2) | |
| H18 | 1.1428 | 0.5541 | 0.5443 | 0.052* | |
| C19 | 1.0363 (9) | 0.4584 (10) | 0.6452 (6) | 0.043 (2) | |
| H19 | 1.0522 | 0.3703 | 0.6243 | 0.051* | |
| C20 | 0.9589 (9) | 0.4815 (9) | 0.7240 (6) | 0.035 (2) | |
| H20 | 0.9230 | 0.4076 | 0.7564 | 0.042* | |
| C21 | 1.0517 (8) | 0.5600 (8) | 1.0069 (5) | 0.0255 (18) | |
| Co1 | 0.83213 (11) | 0.65380 (11) | 0.88211 (7) | 0.0263 (3) | |
| N1 | 0.6258 (7) | 0.6078 (7) | 0.8323 (4) | 0.0302 (16) | |
| N2 | 0.7116 (7) | 0.7170 (7) | 0.9974 (5) | 0.0328 (17) | |
| N3 | 0.8729 (7) | 0.8481 (7) | 0.8254 (5) | 0.0325 (16) | |
| N4 | 0.9334 (7) | 0.6070 (7) | 0.7558 (4) | 0.0269 (15) | |
| O1 | 1.0227 (6) | 0.6668 (6) | 0.9616 (4) | 0.0350 (14) | |
| O2 | 0.8466 (5) | 0.4532 (6) | 0.9359 (4) | 0.0318 (13) | |
| O3 | 0.4860 (7) | 0.8809 (7) | 0.7694 (4) | 0.0521 (18) | |
| O4 | 0.3221 (6) | 1.0289 (6) | 0.6381 (4) | 0.0362 (14) | |
| O5 | 0.3335 (6) | 1.1440 (6) | 0.3892 (4) | 0.0369 (15) | |
| O6 | 0.1293 (7) | 1.1836 (7) | 0.5300 (5) | 0.0540 (18) | |
| O7 | 0.2326 (6) | 0.9278 (6) | 0.4737 (4) | 0.0383 (15) | |
| O8 | 0.5000 | 1.0000 | 0.5000 | 0.0219 (16) | |
| O9 | 0.3985 (6) | 0.7844 (6) | 0.5844 (4) | 0.0342 (14) | |
| O10 | 0.5765 (6) | 0.7561 (6) | 0.4464 (4) | 0.0373 (14) | |
| O11 | 0.2952 (7) | 0.6576 (7) | 0.4139 (5) | 0.0528 (18) | |
| O12 | 0.4107 (6) | 0.8997 (6) | 0.3357 (4) | 0.0338 (14) | |
| W1 | 0.38156 (4) | 0.80202 (4) | 0.45023 (2) | 0.03214 (12) | |
| W2 | 0.49167 (4) | 0.92799 (4) | 0.65534 (2) | 0.03134 (12) | |
| W3 | 0.28645 (4) | 1.10717 (4) | 0.51619 (2) | 0.03216 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.039 (5) | 0.051 (6) | 0.025 (4) | 0.002 (5) | −0.004 (4) | −0.011 (4) |
| C2 | 0.047 (6) | 0.057 (7) | 0.044 (6) | 0.004 (5) | −0.018 (5) | −0.008 (5) |
| C3 | 0.028 (5) | 0.061 (7) | 0.058 (6) | −0.010 (5) | −0.014 (5) | 0.004 (5) |
| C4 | 0.031 (5) | 0.063 (7) | 0.041 (5) | −0.004 (5) | 0.000 (4) | −0.002 (5) |
| C5 | 0.024 (4) | 0.028 (5) | 0.027 (4) | 0.000 (4) | −0.003 (3) | 0.001 (3) |
| C6 | 0.032 (5) | 0.031 (5) | 0.030 (4) | 0.001 (4) | 0.012 (4) | 0.001 (4) |
| C7 | 0.038 (6) | 0.064 (7) | 0.049 (6) | −0.004 (5) | 0.011 (5) | −0.014 (5) |
| C8 | 0.057 (7) | 0.074 (8) | 0.055 (7) | −0.002 (6) | 0.027 (5) | −0.027 (6) |
| C9 | 0.072 (8) | 0.083 (9) | 0.036 (6) | −0.003 (7) | 0.003 (5) | −0.030 (5) |
| C10 | 0.052 (6) | 0.051 (6) | 0.036 (5) | −0.007 (5) | 0.003 (4) | −0.014 (5) |
| C11 | 0.044 (6) | 0.039 (6) | 0.056 (6) | 0.007 (5) | 0.010 (5) | −0.002 (5) |
| C12 | 0.064 (7) | 0.023 (5) | 0.067 (7) | 0.006 (5) | −0.001 (6) | −0.008 (5) |
| C13 | 0.066 (8) | 0.044 (7) | 0.074 (8) | −0.007 (6) | −0.002 (6) | 0.013 (6) |
| C14 | 0.053 (6) | 0.042 (6) | 0.047 (6) | −0.007 (5) | 0.006 (5) | 0.001 (5) |
| C15 | 0.021 (4) | 0.038 (5) | 0.033 (4) | −0.002 (4) | −0.006 (3) | 0.001 (4) |
| C16 | 0.016 (4) | 0.038 (5) | 0.034 (4) | −0.009 (4) | −0.009 (3) | 0.002 (4) |
| C17 | 0.039 (5) | 0.049 (6) | 0.031 (5) | −0.001 (5) | 0.011 (4) | 0.009 (4) |
| C18 | 0.036 (5) | 0.056 (7) | 0.038 (5) | 0.002 (5) | 0.009 (4) | −0.005 (5) |
| C19 | 0.047 (6) | 0.044 (6) | 0.037 (5) | 0.004 (5) | 0.003 (4) | −0.015 (4) |
| C20 | 0.034 (5) | 0.028 (5) | 0.042 (5) | 0.000 (4) | 0.002 (4) | −0.002 (4) |
| C21 | 0.016 (4) | 0.029 (5) | 0.032 (4) | −0.004 (3) | 0.005 (3) | −0.005 (4) |
| Co1 | 0.0243 (6) | 0.0289 (6) | 0.0258 (6) | 0.0001 (5) | 0.0035 (4) | −0.0003 (5) |
| N1 | 0.026 (4) | 0.034 (4) | 0.031 (4) | 0.001 (3) | 0.005 (3) | −0.001 (3) |
| N2 | 0.034 (4) | 0.031 (4) | 0.033 (4) | −0.003 (3) | 0.006 (3) | −0.011 (3) |
| N3 | 0.034 (4) | 0.027 (4) | 0.037 (4) | 0.001 (3) | 0.004 (3) | 0.004 (3) |
| N4 | 0.026 (4) | 0.028 (4) | 0.027 (3) | −0.001 (3) | 0.003 (3) | −0.002 (3) |
| O1 | 0.030 (3) | 0.037 (4) | 0.038 (3) | −0.002 (3) | −0.003 (3) | 0.007 (3) |
| O2 | 0.023 (3) | 0.036 (4) | 0.035 (3) | −0.004 (3) | −0.005 (2) | 0.004 (3) |
| O3 | 0.065 (5) | 0.059 (5) | 0.033 (3) | 0.007 (4) | 0.004 (3) | 0.003 (3) |
| O4 | 0.032 (3) | 0.048 (4) | 0.029 (3) | 0.001 (3) | 0.014 (2) | −0.008 (3) |
| O5 | 0.037 (3) | 0.041 (4) | 0.032 (3) | 0.009 (3) | 0.000 (3) | 0.000 (3) |
| O6 | 0.039 (4) | 0.068 (5) | 0.055 (4) | 0.010 (4) | 0.002 (3) | −0.014 (4) |
| O7 | 0.028 (3) | 0.053 (4) | 0.034 (3) | 0.002 (3) | 0.002 (3) | −0.008 (3) |
| O8 | 0.021 (4) | 0.024 (4) | 0.021 (4) | 0.004 (3) | 0.006 (3) | −0.003 (3) |
| O9 | 0.035 (3) | 0.033 (4) | 0.035 (3) | −0.004 (3) | 0.008 (3) | 0.000 (3) |
| O10 | 0.047 (4) | 0.027 (3) | 0.039 (3) | 0.007 (3) | 0.006 (3) | −0.006 (3) |
| O11 | 0.054 (4) | 0.042 (4) | 0.061 (4) | −0.016 (3) | 0.009 (3) | −0.013 (3) |
| O12 | 0.033 (3) | 0.046 (4) | 0.022 (3) | 0.002 (3) | 0.003 (2) | −0.008 (3) |
| W1 | 0.0353 (2) | 0.0295 (2) | 0.0313 (2) | −0.00686 (16) | 0.00560 (15) | −0.00922 (15) |
| W2 | 0.0381 (2) | 0.0338 (2) | 0.02229 (18) | 0.00114 (16) | 0.00478 (14) | −0.00123 (14) |
| W3 | 0.0274 (2) | 0.0368 (2) | 0.0325 (2) | 0.00719 (16) | 0.00465 (14) | −0.00600 (15) |
Geometric parameters (Å, °)
| C1—N1 | 1.333 (10) | C19—C20 | 1.373 (11) |
| C1—C2 | 1.366 (13) | C19—H19 | 0.9300 |
| C1—H1 | 0.9300 | C20—N4 | 1.340 (10) |
| C2—C3 | 1.355 (13) | C20—H20 | 0.9300 |
| C2—H2 | 0.9300 | C21—O2i | 1.254 (9) |
| C3—C4 | 1.388 (13) | C21—O1 | 1.271 (9) |
| C3—H3 | 0.9300 | C21—C21i | 1.528 (15) |
| C4—C5 | 1.379 (11) | Co1—O1 | 2.104 (6) |
| C4—H4 | 0.9300 | Co1—N1 | 2.101 (7) |
| C5—N1 | 1.360 (9) | Co1—N4 | 2.105 (6) |
| C5—C6 | 1.479 (11) | Co1—N2 | 2.114 (6) |
| C6—N2 | 1.351 (10) | Co1—N3 | 2.119 (7) |
| C6—C7 | 1.370 (11) | Co1—O2 | 2.134 (6) |
| C7—C8 | 1.366 (13) | O2—C21i | 1.254 (9) |
| C7—H7 | 0.9300 | O3—W2 | 1.690 (6) |
| C8—C9 | 1.378 (15) | O4—W2 | 1.919 (6) |
| C8—H8 | 0.9300 | O4—W3 | 1.926 (6) |
| C9—C10 | 1.343 (12) | O5—W3 | 1.904 (5) |
| C9—H9 | 0.9300 | O5—W2ii | 1.935 (5) |
| C10—N2 | 1.346 (10) | O6—W3 | 1.698 (6) |
| C10—H10 | 0.9300 | O7—W3 | 1.915 (6) |
| C11—N3 | 1.325 (11) | O7—W1 | 1.931 (6) |
| C11—C12 | 1.377 (13) | O8—W3ii | 2.3185 (4) |
| C11—H11 | 0.9300 | O8—W3 | 2.3185 (4) |
| C12—C13 | 1.386 (14) | O8—W1 | 2.3240 (4) |
| C12—H12 | 0.9300 | O8—W1ii | 2.3240 (4) |
| C13—C14 | 1.339 (14) | O8—W2ii | 2.3252 (5) |
| C13—H13 | 0.9300 | O8—W2 | 2.3252 (5) |
| C14—C15 | 1.368 (12) | O9—W2 | 1.912 (6) |
| C14—H14 | 0.9300 | O9—W1 | 1.915 (5) |
| C15—N3 | 1.327 (10) | O10—W1 | 1.914 (6) |
| C15—C16 | 1.491 (11) | O10—W3ii | 1.914 (6) |
| C16—N4 | 1.336 (10) | O11—W1 | 1.696 (6) |
| C16—C17 | 1.391 (11) | O12—W1 | 1.922 (5) |
| C17—C18 | 1.367 (12) | O12—W2ii | 1.920 (6) |
| C17—H17 | 0.9300 | W2—O12ii | 1.920 (6) |
| C18—C19 | 1.372 (12) | W2—O5ii | 1.935 (5) |
| C18—H18 | 0.9300 | W3—O10ii | 1.914 (6) |
| N1—C1—C2 | 124.0 (8) | C1—N1—Co1 | 127.4 (5) |
| N1—C1—H1 | 118.0 | C5—N1—Co1 | 114.6 (5) |
| C2—C1—H1 | 118.0 | C10—N2—C6 | 118.9 (7) |
| C3—C2—C1 | 117.5 (9) | C10—N2—Co1 | 126.0 (6) |
| C3—C2—H2 | 121.3 | C6—N2—Co1 | 115.1 (5) |
| C1—C2—H2 | 121.3 | C11—N3—C15 | 118.4 (8) |
| C2—C3—C4 | 121.2 (9) | C11—N3—Co1 | 125.9 (6) |
| C2—C3—H3 | 119.4 | C15—N3—Co1 | 115.7 (6) |
| C4—C3—H3 | 119.4 | C20—N4—C16 | 119.2 (7) |
| C3—C4—C5 | 117.8 (8) | C20—N4—Co1 | 125.2 (6) |
| C3—C4—H4 | 121.1 | C16—N4—Co1 | 115.4 (5) |
| C5—C4—H4 | 121.1 | C21—O1—Co1 | 114.2 (5) |
| N1—C5—C4 | 121.5 (7) | C21i—O2—Co1 | 113.0 (5) |
| N1—C5—C6 | 116.4 (7) | W2—O4—W3 | 117.6 (2) |
| C4—C5—C6 | 122.1 (7) | W3—O5—W2ii | 117.3 (3) |
| N2—C6—C7 | 120.6 (8) | W3—O7—W1 | 117.5 (3) |
| N2—C6—C5 | 115.4 (6) | W3ii—O8—W3 | 180.0 |
| C7—C6—C5 | 124.0 (8) | W3ii—O8—W1 | 89.833 (16) |
| C8—C7—C6 | 120.4 (9) | W3—O8—W1 | 90.167 (16) |
| C8—C7—H7 | 119.8 | W3ii—O8—W1ii | 90.167 (16) |
| C6—C7—H7 | 119.8 | W3—O8—W1ii | 89.833 (16) |
| C7—C8—C9 | 117.8 (9) | W1—O8—W1ii | 180.0 |
| C7—C8—H8 | 121.1 | W3ii—O8—W2ii | 90.173 (13) |
| C9—C8—H8 | 121.1 | W3—O8—W2ii | 89.827 (13) |
| C10—C9—C8 | 120.7 (10) | W1—O8—W2ii | 90.203 (14) |
| C10—C9—H9 | 119.7 | W1ii—O8—W2ii | 89.797 (14) |
| C8—C9—H9 | 119.7 | W3ii—O8—W2 | 89.827 (13) |
| N2—C10—C9 | 121.6 (10) | W3—O8—W2 | 90.173 (13) |
| N2—C10—H10 | 119.2 | W1—O8—W2 | 89.797 (14) |
| C9—C10—H10 | 119.2 | W1ii—O8—W2 | 90.203 (14) |
| N3—C11—C12 | 123.5 (9) | W2ii—O8—W2 | 180.0 |
| N3—C11—H11 | 118.2 | W2—O9—W1 | 118.1 (3) |
| C12—C11—H11 | 118.2 | W1—O10—W3ii | 117.8 (3) |
| C13—C12—C11 | 116.4 (9) | W1—O12—W2ii | 118.0 (3) |
| C13—C12—H12 | 121.8 | O11—W1—O10 | 103.9 (3) |
| C11—C12—H12 | 121.8 | O11—W1—O9 | 104.0 (3) |
| C14—C13—C12 | 120.2 (10) | O10—W1—O9 | 86.9 (2) |
| C14—C13—H13 | 119.9 | O11—W1—O12 | 104.1 (3) |
| C12—C13—H13 | 119.9 | O10—W1—O12 | 86.8 (2) |
| C13—C14—C15 | 119.8 (9) | O9—W1—O12 | 151.9 (2) |
| C13—C14—H14 | 120.1 | O11—W1—O7 | 104.1 (3) |
| C15—C14—H14 | 120.1 | O10—W1—O7 | 152.1 (2) |
| N3—C15—C14 | 121.6 (8) | O9—W1—O7 | 86.5 (2) |
| N3—C15—C16 | 115.2 (7) | O12—W1—O7 | 86.3 (2) |
| C14—C15—C16 | 123.2 (8) | O11—W1—O8 | 180.0 (3) |
| N4—C16—C17 | 121.1 (8) | O10—W1—O8 | 76.11 (17) |
| N4—C16—C15 | 115.8 (7) | O9—W1—O8 | 76.06 (17) |
| C17—C16—C15 | 123.0 (8) | O12—W1—O8 | 75.87 (17) |
| C18—C17—C16 | 119.2 (8) | O7—W1—O8 | 75.96 (18) |
| C18—C17—H17 | 120.4 | O3—W2—O9 | 105.6 (3) |
| C16—C17—H17 | 120.4 | O3—W2—O4 | 103.1 (3) |
| C17—C18—C19 | 119.5 (8) | O9—W2—O4 | 87.0 (2) |
| C17—C18—H18 | 120.2 | O3—W2—O12ii | 102.5 (3) |
| C19—C18—H18 | 120.2 | O9—W2—O12ii | 152.0 (2) |
| C18—C19—C20 | 118.9 (8) | O4—W2—O12ii | 86.6 (2) |
| C18—C19—H19 | 120.6 | O3—W2—O5ii | 104.7 (3) |
| C20—C19—H19 | 120.6 | O9—W2—O5ii | 86.7 (2) |
| N4—C20—C19 | 122.1 (8) | O4—W2—O5ii | 152.2 (2) |
| N4—C20—H20 | 118.9 | O12ii—W2—O5ii | 86.3 (2) |
| C19—C20—H20 | 118.9 | O3—W2—O8 | 178.2 (2) |
| O2i—C21—O1 | 125.8 (7) | O9—W2—O8 | 76.08 (16) |
| O2i—C21—C21i | 117.7 (9) | O4—W2—O8 | 76.11 (15) |
| O1—C21—C21i | 116.4 (9) | O12ii—W2—O8 | 75.88 (15) |
| O1—Co1—N1 | 164.8 (2) | O5ii—W2—O8 | 76.08 (16) |
| O1—Co1—N4 | 93.3 (2) | O6—W3—O5 | 104.3 (3) |
| N1—Co1—N4 | 96.6 (2) | O6—W3—O7 | 103.2 (3) |
| O1—Co1—N2 | 92.9 (2) | O5—W3—O7 | 87.2 (2) |
| N1—Co1—N2 | 78.4 (3) | O6—W3—O10ii | 104.2 (3) |
| N4—Co1—N2 | 172.1 (3) | O5—W3—O10ii | 87.1 (2) |
| O1—Co1—N3 | 90.4 (2) | O7—W3—O10ii | 152.6 (2) |
| N1—Co1—N3 | 103.0 (3) | O6—W3—O4 | 102.7 (3) |
| N4—Co1—N3 | 77.4 (3) | O5—W3—O4 | 153.0 (2) |
| N2—Co1—N3 | 97.7 (3) | O7—W3—O4 | 86.6 (2) |
| O1—Co1—O2 | 78.3 (2) | O10ii—W3—O4 | 86.3 (2) |
| N1—Co1—O2 | 89.4 (2) | O6—W3—O8 | 178.8 (2) |
| N4—Co1—O2 | 94.3 (2) | O5—W3—O8 | 76.81 (16) |
| N2—Co1—O2 | 91.7 (2) | O7—W3—O8 | 76.38 (16) |
| N3—Co1—O2 | 165.6 (2) | O10ii—W3—O8 | 76.26 (16) |
| C1—N1—C5 | 118.0 (7) | O4—W3—O8 | 76.16 (15) |
Symmetry codes: (i) −x+2, −y+1, −z+2; (ii) −x+1, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: WM2358).
References
- Bruker (2001). SAINT-Plus and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2004). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Li, P.-Z. & Xu, Q. (2009). Acta Cryst. E65, m508. [DOI] [PMC free article] [PubMed]
- Meng, F. X., Liu, K. & Chen, Y. G. (2006). Chin. J. Struct. Chem.25, 837–843.
- Pope, M. T. & Müller, A. (1991). Angew. Chem. Int. Ed.30, 34–38.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhang, X. T., Dou, J. M., Wei, P. H., Li, D. C., Li, B., Shi, C. W. & Hu, B. (2009). Inorg. Chim. Acta, 362, 3325–3332.
- Zhang, X., Wei, P., Shi, C., Li, B. & Hu, B. (2010). Acta Cryst. E66, m26–m27. [DOI] [PMC free article] [PubMed]
- Zhang, X. T., Wei, P. H., Sun, D. F., Ni, Z. H., Dou, J. M., Li, B., Shi, C. W. & Hu, B. (2009). Cryst. Growth Des.9, 4424–4428.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810023007/wm2358sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810023007/wm2358Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report