Abstract
Biallelic inactivation of the von Hippel–Lindau tumor suppressor gene (VHL) is linked to the development of hereditary (VHL-associated) and sporadic clear-cell renal carcinomas as well as other abnormalities. The VHL gene product, pVHL, is part of an E3 ubiquitin ligase complex that targets the α subunits of the heterodimeric transcription factor HIF (hypoxia-inducible factor) for degradation in the presence of oxygen. Here we report that a HIF2α variant lacking both of its two prolyl hydroxylation/pVHL-binding sites prevents tumor inhibition by pVHL in a DNA-binding dependent manner. Conversely, downregulation of HIF2α with short hairpin RNAs is sufficient to suppress tumor formation by pVHL-defective renal carcinoma cells. These results establish that tumor suppression by pVHL is linked to regulation of HIF target genes.
Specific downregulation of the transcription factor HIF2α is sufficient to suppress tumor formation by cells lacking the functional tumor suppressor (pVHL), demonstrating that tumor suppression by pVHL is linked to regulation of HIF target genes
Introduction
von Hippel–Lindau (VHL) disease is caused by heterozygous germline inactivation of the VHL tumor suppressor gene, which resides on chromosome 3p25 (Kaelin 2002). The cardinal feature of this hereditary cancer syndrome is the development of multiple vascular tumors, called hemangioblastomas, in the central nervous system and retina, as well as an increased risk of clear-cell carcinoma of the kidney and pheochromocytoma. Tumor development in VHL disease is linked to somatic inactivation or loss of the remaining wild-type VHL allele, leading to loss of the wild-type VHL gene product, pVHL. In the kidney, this event occurs very early, as it has been documented in epithelial cells lining premalignant renal cysts (Zhuang et al. 1995; Lubensky et al. 1996; Mandriota et al. 2002). Consistent with Knudson's two-hit model, somatic VHL mutations are also common in sporadic clear-cell renal carcinomas and hemangioblastomas. Conversely, restoration of pVHL function is sufficient to suppress tumor formation by pVHL-defective renal carcinoma cells in vivo (Iliopoulos et al. 1995; Gnarra et al. 1996; Schoenfeld et al. 1998).
pVHL is the substrate recognition module of an E3 ubiquitin ligase complex that contains elongin B, elongin C, Cul2, and Rbx1 (also called ROC1 or Hrt1) (Kaelin 2002). This complex targets the α subunits of the heterodimeric transcription factor HIF (hypoxia-inducible factor) for polyubiquitination and hence proteasomal degradation. There are three human HIFαproteins (HIF1α, HIF2α, and HIF3α). Enzymatic hydroxylation of conserved prolyl residues within these proteins by members of the egg-laying-defective nine (EGLN) family is required for their recognition by pVHL (Kaelin 2002). This posttranslational modification is inherently oxygen-dependent. Accordingly, HIFα subunits are normally unstable in the presence of oxygen, but are stabilized under low-oxygen (hypoxic) conditions. In contrast, cells lacking wild-type pVHL fail to degrade HIFα subunits in the presence of oxygen, and thus hypoxia-inducible gene products are constitutively overproduced. Among these proteins are vascular endothelial growth factor (VEGF) and platelet-derived growth factor B, implicated in angiogenesis; phosphoglycerate kinase and glucose transporter 1 (GLUT1), involved in glucose uptake and metabolism; and transforming growth factor α (TGFα), which can establish a mitogenic autocrine loop with the epidermal growth factor (EGF) receptor (EGFR) (Iliopoulos et al. 1996; Knebelmann et al. 1998; Maxwell et al. 1999; de Paulsen et al. 2001).
Tumor-derived pVHL mutants are typically defective with respect to HIF polyubiquitination in vivo, and the HIF target genes cited above are implicated in tumorigenesis. Thus, correlative data and biological plausibility support a role for HIF in pVHL-defective tumor formation. Nonetheless, emerging genotype–phenotype correlations in VHL disease suggest that pVHL has multiple functions. For example, pVHL mutants associated with a low risk (type 2A VHL disease) and high risk (type 2B disease) of renal cell carcinoma are similarly defective with respect to HIF regulation (Clifford et al. 2001; Hoffman et al. 2001). Interestingly, individuals with Chuvash polycythemia are homozygous for a hypomorphic VHL allele that is quantitatively defective with respect to HIF regulation, which leads to overproduction of erythropoietin in vivo but not tumor formation (Ang et al. 2002). Moreover, forced activation of HIF target genes has not led to tumor formation in the animal models tested so far (Vincent et al. 2000; Elson et al. 2001; Rebar et al. 2002). Conversely, some pVHL mutants that retain the ability to regulate HIF are linked to familial pheochromocytoma (type 2C VHL disease) (Clifford et al. 2001; Hoffman et al. 2001). Collectively, these findings suggest that tumor formation following pVHL inactivation reflects the loss of multiple pVHL functions in a context-dependent manner.
In this report we provide data that strengthen our earlier conclusion that inhibition of HIF2α is necessary for pVHL-dependent suppression of renal carcinoma tumor formation in vivo (Kondo et al. 2002). Moreover, we provide evidence that inhibition of HIF2α is likewise sufficient to suppress tumor formation by VHL(−/−) renal carcinoma cells in vivo. Collectively, these results indicate that HIF2α is a critical downstream target of pVHL with respect to suppression of renal carcinogenesis.
Results and Discussion
Inhibition of HIF2α Target Genes Is Necessary for Tumor Suppression by pVHL
Hydroxylation of HIF1α Pro564 or HIF2α Pro531 generates a pVHL-binding site (Ivan et al. 2001; Jaakkola et al. 2001; Yu et al. 2001). We previously showed that a HIF2α variant in which Pro531 was replaced by alanine (HIF2α P531A) escaped recognition by pVHL and induced the expression of HIF target genes in vivo (Kondo et al. 2002). Moreover, HIF2α P531A abrogated pVHL-dependent tumor suppression in vivo, implying that HIF is functionally downstream of pVHL and that inhibition of HIF is necessary for tumor suppression by pVHL (Kondo et al. 2002). Shortly thereafter, it was shown that hydroxylation of HIF1α Pro404 (corresponding to HIF2α Pro405) creates a second potential pVHL-binding site within HIF1α (Masson et al. 2001). Although we could not detect a physical interaction between pVHL and HIF2α P531A (Kondo et al. 2002), the identification of a second potential pVHL-binding site left open the possibility that the biological effects of HIF2α P531A were due, at least partly, to perturbation of pVHL function as a result of direct binding. If true, this would undermine the conclusions described above. Moreover, we had not established whether the biological effects of HIF2α P531A required that it bind to DNA, as would be expected if its oncogenic effects were due to transcriptional activation of specific hypoxia-inducible promoters. To this end, we repeated our earlier experiments using retroviral vectors encoding HIF2α P405A;P531A or HIF2α P405A;P531A;bHLH*. The latter contains a five amino acid substitution within the HIF2α basic helix–loop–helix (bHLH) domain that leads to loss of DNA-binding capability (Kondo et al. 2002).
786-O renal carcinoma cells lack wild-type pVHL and overproduce HIF2α (Iliopoulos et al. 1995; Maxwell et al. 1999). HIF1α is not detectable in these cells (Maxwell et al. 1999). Reintroduction of wild-type pVHL into 786-O cells by stable transfection does not affect cell growth in vitro under standard serum-rich growth conditions, but leads to downregulation of HIF2α protein levels, suppression of hypoxia-inducible gene expression, and impaired tumorigenesis in vivo (Iliopoulos et al. 1995, 1996; Gnarra et al. 1996; Schoenfeld et al. 1998; Maxwell et al. 1999; Davidowitz et al. 2001). A 786-O subclone stably transfected to produce wild-type pVHL (WT8) (Iliopoulos et al. 1995) was infected with a retrovirus encoding HIF2α P405A;P531A or HIF2α P405A;P531A;bHLH* and grown under hypoxic (1% oxygen) or normoxic (21% oxygen) conditions. As expected, HIF2α was only detectable under hypoxic conditions in WT8 cells and in WT8 cells infected with an empty retrovirus (Figure 1A). In contrast, HIF2α was readily detectable under both hypoxic and normoxic conditions in WT8 cells infected to produce either of the two HIF2α P405A;P531A variants. Indeed, the levels of HIF2α present in these cells approximated those seen in a 786-O subclone (PRC3) (Iliopoulos et al. 1995) that, unlike WT8 cells, was stably transfected with an empty expression plasmid and hence still lacks wild-type pVHL.
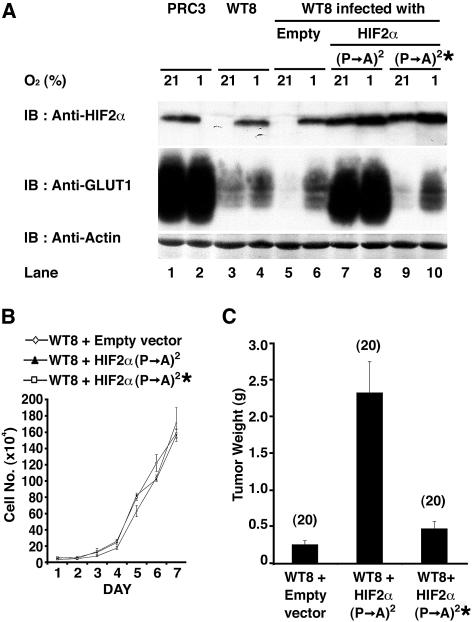
Figure 1. HIF2α Overrides Tumor Suppression by pVHL.
(A) 786-O subclones that were transfected to produce wild-type pVHL (WT8) or with an empty plasmid (PRC3) cells, as well as WT8 cells infected with an empty retrovirus (Empty) or retroviruses encoding the indicated HIF2α variants [ (P→A)2 = P405A;P531A and * = bHLH mutation] were grown in the presence of 21% or 1% oxygen and immunoblotted (IB) with the indicated antibodies.
(B) In vitro proliferation of WT8 cells infected with the indicated retroviruses.
(C) Tumor weights approximately 9 wk after subcutaneous implantation of WT8 cells infected with the indicated retroviruses in nude mice. Number of tumors analyzed is shown in parentheses. Error bars = one standard error.
Neither HIF2α P405A;P531A nor HIF2α P405A;P531A;bHLH* affected the proliferation of WT8 cells in vitro under standard cell culture conditions (Figure 1B). In contrast, but in keeping with our earlier results with HIF2α P531A (Kondo et al. 2002), HIF2α P405A;P531A restored the ability of WT8 cells to form large tumors in vivo in nude mouse xenograft assays (Figure 1C). HIF2α P405A;P531A;bHLH* did not promote tumor formation by WT8 cells, implying that tumor promotion by HIF2α P405A;P531A is linked to its ability to act as a sequence-specific DNA-binding transcriptional regulator. These results, together with our earlier findings, indicate that inhibition of HIF2α is necessary for tumor suppression by pVHL.
Loss of HIF2α Is Sufficient to Suppress pVHL-Defective Tumor Growth In Vivo
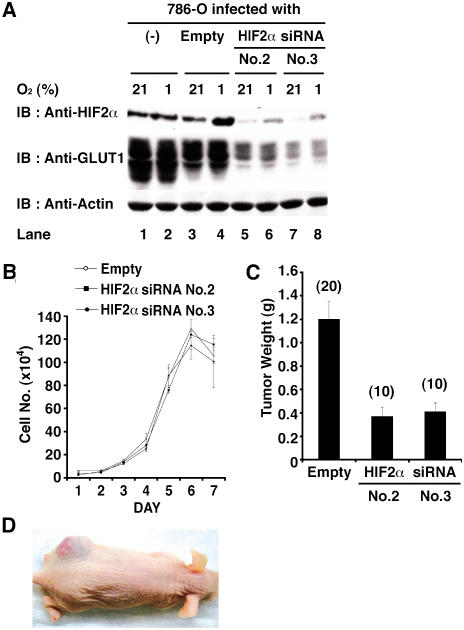
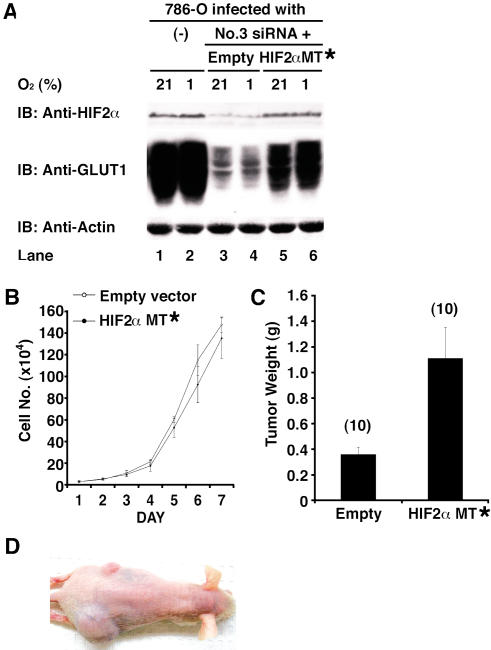
To ask whether inhibition of HIF2α is likewise sufficient for tumor suppression by pVHL, we set out to inhibit HIF2α in VHL(−/−) renal carcinoma cells using short hairpin RNAs (shRNA). We tested five HIF2α shRNAs based on 19mer sequences that are unique to HIF2α according to GenBank. Two such shRNAs (#2 and #3) decreased HIF2α protein levels, as determined by anti-HIF2α immunoblot analysis and by diminished activity of a cotransfected HRE–luciferase reporter plasmid, when transiently introduced into 786-O cells (data not shown). Infection of 786-O cells with retroviruses encoding shRNA #2 or #3, but not the parental retrovirus, led to decreased steady-state levels of HIF2α protein as well as decreased levels of GLUT1, which is encoded by a HIF-responsive gene (Figure 2A). Downregulation of HIF2α did not affect cell growth in vitro, but was sufficient to impair tumor growth in vivo (Figure 2B–2D). The former observation is consistent with the finding that pVHL does not inhibit cell proliferation under standard cell culture conditions and argues against the idea that the latter was due to nonspecific toxicity. Moreover, these in vivo effects could be prevented by coadministration of a retrovirus encoding an HIF2α mRNA with silent third-base mutations within the shRNA recognition site (Figure 3) and were not observed with retroviruses encoding a scrambled HIF2α shRNA or luciferase shRNA (data not shown). Thus, tumor suppression by the HIF2α shRNA was unlikely to reflect a spurious interaction with an unintended target.
Figure 2. Downregulation of HIF2α Is Sufficient to Suppress Tumor Growth by pVHL-Defective Renal Carcinoma Cells.
(A) Parental 786-O cells (VHL[−/−]) and 786-O cells infected with an empty retrovirus (Empty) or retroviruses encoding HIF2α shRNAs (sequence #2 or #3) were grown in the presence of 21% or 1% oxygen and immunoblotted (IB) with the indicated antibodies.
(B) In vitro proliferation of 786-O cells infected with the indicated retroviruses.
(C) Tumor weights approximately 9 wk after subcutaneous implantation of 786-O cells infected with the indicated retroviruses in nude mice. Number of tumors analyzed is shown in parentheses. Error bars = one standard error.
(D) Representative photograph of nude mouse 9 wk after subcutaneous injection of 786-O cells in left (upper) flank and 786-O cells infected with HIF2α shRNA (#3) retrovirus on right (lower) flank.
Figure 3. Effect of HIF2α shRNA Is Specifically Due to Downregulation of HIF2α.
(A) Parental 786-O cells (VHL[−/−]) and 786-O cells stably producing HIF2α shRNA #3 that were coinfected with an empty retrovirus (Empty) or a retrovirus encoding a HIF2α mRNA with three silent mutations in the #3 recognition site (MT*) were grown in the presence of 21% or 1% oxygen and immunoblotted (IB) with the indicated antibodies.
(B) In vitro proliferation of 786-O HIF2α shRNA #3 cells infected with the indicated retroviruses.
(C) Tumor weights approximately 9 wk after subcutaneous implantation of 786-O HIF2α shRNA cells infected with the indicated retroviruses in nude mice. Number of tumors analyzed is shown in parentheses. Error bars = one standard error.
(D) Representative photograph of nude mouse 9 wk after subcutaneous injection of 786-O HIF2α shRNA #3 cells in left (upper) flank and 786-O HIF2α shRNA #3 cells infected with retrovirus encoding HIF2α MT* mRNA on right (lower) flank.
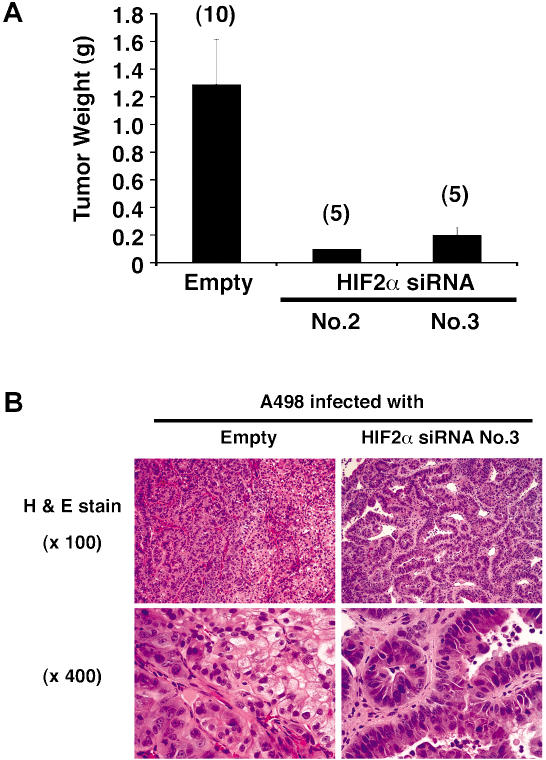
To ask whether these findings could be extended to other VHL(−/−) renal carcinoma cell lines, we repeated these experiments in A498 VHL(−/−) renal carcinoma cells. Tumor formation by these cells in nude mice is diminished following restoration of pVHL function (Lonergan et al. 1998). In keeping with the results obtained with 786-O cells, downmodulation of HIF2α levels with shRNA did not affect A498 cell growth in vitro (data not shown), but dramatically inhibited tumor growth in vivo (Figure 4A). It is noteworthy that both 786-O cells and A498 cells produce HIF2α and not HIF1α (Maxwell et al. 1999). It will be important in the future to ask whether disruption of HIF2α is sufficient to suppress tumor formation by pVHL-defective cells that produce both HIFα paralogs. In this regard, studies of renal precursor lesions in VHL patients suggest that HIF2α is more oncogenic than HIF1α (Mandriota et al. 2002). It is tempting to speculate that loss of HIF1α expression in some pVHL-defective renal carcinoma cells confers a selective advantage in vivo, perhaps related to the ability of HIF1α to induce apoptosis in some settings (Carmeliet et al. 1998).
Figure 4. Tumor Suppression by HIF2α shRNA Is Not Restricted to a Single Cell Line.
(A) Tumor weights approximately 8 wk after subcutaneous implantation of A498 cells infected with the indicated retroviruses in nude mice. Number of tumors analyzed is shown in parentheses. Error bars = one standard error.
(B) Representative histological sections after staining with hematoxylin and eosin of tumors formed by A498 cells infected with the indicated retroviruses.
Several histological renal carcinoma variants have been recognized, including clear-cell carcinoma and papillary (chromophil) carcinoma. VHL mutations are common in the former, but not in the latter (Gnarra et al. 1994; Takahashi et al. 2002). Interestingly, the small A498 tumors that did form in the presence of HIF2α shRNA consisted of malignant cells forming tubulopapillary structures, corresponding to papillary (chromophil) renal carcinoma histology, whereas the empty vector tumors consisted primarily of sheets of clear cells, as would be seen in typical clear-cell renal carcinoma, with interspersed areas displaying papillary features (Figure 4B). This suggests that dysregulation of HIF2α is causally linked to the clear-cell pattern and is consistent with the tight linkage between VHL mutations and this renal carcinoma subtype.
Most of the work performed so far with respect to the oncogenic effects of HIF has focused exclusively on HIF1α, where both prooncogenic and antioncogenic effects have been reported (Maxwell et al. 1997; Carmeliet et al. 1998; Ryan et al. 1998, 2000; Hopfl et al. 2002). Likewise, loss of pVHL is prooncogenic in a restricted subset of human tissues (Kaelin 2002). In the mouse, loss of pVHL promotes hemangioma development in the liver, but inhibits tumor formation by embryonic stem cells (Haase et al. 2001; Mack et al. 2003). These observations conform to the emerging paradigm that the same mutation can be either prooncogenic or antioncogenic, depending on the molecular and cellular context. Therefore, one must be cautious in extrapolating our findings beyond human clear-cell renal carcinomas.
Loss of pVHL in the human kidney gives rise to premaligant renal cysts (Zhuang et al. 1995; Lubensky et al. 1996; Mandriota et al. 2002). It is presumed that additional mutations at non-VHL loci are required for conversion to frank renal cell carcinomas. It will therefore be of interest to determine whether dysregulation of HIF is sufficient to produce renal cysts. In this regard, TGFα, which is encoded by a HIF target gene, is a potent renal mitogen and is sufficient to induce renal cysts in the mouse (Lowden et al. 1994; Chailler and Briere 1998; Ramp et al. 2000; de Paulsen et al. 2001). On the other hand, our data do not exclude the possibility that the development of renal pathology following pVHL loss in humans reflects a complex interplay between dysregulated HIF2α and loss of a second pVHL function. pVHL has been implicated in control of cell-cycle, differentiation, and extracellular matrix formation, although the extent to which these activities are due to control of HIF is not known (Kaelin 2002). A number of non-HIF pVHL-binding partners have, however, been reported, including atypical protein kinase C members, VDU1, SP1, and fibronectin (Kaelin 2002).
Therapeutic Implications
Our findings strengthen the notion that inhibition of HIF2α might be therapeutically useful in pVHL-defective clear-cell renal carcinoma. On the other hand, sequence-specific DNA-binding transcription factors have not proven to be attractive drug targets to date. For this reason, it will be important to determine which HIF2α target genes are necessary for its oncogenic activity. Among the known HIF targets, the abovementioned TGFα and its cognate receptor, EGFR, are frequently overproduced in renal carcinoma and are suspected to establish an autocrine loop (Mydlo et al. 1989; Lager et al. 1994; Knebelmann et al. 1998; de Paulsen et al. 2001). A number of EGFR are presently in clinical trials (Fabbro et al. 2002). Likewise, overproduction of VEGF is common in renal cell carcinoma and likely contributes to tumor angiogenesis in this setting (Walke et al. 1991; Brown et al. 1993; Takahashi et al. 1994; Nicol et al. 1997; Ramp et al. 1997). Drugs directed against VEGF or its receptors are also being tested in humans (Fabbro et al. 2002). In a recent Phase II study, a neutralizing VEGF antibody was shown to delay disease progression in metastatic renal carcinoma (Yang et al. 2003) and offers hope that rational combinations of small molecules directed against HIF targets will alter the natural history of this disease.
Materials and Methods
Plasmids
pBABE-puro-HA-HIF2α P405A;P531A was generated by two-step PCR. The pcDNA3.0-HA-HIF2α P531A (Kondo et al. 2002) insert was first amplified with primer A (5′-GCGCGGATCCGCCACCATGACA-3′) and primer B (5′-TCCTGGGGTAGCAGCCAGCTG-3′) or primer C (5′-CAGCTGGCTGCTACCCCAGGA-3′) and primer D (5′-GCGCCAATTGTCAGGTGGCCTGGTC-3′). Aliquots of these two PCRs were then mixed and amplified with primers A and D. The resulting PCR product was digested with BamHI and MunI and ligated into pBABE-puro-HA vector cut with BamHI and EcoRI. In parallel, similar reactions were carried out with pcDNA3.0-HA-HIF2α P531A/bHLH* (Kondo et al. 2002) as the PCR template to make pBABE-puro HA-HIF2α P405A;P531A;bHLH* (conversion of amino acids residues 24–29, RCRRSK to ACAASA).
Short interfering RNAs (siRNAs) corresponding to two unique HIF2α 19mer sequences (#2, 5′-GACAAGGTCTGCAAAGGGT-3′ and #3, 5′-GGAGACGGAGGTGTTCTAT-3′) downregulated HIF2α protein levels and HIF-dependent transcriptional activity. Synthetic oligonucleotides spanning the #2 siRNA sequence (5′-GATCCCCGACAAGGTCTGCAAAGGGTTTCAAGAGAACCCTTTGCAGACCTTGTCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGACAAGGTCTGCAAAGGGTTCTCTTGAAACCCTTTGCAGACCTTGTCGGG-3′) or the #3 sequence (5′-GATCCCCGGAGACGGAGGTGTTCTATTTCAAGAGAATAGAACACCTCCGTCTCCTTTTTGGAAA and 5′-AGCTTTTCCAAAAAGGAGACGGAGGTGTTCTATTCTCTTGAAATAGAACACCTCCGTCTCCGGG-3′) were annealed by incubation in 30 mM HEPES–KOH (pH 7.4), 100 mM potassium acetate, 30 mM HEPES–KOH, 2 mM Mg–acetate for 4 min at 95°C followed by 10 min at 70°C. The resulting duplex oligonucleotides were phosphorylated with T4 polynucleotide kinase (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's protocol and ligated into pRETRO-SUPER vector (Brummelkamp et al. 2002) cut with BglII and HindIII.
The pBABE-hygro-HA-HIF2α siRNA recognition site mutant for #3 siRNA (5′-GGAGACCGAAGTCTTCTAT-3′) (called HIF2α MT) was generated by two-step PCR. The pcDNA3.0-HA-HIF2α insert was first amplified was first amplified with primer A and primer E (5′-GTCTCCTTGCTCCGCCG-3′) or primer F (5′-CGAAGTCTTCTATGAGCTGGCCCATG-3′) and primer D. Aliquots of these two PCRs were then mixed and amplified with primers A and D. The resulting PCR product was digested with BamHI and MunI and ligated into pBABE-hygro-HA vector cut with BamHI and EcoRI.
All plasmids were authenticated by DNA sequencing. pGL2-VEGF promoter plasmid was a kind gift from Dr. Deb Mukhopadhyay (Harvard Medical School, Boston, Massachusetts, United States). pSV-β-Gal plasmid was purchased from Promega Corporation (Madison, Wisconsin, United States).
Cell culture
Renal carcinoma cell lines (786-O and A498) and Phoenix cells (a generous gift of Dr. Gary Nolan, Department of Molecular Pharmacology, Stanford University, Stanford, California, United States) were grown in Dulbecco's modified Eagle's medium containing 10% fetal clone I (Hyclone, Logan Utah, United States) in presence of 10% CO2 at 37°C. 786-O renal cell carcinoma subclones stably transfected with either pRc/CMV empty vector (PRC3) or pRc/CMV-HA-VHL (WT8) (Iliopoulos et al. 1995) were grown in the same media supplemented with 1 mg/ml G418. Retrovirally infected cells were selected and maintained in the presence of puromycin (1.5 μg/ml for pBABE-puro retroviruses or 1.0 μg/ml for pSUPER retroviruses) or hygromycin 0.5 μg/ml for pBABE-hygro retrovirus.
Retroviruses.
Retroviral plasmids were transfected into the Phoenix packaging cell line using FuGene (Roche Molecular Biochemicals) according to the manufacturer's instructions. Tissue culture supernatant was harvested 48 h later, passed though a 0.45-μm filter, and added to cells in the presence of 4 μg/ml polybrene.
Immunoblot analysis
Cells were lysed in EBC lysis buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40) supplemented with complete protease inhibitor cocktail (Roche Molecular Biochemicals). Approximately 300 μg of cell extract per lane, as determined by the Bradford method, was resolved by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, California, United States). After blocking in Tris-buffered saline (TBS) with 4% nonfat milk, the membranes were probed with anti-HA rabbit polyclonal antibody (Y-11; Santa Cruz Biotechnology, Santa Cruz, California, United States), anti-HIF2α mouse monoclonal antibody (NB100–132; Novus Biologicals, Littleton, Colorado, United States), anti-GLUT1 rabbit polyclonal antibody (GT11-A; Alpha Diagnostic, San Antonio, Texas, United States), or anti-actin goat polyclonal antibody (sc-1615; Santa Cruz Biotechnology) diluted in TBS with 4% bovine serum albumin. Bound antibody was detected with horseradish peroxidase-conjugated goat anti-rabbit IgG, goat anti-mouse IgG, or rabbit anti-goat IgG (Pierce, Rockford, Illinois, United States) and SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's instructions.
In vitro proliferation assays
Approximately 5,000 cells were plated per well in 6-well plates and grown under the cell culture conditions described above. At various timepoints thereafter, cells were released by trypsinization, resuspended in phosphate-buffered saline (PBS), and stained with trypan blue. Viable cells, as determined by trypan blue exclusion, were counted using a hemocytometer.
Nude mouse xenograft assays
Nude mouse xenograft assays were performed as described elsewhere (Iliopoulos et al. 1995; Kondo et al. 2002). In brief, cells were released by trypsinization and resuspended in PBS. Viable cells (107), as determined by trypan blue staining, were injected subcutaneously into the flanks of nude mice. Both flanks were used for each mouse. The animals were sacrificed 8–10 wk after injection. Autopsy was performed by animal care technicians who were unaware of the HIF status of the tumors. Tumors were weighed, cut in half, and fixed in either formalin or frozen in optimal cutting temperature compound.
Acknowledgments
We thank members of the Kaelin Laboratory for useful discussions and Rene Bernards for generously providing the pSUPER-retro plasmid prior to publication. WGK is a Howard Hughes Medical Institute investigator. This work was sponsored by the National Cancer Institute and the Murray Foundation.
Abbreviations
- bHLH
basic helix–loop–helix
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EGLN
egg-laying-defective nine
- GLUT1
glucose transporter 1
- HIF
hypoxia-inducible factor
- PBS
phosphate-buffered saline
- pVHL
VHL gene product
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- TBS
Tris-buffered saline
- TGF
transforming growth factor
- VEGF
vascular endothelial growth factor
- VHL
von Hippel–Lindau
Conflicts of interest. The authors have declared that no conflicts of interest exist.
Author contributions. KK and WGK conceived and designed the experiments. KK, WYK, and ML performed the experiments. KK, ML, and WGK analyzed the data. KK contributed reagents/materials/analysis tools. WYK and WGK wrote the paper.
Academic Editor: Christopher Kemp, Fred Hutchinson Cancer Research Center
References
- Ang SO, Chen H, Hirota K, Gordeuk VR, Jelinek J, et al. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat Genet. 2002;32:614–621. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- Brown L, Berse B, Jackman R, Tognazzi K, Manseau E, et al. Increased expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in kidney and bladder carcinomas. Am J Pathol. 1993;143:1255–1262. [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Chailler P, Briere N. Mitogenic effects of EGF/TGF alpha and immunolocalization of cognate receptors in human fetal kidneys. Biofactors. 1998;7:323–335. doi: 10.1002/biof.5520070404. [DOI] [PubMed] [Google Scholar]
- Clifford S, Cockman M, Smallwood A, Mole D, Woodward E, et al. Contrasting effects on HIF-1α regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel–Lindau disease. Hum Mol Genet. 2001;10:1029–1038. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- Davidowitz E, Schoenfeld A, Burk R. VHL induces renal cell differentiation and growth arrest through integration of cell–cell and cell–extracellular matrix signaling. Mol Cell Biol. 2001;21:865–874. doi: 10.1128/MCB.21.3.865-874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paulsen N, Brychzy A, Fournier MC, Klausner RD, Gnarra JR, et al. Role of transforming growth factor-alpha in VHL−/− clear cell renal carcinoma cell proliferation: A possible mechanism coupling von Hippel–Lindau tumor suppressor inactivation and tumorigenesis. Proc Natl Acad Sci U S A. 2001;98:1387–1392. doi: 10.1073/pnas.031587498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson D, Thurston G, Huang L, Ginzinger D, McDonald D, et al. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1α. Genes Dev. 2001;15:2520–2532. doi: 10.1101/gad.914801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbro D, Parkinson D, Matter A. Protein tyrosine kinase inhibitors: New treatment modalities? Curr Opin Pharmacol. 2002;2:374–381. doi: 10.1016/s1471-4892(02)00179-0. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Zhou S, Merrill MJ, Wagner J, Krumm A, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the VHL tumor suppressor gene product. Proc Natl Acad Sci U S A. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase V, Glickman J, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel–Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Ohh M, Yang H, Klco J, Ivan M, et al. von Hippel–Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10:1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- Hopfl G, Wenger RH, Ziegler U, Stallmach T, Gardelle O, et al. Rescue of hypoxia-inducible factor-1α-deficient tumor growth by wild-type cells is independent of vascular endothelial growth factor. Cancer Res. 2002;62:2962–2970. [PubMed] [Google Scholar]
- Iliopoulos O, Kibel A, Gray S, Kaelin WG. Tumor suppression by the human von Hippel–Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- Iliopoulos O, Jiang C, Levy AP, Kaelin WG, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel–Lindau protein. Proc Natl Acad Sci U S A. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole D, Tian Y, Wilson M, Gielbert J, et al. Targeting of HIF-alpha to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- Knebelmann B, Ananth S, Cohen H, Sukhatme V. Transforming growth factor alpha is a target for the von Hippel–Lindau tumor suppressor. Cancer Res. 1998;58:226–231. [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG. Inhibition of HIF is necessary for tumor suppression by the von Hippel–Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Lager D, Slagel D, Palechek P. The expression of epidermal growth factor receptor and transforming growth factor alpha in renal cell carcinoma. Mod Pathol. 1994;7:544–548. [PubMed] [Google Scholar]
- Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, et al. Regulation of hypoxia-inducible mRNAs by the von Hippel–Lindau protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden D, Lindemann G, Merlino G, Barash B, Calvet J, et al. Renal cysts in transgenic mice expressing transforming growth factor-alpha. J Lab Clin Med. 1994;124:386–394. [PubMed] [Google Scholar]
- Lubensky IA, Gnarra JR, Bertheau P, Walther MM, Linehan WM, et al. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel–Lindau disease patients. Am J Pathol. 1996;149:2089–2094. [PMC free article] [PubMed] [Google Scholar]
- Mack FA, Rathmell WK, Arsham AM, Gnarra J, Keith B, et al. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell. 2003;3:75–88. doi: 10.1016/s1535-6108(02)00240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, et al. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell P, Pugh C, Ratcliffe P. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P, Dachs G, Gleadle J, Nicholls L, Harris A, et al. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P, Weisner M, Chang G-W, Clifford S, Vaux E, et al. The von Hippel–Lindau gene product is necessary for oxgyen-dependent proteolysis of hypoxia-inducible factor α subunits. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Mydlo J, Michaeli J, Cordon-Cardo C, Goldenberg A, Heston W, et al. Expression of transforming growth factor alpha and epidermal growth factor receptor messenger RNA in neoplastic and non-neoplastic human kidney tissue. Cancer Res. 1989;49:3407–3411. [PubMed] [Google Scholar]
- Nicol D, Hii SI, Walsh M, Teh B, Thompson L, et al. Vascular endothelial growth factor expression is increased in renal cell carcinoma. J Urol. 1997;157:1482–1486. [PubMed] [Google Scholar]
- Ramp U, Jaquet K, Reinecke P, Schardt C, Friebe U, et al. Functional intactness of stimulatory and inhibitory autocrine loops in human renal carcinoma cell lines of the clear cell type. J Urol. 1997;157:2345–2350. [PubMed] [Google Scholar]
- Ramp U, Reinecke P, Gabbert H, Gerharz C. Differential response to transforming growth factor (TGF)-alpha and fibroblast growth factor (FGF) in human renal cell carcinomas of the clear cell and papillary types. Eur J Cancer. 2000;36:932–941. doi: 10.1016/s0959-8049(00)00030-7. [DOI] [PubMed] [Google Scholar]
- Rebar EJ, Huang Y, Hickey R, Nath AK, Meoli D, et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- Ryan H, Lo J, Johnson R. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan H, Poloni M, McNulty W, Elson D, Gassmann M, et al. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Schoenfeld A, Davidowitz E, Burk R. A second major native von Hippel–Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci U S A. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Sasaki H, Kim S, Tobisu K, Kakizoe T, et al. Markedly increased amounts of messenger RNAs for vascular endothelial growth factor and placenta growth factor in renal cell carcinoma associated with angiogenesis. Cancer Res. 1994;54:4233–4237. [PubMed] [Google Scholar]
- Takahashi M, Kahnoski R, Gross D, Nicol D, Teh BT. Familial adult renal neoplasia. J Med Genet. 2002;39:1–5. doi: 10.1136/jmg.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent K, Shyu K, Luo Y, Magner M, Tio R, et al. Angiogenesis is induced in a rabbit model of hindlimb ischemia by naked DNA encoding an HIF-1α/VP16 hybrid transcription factor. Circulation. 2000;102:2255–2261. doi: 10.1161/01.cir.102.18.2255. [DOI] [PubMed] [Google Scholar]
- Walke C, Everitt J, Freed J, Knudson AJ, Whiteley L. Altered expression of transforming growth factor-alpha in hereditary rat renal cell carcinoma. Cancer Res. 1991;51:2973–2978. [PubMed] [Google Scholar]
- Yang JC, Haworth L, Sherry RM, Hwu P, Schwartzentruber DJ, et al. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med. 2003;349:427–434. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, White S, Zhao Q, Lee F. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci U S A. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Bertheau P, Emmert-Buck M, Liotta L, Gnarra J, et al. A microscopic dissection technique for archival DNA analysis of specific cell populations in lesions <1 mm in size. Am J Pathol. 1995;146:620–625. [PMC free article] [PubMed] [Google Scholar]