Abstract
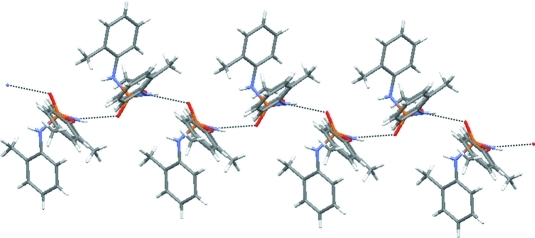
In the title compound, C21H23N2O2P, the P atom has a distorted tetrahedral configuration. The O atom of the OC6H4-4-CH3 group and the N atoms show sp 2 character. In the crystal, adjacent molecules are linked by N—H⋯O hydrogen bonds into helical chains parallel to the b axis.
Related literature
For a related structure, see: Pourayoubi et al. (2009 ▶).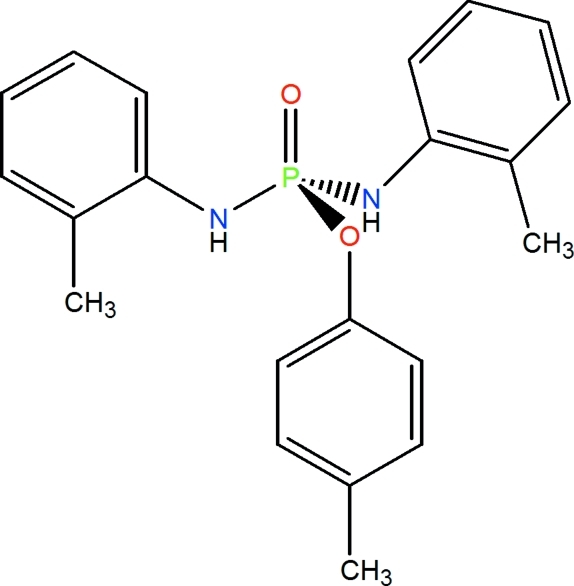
Experimental
Crystal data
C21H23N2O2P
M r = 366.38
Monoclinic,
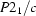
a = 12.157 (3) Å
b = 8.978 (2) Å
c = 18.080 (5) Å
β = 101.569 (1)°
V = 1933.3 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.16 mm−1
T = 293 K
0.6 × 0.54 × 0.47 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (Blessing, 1995 ▶) T min = 0.860, T max = 0.968
23372 measured reflections
4402 independent reflections
3097 reflections with I > 2σ(I)
R int = 0.048
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.128
S = 1.06
4402 reflections
257 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.18 e Å−3
Δρmin = −0.36 e Å−3
Data collection: COLLECT (Nonius, 2001 ▶); cell refinement: HKL SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: HKL DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810023512/ng2779sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810023512/ng2779Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.91 (2) | 2.02 (2) | 2.8963 (19) | 161 (2) |
Symmetry code: (i) 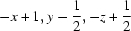 .
.
Acknowledgments
Support of this investigation by Islamic Azad University-Zanjan Branch is gratefully acknowledged.
supplementary crystallographic information
Comment
In the previous work, the structure determination of p-tolyl bis(p-tolylamido)phosphate (Pourayoubi et al., 2009) has been investigated; we report here on the crystal structure of title compound (Fig. 1). The title compound was synthesized from the reaction of (4-tolyl)-dichlorophosphate with an excess amount of ortho-toluidine (1:4 mole ratio). Single crystals were obtained from CHCl3/n-C6H14 at room temperature. Molecular structure of [4-H3C—C6H4O]P(O)[NHC6H4-2-CH3]2 is shown in Fig. 1. The phosphorus atom has a distorted tetrahedral configuration. The bond angles around P atom are in the range of 96.87 (7)° to 118.95 (8)°. The oxygen atom of OC6H4-4-CH3 moiety and the nitrogen atoms show sp2 character (the C15—O2—P1 angle is 124.67 (11)°, the C1—N1—P1 and C8—N2—P1 are 123.77 (12)° and 127.71 (12)°, respectively. In the crystal structure, molecules are linked via N—H···O hydrogen bonds (N1···O1 = 2.8963 (19) Å) into an extended chain (Fig. 2) parallel to the b axis.
Experimental
To a solution of (4-tolyl)-dichlorophosphate (2.250 g, 10 mmol) in 15 ml dry acetonitrile, a solution of ortho-toluidine (4.286 g, 40 mmol) in 30 ml acetonitrile was added at 0°C. After 4 h stirring, the solvent was evaporated in vacuum. The solid was washed with distilled water. Single crystals of the product were obtained from a solution of CHCl3/n-C6H14 at room temperature.
Refinement
H atoms of both nitrogen were found by Fourier differences, it was necessary to restrain distances setting the NH as 1.01 Å instead of 0.86 Å as the ideal would be, but under this proposal to refine, both distances are obtained, 0.9119 (152) Å for N1—H1 and 0.8982 (153) Å for N2—H21,respectively, which are more realistic. The difference can be due to the effect of hydrogen bond generates by N1—H1—O1. All other hydrogen atoms were placed geometrically.
Figures
Fig. 1.
A general view of the title compound, showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
A view of N—H···O hydrogen bond.
Crystal data
| C21H23N2O2P | F(000) = 776 |
| Mr = 366.38 | Dx = 1.259 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 600 reflections |
| a = 12.157 (3) Å | θ = 1–14° |
| b = 8.978 (2) Å | µ = 0.16 mm−1 |
| c = 18.080 (5) Å | T = 293 K |
| β = 101.569 (1)° | Priem, colourless |
| V = 1933.3 (8) Å3 | 0.6 × 0.54 × 0.47 mm |
| Z = 4 |
Data collection
| Nonius KappaCCD diffractometer | 4402 independent reflections |
| Radiation source: Enraf Nonius FR590 | 3097 reflections with I > 2σ(I) |
| graphite | Rint = 0.048 |
| Detector resolution: 9 pixels mm-1 | θmax = 27.5°, θmin = 2.6° |
| CCD rotation images, thick slices scans | h = −15→15 |
| Absorption correction: multi-scan (Blessing, 1995) | k = −10→11 |
| Tmin = 0.860, Tmax = 0.968 | l = −19→23 |
| 23372 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.128 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0666P)2 + 0.2653P] where P = (Fo2 + 2Fc2)/3 |
| 4402 reflections | (Δ/σ)max = 0.015 |
| 257 parameters | Δρmax = 0.18 e Å−3 |
| 2 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.28763 (13) | 0.18689 (18) | 0.26516 (10) | 0.0359 (4) | |
| C2 | 0.25191 (16) | 0.3029 (2) | 0.21582 (11) | 0.0464 (4) | |
| H2 | 0.3036 | 0.3535 | 0.1936 | 0.056* | |
| C3 | 0.14023 (18) | 0.3444 (3) | 0.19930 (13) | 0.0606 (6) | |
| H3 | 0.1171 | 0.4232 | 0.1665 | 0.073* | |
| C4 | 0.06346 (18) | 0.2691 (3) | 0.23138 (15) | 0.0693 (7) | |
| H4 | −0.0121 | 0.2951 | 0.2197 | 0.083* | |
| C5 | 0.09918 (17) | 0.1546 (3) | 0.28107 (14) | 0.0620 (6) | |
| H5 | 0.0466 | 0.1044 | 0.3027 | 0.074* | |
| C6 | 0.21099 (15) | 0.1116 (2) | 0.29999 (11) | 0.0446 (4) | |
| C7 | 0.24895 (19) | −0.0078 (2) | 0.35748 (13) | 0.0628 (6) | |
| H7A | 0.1847 | −0.0521 | 0.3722 | 0.094* | |
| H7B | 0.2969 | 0.0351 | 0.401 | 0.094* | |
| H7C | 0.2896 | −0.0827 | 0.3361 | 0.094* | |
| C8 | 0.43487 (15) | 0.36933 (19) | 0.41804 (9) | 0.0395 (4) | |
| C9 | 0.47389 (18) | 0.2563 (2) | 0.46833 (11) | 0.0518 (5) | |
| H9 | 0.5289 | 0.1915 | 0.4585 | 0.062* | |
| C10 | 0.4311 (2) | 0.2392 (3) | 0.53349 (12) | 0.0656 (6) | |
| H10 | 0.4559 | 0.1615 | 0.5666 | 0.079* | |
| C11 | 0.3524 (2) | 0.3374 (3) | 0.54873 (13) | 0.0687 (7) | |
| H11 | 0.3252 | 0.3283 | 0.5931 | 0.082* | |
| C12 | 0.31363 (18) | 0.4494 (3) | 0.49840 (12) | 0.0614 (6) | |
| H12 | 0.2603 | 0.5155 | 0.5095 | 0.074* | |
| C13 | 0.35181 (15) | 0.4668 (2) | 0.43141 (10) | 0.0464 (5) | |
| C14 | 0.3050 (2) | 0.5866 (3) | 0.37677 (16) | 0.0643 (6) | |
| C15 | 0.71094 (14) | 0.1945 (2) | 0.36813 (10) | 0.0399 (4) | |
| C16 | 0.76182 (16) | 0.1141 (2) | 0.43050 (12) | 0.0530 (5) | |
| H16 | 0.7234 | 0.0378 | 0.4492 | 0.064* | |
| C17 | 0.87118 (17) | 0.1486 (3) | 0.46506 (13) | 0.0607 (6) | |
| H17 | 0.9056 | 0.0944 | 0.5072 | 0.073* | |
| C18 | 0.93031 (17) | 0.2605 (3) | 0.43883 (13) | 0.0575 (5) | |
| C19 | 0.87634 (17) | 0.3392 (3) | 0.37669 (13) | 0.0633 (6) | |
| H19 | 0.9143 | 0.4163 | 0.3583 | 0.076* | |
| C20 | 0.76747 (16) | 0.3071 (2) | 0.34082 (12) | 0.0539 (5) | |
| H20 | 0.7331 | 0.3613 | 0.2987 | 0.065* | |
| C21 | 1.0505 (2) | 0.2947 (4) | 0.47617 (17) | 0.0897 (9) | |
| H21A | 1.0534 | 0.3242 | 0.5276 | 0.135* | |
| H21B | 1.0784 | 0.3741 | 0.4495 | 0.135* | |
| H21C | 1.0959 | 0.2075 | 0.4751 | 0.135* | |
| N1 | 0.40296 (12) | 0.14475 (16) | 0.28185 (9) | 0.0387 (3) | |
| N2 | 0.48102 (13) | 0.38993 (16) | 0.35183 (8) | 0.0393 (3) | |
| O1 | 0.53434 (10) | 0.33484 (13) | 0.22624 (7) | 0.0420 (3) | |
| O2 | 0.60148 (10) | 0.15191 (13) | 0.33463 (7) | 0.0444 (3) | |
| P1 | 0.50634 (4) | 0.26305 (4) | 0.29312 (2) | 0.03385 (15) | |
| H1 | 0.4230 (17) | 0.0477 (18) | 0.2912 (12) | 0.062 (6)* | |
| H21 | 0.4928 (17) | 0.4862 (18) | 0.3421 (12) | 0.061 (6)* | |
| H14C | 0.267 (3) | 0.548 (3) | 0.3309 (19) | 0.104 (10)* | |
| H14B | 0.249 (3) | 0.645 (4) | 0.3955 (18) | 0.123 (11)* | |
| H14A | 0.363 (3) | 0.660 (3) | 0.3641 (17) | 0.105 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0343 (9) | 0.0320 (9) | 0.0410 (9) | −0.0007 (7) | 0.0063 (7) | −0.0066 (7) |
| C2 | 0.0451 (11) | 0.0446 (10) | 0.0496 (11) | 0.0036 (8) | 0.0099 (9) | 0.0016 (8) |
| C3 | 0.0508 (12) | 0.0606 (13) | 0.0673 (13) | 0.0155 (10) | 0.0046 (10) | 0.0094 (11) |
| C4 | 0.0381 (11) | 0.0708 (15) | 0.0969 (19) | 0.0124 (10) | 0.0085 (12) | 0.0035 (13) |
| C5 | 0.0429 (12) | 0.0608 (13) | 0.0875 (16) | −0.0049 (10) | 0.0255 (11) | −0.0025 (12) |
| C6 | 0.0434 (10) | 0.0400 (10) | 0.0524 (11) | −0.0032 (8) | 0.0142 (8) | −0.0028 (8) |
| C7 | 0.0634 (13) | 0.0583 (13) | 0.0724 (14) | −0.0046 (11) | 0.0272 (11) | 0.0159 (11) |
| C8 | 0.0440 (10) | 0.0391 (9) | 0.0351 (9) | −0.0131 (7) | 0.0074 (7) | −0.0069 (7) |
| C9 | 0.0580 (12) | 0.0516 (12) | 0.0446 (11) | −0.0084 (9) | 0.0072 (9) | 0.0031 (9) |
| C10 | 0.0774 (16) | 0.0723 (15) | 0.0447 (12) | −0.0274 (13) | 0.0066 (11) | 0.0083 (10) |
| C11 | 0.0781 (16) | 0.0866 (17) | 0.0469 (12) | −0.0379 (14) | 0.0255 (11) | −0.0122 (12) |
| C12 | 0.0569 (12) | 0.0702 (15) | 0.0627 (13) | −0.0206 (11) | 0.0254 (10) | −0.0215 (12) |
| C13 | 0.0456 (10) | 0.0465 (11) | 0.0488 (10) | −0.0140 (8) | 0.0137 (8) | −0.0136 (8) |
| C14 | 0.0639 (15) | 0.0566 (14) | 0.0739 (16) | 0.0124 (12) | 0.0176 (13) | −0.0040 (12) |
| C15 | 0.0335 (9) | 0.0392 (10) | 0.0470 (10) | 0.0002 (7) | 0.0080 (8) | −0.0031 (8) |
| C16 | 0.0428 (11) | 0.0542 (12) | 0.0610 (12) | 0.0002 (9) | 0.0085 (9) | 0.0113 (10) |
| C17 | 0.0464 (12) | 0.0712 (14) | 0.0597 (13) | 0.0073 (10) | −0.0009 (10) | 0.0060 (11) |
| C18 | 0.0394 (11) | 0.0678 (14) | 0.0626 (13) | −0.0017 (9) | 0.0039 (10) | −0.0080 (11) |
| C19 | 0.0461 (12) | 0.0682 (14) | 0.0757 (15) | −0.0163 (10) | 0.0125 (11) | 0.0062 (12) |
| C20 | 0.0420 (11) | 0.0583 (12) | 0.0592 (12) | −0.0062 (9) | 0.0052 (9) | 0.0133 (10) |
| C21 | 0.0464 (14) | 0.108 (2) | 0.105 (2) | −0.0113 (14) | −0.0079 (14) | −0.0065 (18) |
| N1 | 0.0352 (8) | 0.0277 (7) | 0.0527 (9) | 0.0005 (6) | 0.0075 (6) | −0.0013 (6) |
| N2 | 0.0519 (9) | 0.0282 (8) | 0.0399 (8) | −0.0047 (6) | 0.0144 (7) | −0.0016 (6) |
| O1 | 0.0477 (7) | 0.0385 (7) | 0.0428 (7) | −0.0021 (5) | 0.0159 (5) | 0.0007 (5) |
| O2 | 0.0335 (6) | 0.0342 (6) | 0.0627 (8) | −0.0016 (5) | 0.0025 (6) | 0.0045 (6) |
| P1 | 0.0341 (2) | 0.0287 (2) | 0.0391 (3) | −0.00105 (17) | 0.00834 (18) | −0.00115 (17) |
Geometric parameters (Å, °)
| C1—C2 | 1.383 (3) | C13—C14 | 1.494 (3) |
| C1—C6 | 1.399 (2) | C14—H14C | 0.93 (3) |
| C1—N1 | 1.425 (2) | C14—H14B | 0.97 (3) |
| C2—C3 | 1.381 (3) | C14—H14A | 1.02 (3) |
| C2—H2 | 0.93 | C15—C20 | 1.369 (3) |
| C3—C4 | 1.372 (3) | C15—C16 | 1.377 (3) |
| C3—H3 | 0.93 | C15—O2 | 1.400 (2) |
| C4—C5 | 1.377 (3) | C16—C17 | 1.386 (3) |
| C4—H4 | 0.93 | C16—H16 | 0.93 |
| C5—C6 | 1.388 (3) | C17—C18 | 1.374 (3) |
| C5—H5 | 0.93 | C17—H17 | 0.93 |
| C6—C7 | 1.500 (3) | C18—C19 | 1.377 (3) |
| C7—H7A | 0.96 | C18—C21 | 1.513 (3) |
| C7—H7B | 0.96 | C19—C20 | 1.382 (3) |
| C7—H7C | 0.96 | C19—H19 | 0.93 |
| C8—C9 | 1.382 (3) | C20—H20 | 0.93 |
| C8—C13 | 1.393 (3) | C21—H21A | 0.96 |
| C8—N2 | 1.432 (2) | C21—H21B | 0.96 |
| C9—C10 | 1.388 (3) | C21—H21C | 0.96 |
| C9—H9 | 0.93 | N1—P1 | 1.6268 (15) |
| C10—C11 | 1.369 (4) | N1—H1 | 0.911 (15) |
| C10—H10 | 0.93 | N2—P1 | 1.6279 (15) |
| C11—C12 | 1.375 (3) | N2—H21 | 0.899 (15) |
| C11—H11 | 0.93 | O1—P1 | 1.4692 (12) |
| C12—C13 | 1.390 (3) | O2—P1 | 1.5964 (13) |
| C12—H12 | 0.93 | ||
| C2—C1—C6 | 120.19 (16) | C13—C14—H14B | 111.0 (19) |
| C2—C1—N1 | 120.32 (15) | H14C—C14—H14B | 105 (3) |
| C6—C1—N1 | 119.48 (16) | C13—C14—H14A | 114.9 (17) |
| C3—C2—C1 | 120.67 (18) | H14C—C14—H14A | 106 (2) |
| C3—C2—H2 | 119.7 | H14B—C14—H14A | 107 (2) |
| C1—C2—H2 | 119.7 | C20—C15—C16 | 120.46 (17) |
| C4—C3—C2 | 119.9 (2) | C20—C15—O2 | 123.22 (17) |
| C4—C3—H3 | 120.1 | C16—C15—O2 | 116.30 (16) |
| C2—C3—H3 | 120.1 | C15—C16—C17 | 119.01 (19) |
| C3—C4—C5 | 119.4 (2) | C15—C16—H16 | 120.5 |
| C3—C4—H4 | 120.3 | C17—C16—H16 | 120.5 |
| C5—C4—H4 | 120.3 | C18—C17—C16 | 121.9 (2) |
| C4—C5—C6 | 122.25 (19) | C18—C17—H17 | 119 |
| C4—C5—H5 | 118.9 | C16—C17—H17 | 119 |
| C6—C5—H5 | 118.9 | C17—C18—C19 | 117.39 (19) |
| C5—C6—C1 | 117.52 (18) | C17—C18—C21 | 121.3 (2) |
| C5—C6—C7 | 121.31 (18) | C19—C18—C21 | 121.3 (2) |
| C1—C6—C7 | 121.15 (17) | C18—C19—C20 | 122.0 (2) |
| C6—C7—H7A | 109.5 | C18—C19—H19 | 119 |
| C6—C7—H7B | 109.5 | C20—C19—H19 | 119 |
| H7A—C7—H7B | 109.5 | C15—C20—C19 | 119.2 (2) |
| C6—C7—H7C | 109.5 | C15—C20—H20 | 120.4 |
| H7A—C7—H7C | 109.5 | C19—C20—H20 | 120.4 |
| H7B—C7—H7C | 109.5 | C18—C21—H21A | 109.5 |
| C9—C8—C13 | 120.81 (18) | C18—C21—H21B | 109.5 |
| C9—C8—N2 | 120.30 (17) | H21A—C21—H21B | 109.5 |
| C13—C8—N2 | 118.87 (16) | C18—C21—H21C | 109.5 |
| C8—C9—C10 | 120.1 (2) | H21A—C21—H21C | 109.5 |
| C8—C9—H9 | 119.9 | H21B—C21—H21C | 109.5 |
| C10—C9—H9 | 119.9 | C1—N1—P1 | 123.77 (12) |
| C11—C10—C9 | 119.7 (2) | C1—N1—H1 | 120.6 (13) |
| C11—C10—H10 | 120.1 | P1—N1—H1 | 115.4 (13) |
| C9—C10—H10 | 120.1 | C8—N2—P1 | 127.71 (12) |
| C10—C11—C12 | 119.9 (2) | C8—N2—H21 | 113.0 (14) |
| C10—C11—H11 | 120.1 | P1—N2—H21 | 119.2 (14) |
| C12—C11—H11 | 120.1 | C15—O2—P1 | 124.67 (11) |
| C11—C12—C13 | 121.9 (2) | O1—P1—O2 | 113.30 (7) |
| C11—C12—H12 | 119 | O1—P1—N1 | 118.95 (8) |
| C13—C12—H12 | 119 | O2—P1—N1 | 96.87 (7) |
| C12—C13—C8 | 117.46 (19) | O1—P1—N2 | 109.57 (7) |
| C12—C13—C14 | 120.5 (2) | O2—P1—N2 | 110.16 (8) |
| C8—C13—C14 | 122.06 (18) | N1—P1—N2 | 107.23 (7) |
| C13—C14—H14C | 112.0 (18) | ||
| C6—C1—C2—C3 | −1.1 (3) | C15—C16—C17—C18 | −0.1 (3) |
| N1—C1—C2—C3 | 179.94 (17) | C16—C17—C18—C19 | 0.6 (3) |
| C1—C2—C3—C4 | −0.7 (3) | C16—C17—C18—C21 | −178.7 (2) |
| C2—C3—C4—C5 | 1.3 (4) | C17—C18—C19—C20 | −0.8 (3) |
| C3—C4—C5—C6 | −0.2 (4) | C21—C18—C19—C20 | 178.5 (2) |
| C4—C5—C6—C1 | −1.5 (3) | C16—C15—C20—C19 | 0.0 (3) |
| C4—C5—C6—C7 | 176.6 (2) | O2—C15—C20—C19 | −178.57 (19) |
| C2—C1—C6—C5 | 2.1 (3) | C18—C19—C20—C15 | 0.5 (3) |
| N1—C1—C6—C5 | −178.90 (17) | C2—C1—N1—P1 | 39.1 (2) |
| C2—C1—C6—C7 | −175.95 (18) | C6—C1—N1—P1 | −139.85 (15) |
| N1—C1—C6—C7 | 3.0 (3) | C9—C8—N2—P1 | 45.5 (2) |
| C13—C8—C9—C10 | −0.6 (3) | C13—C8—N2—P1 | −135.66 (15) |
| N2—C8—C9—C10 | 178.22 (17) | C20—C15—O2—P1 | −33.5 (2) |
| C8—C9—C10—C11 | −1.8 (3) | C16—C15—O2—P1 | 147.94 (14) |
| C9—C10—C11—C12 | 2.0 (3) | C15—O2—P1—O1 | 62.76 (15) |
| C10—C11—C12—C13 | 0.2 (3) | C15—O2—P1—N1 | −171.60 (13) |
| C11—C12—C13—C8 | −2.4 (3) | C15—O2—P1—N2 | −60.38 (15) |
| C11—C12—C13—C14 | 177.9 (2) | C1—N1—P1—O1 | −75.73 (15) |
| C9—C8—C13—C12 | 2.6 (3) | C1—N1—P1—O2 | 162.81 (14) |
| N2—C8—C13—C12 | −176.18 (16) | C1—N1—P1—N2 | 49.18 (16) |
| C9—C8—C13—C14 | −177.7 (2) | C8—N2—P1—O1 | 169.91 (14) |
| N2—C8—C13—C14 | 3.5 (3) | C8—N2—P1—O2 | −64.80 (17) |
| C20—C15—C16—C17 | −0.2 (3) | C8—N2—P1—N1 | 39.51 (17) |
| O2—C15—C16—C17 | 178.44 (18) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.91 (2) | 2.02 (2) | 2.8963 (19) | 161 (2) |
Symmetry codes: (i) −x+1, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG2779).
References
- Blessing, R. H. (1995). Acta Cryst. A51, 33–38. [DOI] [PubMed]
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Nonius (2001). COLLECT. Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Pourayoubi, M., Ghadimi, S., Ebrahimi Valmoozi, A. A. & Banan, A. R. (2009). Acta Cryst. E65, o1973. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810023512/ng2779sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810023512/ng2779Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report