Abstract
The title compound, C14H11N3O3, adopts an E or trans configuration with respect to the C=N bond. In the molecule there is an intramolecular O—H⋯N hydrogen bond involving the hydroxy substituent at the 2-positon of the naphthalene ring and the adjacent methyleneamino N atom. The molecule is roughly planar, the dihedral angle between the naphthalene and imidazolidine-2,4-dione mean planes being 8.4 (1)°. In the crystal, pairs of N—H⋯O hydrogen bonds link molecules into inversion dimers. These dimers are futher linked via C—H⋯O interactions, forming a three-dimensional network.
Related literature
For the naphthalene group as a fluorophore, see: Li et al. (2010 ▶); Iijima et al. (2010 ▶). For a related structure, see: Xu et al. (2009 ▶). For bond-length data, see: Allen et al. (1987 ▶).
Experimental
Crystal data
C14H11N3O3
M r = 269.26
Monoclinic,
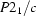
a = 11.5122 (7) Å
b = 6.0233 (3) Å
c = 17.9955 (10) Å
β = 96.773 (5)°
V = 1239.13 (12) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 293 K
0.40 × 0.30 × 0.30 mm
Data collection
Oxford Diffraction Gemini S Ultra diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2009 ▶) T min = 0.959, T max = 0.969
4288 measured reflections
2136 independent reflections
1053 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.067
S = 0.72
2136 reflections
186 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.11 e Å−3
Δρmin = −0.13 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810020118/su2180sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810020118/su2180Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3N⋯O1i | 0.890 (18) | 1.962 (18) | 2.851 (2) | 177.9 (19) |
| O3—H4⋯N1 | 0.82 | 1.91 | 2.622 (2) | 145 |
| C6—H6⋯O2ii | 0.93 | 2.55 | 3.469 (2) | 169 |
| C14—H14B⋯O2ii | 0.97 | 2.36 | 3.038 (2) | 127 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was supported by the Key Project of Science and Technology of Anhui, (grant No. 08010302218), the Natural Science Foundation of Anhui Provincial University (grant No. KJ2009A127) and the National Natural Science Foundation of China (grant No. 20971024).
supplementary crystallographic information
Comment
The naphthalene group as a fluorophore has been studied extensively due to its characteristic photophysical properties and the competitive stability in the environment (Li et al., 2010; Iijima et al., 2010). As part of an ongoing study of Schiff bases incorporating the naphthalene group (Xu et al., 2009), we report here on the crystal structure of the title compound.
The molecular structure of the title compound is shown in Fig. 1. It can be seen to display a trans configuration about the C\bN bond. The bond lengths are normal (Allen et al., 1987). There is an intramolecular O-H···N hydrogen bond (Table 1) and the molecule is relatively planar; dihedral angle involving the naphthalene mean plane and the imidazolidine-2,4-dione group mean plane being 8.4 (1)°. In the crystal structure of the title compound N—H···O intermolecular hydrongen bonds (Table 1) link two molecules to form dimmers situated about an inversion center. The molecules are further linked by weak C-H···O interactions to form a three-dimensional network (Fig. 2).
Experimental
The solution of 1-Aminohydantoin hydrochloride (0.151 g, 1 mmol) in 5 ml of ethanol was added slowly to a solution containing 2-hydro-1- naphthaldehyde (0.172 g, 1 mmol) in 15 ml of absolute ethanol under heating and stirring. The mixture was then refluxed for 2 h. Afterwards the mixture was cooled to rt and the resulting solution to stand in air. After 15 days yellow block-shaped crystals were formed, on slow evaporation of the solvent.
Refinement
The N3 H-atom was located in a difference Fourier map and freely refined: N-H = 0.89 (2) Å. All other H-atoms were placed in calculated positions and treated as riding: O—H = 0.82 Å, C—H = 0.93 or 0.97 Å, with Uiso(H) = 1.5 Ueq(parent O-atom) and = 1.2Ueq(parent C-atom).
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% probability displacement ellipsoids. The intramolecular O-H···N hydrogen bond is shown as a dashed line (see Table 1 for details).
Fig. 2.
Crystal packing viewed along the b-axis of the title compound. The intra- and intermolecular hydrogen bonds are shown as dashed lines (see Table 1 for details).
Crystal data
| C14H11N3O3 | F(000) = 560 |
| Mr = 269.26 | Dx = 1.443 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 1596 reflections |
| a = 11.5122 (7) Å | θ = 3.1–29.0° |
| b = 6.0233 (3) Å | µ = 0.11 mm−1 |
| c = 17.9955 (10) Å | T = 293 K |
| β = 96.773 (5)° | Block, yellow |
| V = 1239.13 (12) Å3 | 0.40 × 0.30 × 0.30 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Gemini S Ultra diffractometer | 2136 independent reflections |
| Radiation source: fine-focus sealed tube | 1053 reflections with I > 2σ(I) |
| graphite | Rint = 0.031 |
| Detector resolution: 15.9149 pixels mm-1 | θmax = 25.0°, θmin = 3.6° |
| ω scans | h = −13→13 |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2009) | k = 0→6 |
| Tmin = 0.959, Tmax = 0.969 | l = 0→21 |
| 4288 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.067 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.72 | w = 1/[σ2(Fo2) + (0.0266P)2] where P = (Fo2 + 2Fc2)/3 |
| 2136 reflections | (Δ/σ)max = 0.002 |
| 186 parameters | Δρmax = 0.11 e Å−3 |
| 0 restraints | Δρmin = −0.13 e Å−3 |
Special details
| Experimental. Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm CrysAlisPro (Oxford Diffraction, 2009). |
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.59888 (11) | 0.25607 (18) | 0.48396 (8) | 0.0491 (5) | |
| O2 | 0.40014 (13) | 0.1526 (2) | 0.68676 (8) | 0.0710 (7) | |
| O3 | 0.80499 (13) | 0.8397 (2) | 0.49453 (8) | 0.0645 (6) | |
| N1 | 0.67397 (13) | 0.6177 (2) | 0.57838 (9) | 0.0420 (6) | |
| N2 | 0.59925 (13) | 0.4517 (2) | 0.59460 (9) | 0.0425 (6) | |
| N3 | 0.48790 (15) | 0.1552 (3) | 0.57835 (10) | 0.0454 (7) | |
| C1 | 0.79653 (19) | 1.2321 (3) | 0.80751 (14) | 0.0665 (10) | |
| C2 | 0.8627 (2) | 1.4184 (4) | 0.79511 (16) | 0.0705 (10) | |
| C3 | 0.90328 (19) | 1.4454 (3) | 0.72772 (16) | 0.0626 (9) | |
| C4 | 0.87808 (17) | 1.2884 (3) | 0.66974 (14) | 0.0504 (9) | |
| C5 | 0.80840 (16) | 1.1006 (3) | 0.68128 (12) | 0.0434 (8) | |
| C6 | 0.77027 (17) | 1.0762 (3) | 0.75298 (13) | 0.0549 (9) | |
| C7 | 0.91986 (18) | 1.3147 (3) | 0.59955 (15) | 0.0606 (10) | |
| C8 | 0.89532 (17) | 1.1653 (4) | 0.54362 (13) | 0.0589 (9) | |
| C9 | 0.82579 (17) | 0.9788 (3) | 0.55419 (13) | 0.0495 (9) | |
| C10 | 0.78023 (16) | 0.9449 (3) | 0.62117 (12) | 0.0413 (8) | |
| C11 | 0.70303 (15) | 0.7611 (3) | 0.63076 (11) | 0.0436 (7) | |
| C12 | 0.56648 (16) | 0.2867 (3) | 0.54520 (12) | 0.0398 (8) | |
| C13 | 0.46509 (18) | 0.2336 (3) | 0.64613 (12) | 0.0503 (9) | |
| C14 | 0.53798 (18) | 0.4406 (3) | 0.66051 (11) | 0.0496 (8) | |
| H1 | 0.76960 | 1.21310 | 0.85380 | 0.0800* | |
| H2 | 0.87910 | 1.52400 | 0.83260 | 0.0850* | |
| H3 | 0.94840 | 1.56920 | 0.71970 | 0.0750* | |
| H3N | 0.4590 (16) | 0.028 (3) | 0.5590 (11) | 0.065 (7)* | |
| H4 | 0.76770 | 0.73180 | 0.50630 | 0.0970* | |
| H6 | 0.72670 | 0.95210 | 0.76300 | 0.0660* | |
| H7 | 0.96540 | 1.43790 | 0.59150 | 0.0730* | |
| H8 | 0.92460 | 1.18630 | 0.49810 | 0.0710* | |
| H11 | 0.67310 | 0.74520 | 0.67630 | 0.0520* | |
| H14A | 0.48920 | 0.57040 | 0.66440 | 0.0600* | |
| H14B | 0.59220 | 0.42690 | 0.70580 | 0.0600* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0538 (10) | 0.0529 (8) | 0.0415 (10) | −0.0044 (7) | 0.0094 (8) | −0.0084 (7) |
| O2 | 0.0901 (13) | 0.0776 (10) | 0.0488 (11) | −0.0257 (9) | 0.0233 (9) | 0.0057 (9) |
| O3 | 0.0725 (12) | 0.0686 (10) | 0.0544 (11) | −0.0094 (8) | 0.0156 (9) | −0.0054 (9) |
| N1 | 0.0401 (10) | 0.0373 (9) | 0.0473 (12) | −0.0016 (8) | 0.0000 (9) | −0.0017 (9) |
| N2 | 0.0467 (11) | 0.0387 (10) | 0.0424 (12) | −0.0071 (9) | 0.0067 (9) | −0.0046 (9) |
| N3 | 0.0522 (12) | 0.0420 (11) | 0.0421 (13) | −0.0091 (9) | 0.0061 (10) | −0.0013 (10) |
| C1 | 0.0649 (16) | 0.0670 (16) | 0.0662 (18) | −0.0015 (14) | 0.0014 (14) | −0.0187 (14) |
| C2 | 0.0648 (18) | 0.0583 (16) | 0.083 (2) | 0.0033 (13) | −0.0137 (16) | −0.0216 (15) |
| C3 | 0.0477 (15) | 0.0391 (13) | 0.095 (2) | −0.0026 (11) | −0.0162 (15) | −0.0068 (15) |
| C4 | 0.0383 (14) | 0.0375 (13) | 0.0718 (18) | 0.0023 (11) | −0.0083 (12) | 0.0001 (13) |
| C5 | 0.0343 (12) | 0.0361 (12) | 0.0576 (16) | 0.0059 (10) | −0.0042 (11) | −0.0009 (11) |
| C6 | 0.0523 (15) | 0.0508 (14) | 0.0602 (17) | −0.0031 (11) | 0.0014 (13) | −0.0107 (13) |
| C7 | 0.0455 (15) | 0.0441 (14) | 0.090 (2) | −0.0063 (11) | −0.0017 (15) | 0.0105 (14) |
| C8 | 0.0478 (14) | 0.0590 (15) | 0.0704 (18) | −0.0027 (12) | 0.0097 (13) | 0.0143 (14) |
| C9 | 0.0432 (14) | 0.0457 (13) | 0.0586 (17) | 0.0027 (11) | 0.0013 (12) | −0.0004 (12) |
| C10 | 0.0328 (12) | 0.0372 (12) | 0.0532 (15) | −0.0012 (10) | 0.0017 (11) | 0.0029 (11) |
| C11 | 0.0457 (13) | 0.0405 (11) | 0.0441 (14) | 0.0024 (11) | 0.0037 (11) | −0.0020 (11) |
| C12 | 0.0377 (13) | 0.0386 (13) | 0.0417 (14) | 0.0016 (10) | −0.0010 (11) | 0.0013 (11) |
| C13 | 0.0601 (16) | 0.0494 (14) | 0.0406 (15) | −0.0026 (12) | 0.0029 (12) | 0.0028 (12) |
| C14 | 0.0585 (15) | 0.0488 (13) | 0.0410 (14) | −0.0022 (11) | 0.0036 (12) | −0.0052 (10) |
Geometric parameters (Å, °)
| O1—C12 | 1.219 (3) | C5—C10 | 1.440 (3) |
| O2—C13 | 1.209 (3) | C5—C6 | 1.419 (3) |
| O3—C9 | 1.361 (3) | C7—C8 | 1.355 (3) |
| O3—H4 | 0.8200 | C8—C9 | 1.405 (3) |
| N1—N2 | 1.3726 (19) | C9—C10 | 1.385 (3) |
| N1—C11 | 1.293 (2) | C10—C11 | 1.443 (3) |
| N2—C12 | 1.357 (2) | C13—C14 | 1.508 (3) |
| N2—C14 | 1.451 (3) | C1—H1 | 0.9300 |
| N3—C13 | 1.362 (3) | C2—H2 | 0.9300 |
| N3—C12 | 1.389 (3) | C3—H3 | 0.9300 |
| N3—H3N | 0.890 (18) | C6—H6 | 0.9300 |
| C1—C6 | 1.366 (3) | C7—H7 | 0.9300 |
| C1—C2 | 1.389 (3) | C8—H8 | 0.9300 |
| C2—C3 | 1.360 (4) | C11—H11 | 0.9300 |
| C3—C4 | 1.413 (3) | C14—H14A | 0.9700 |
| C4—C7 | 1.413 (4) | C14—H14B | 0.9700 |
| C4—C5 | 1.416 (3) | ||
| C9—O3—H4 | 109.00 | N2—C12—N3 | 106.33 (17) |
| N2—N1—C11 | 116.51 (16) | O1—C12—N2 | 127.76 (17) |
| N1—N2—C14 | 125.77 (14) | O1—C12—N3 | 125.91 (18) |
| C12—N2—C14 | 112.17 (15) | O2—C13—N3 | 126.88 (18) |
| N1—N2—C12 | 121.80 (16) | O2—C13—C14 | 126.97 (19) |
| C12—N3—C13 | 113.02 (17) | N3—C13—C14 | 106.15 (17) |
| C13—N3—H3N | 123.1 (13) | N2—C14—C13 | 102.24 (15) |
| C12—N3—H3N | 123.7 (13) | C2—C1—H1 | 119.00 |
| C2—C1—C6 | 121.3 (2) | C6—C1—H1 | 119.00 |
| C1—C2—C3 | 119.5 (2) | C1—C2—H2 | 120.00 |
| C2—C3—C4 | 121.1 (2) | C3—C2—H2 | 120.00 |
| C3—C4—C5 | 119.8 (2) | C2—C3—H3 | 119.00 |
| C3—C4—C7 | 121.57 (18) | C4—C3—H3 | 119.00 |
| C5—C4—C7 | 118.64 (19) | C1—C6—H6 | 119.00 |
| C4—C5—C10 | 119.44 (19) | C5—C6—H6 | 119.00 |
| C4—C5—C6 | 117.23 (19) | C4—C7—H7 | 119.00 |
| C6—C5—C10 | 123.33 (17) | C8—C7—H7 | 119.00 |
| C1—C6—C5 | 121.07 (18) | C7—C8—H8 | 120.00 |
| C4—C7—C8 | 121.78 (19) | C9—C8—H8 | 120.00 |
| C7—C8—C9 | 120.2 (2) | N1—C11—H11 | 119.00 |
| C8—C9—C10 | 121.05 (19) | C10—C11—H11 | 119.00 |
| O3—C9—C10 | 123.12 (17) | N2—C14—H14A | 111.00 |
| O3—C9—C8 | 115.82 (19) | N2—C14—H14B | 111.00 |
| C5—C10—C9 | 118.88 (17) | C13—C14—H14A | 111.00 |
| C5—C10—C11 | 119.83 (18) | C13—C14—H14B | 111.00 |
| C9—C10—C11 | 121.27 (18) | H14A—C14—H14B | 109.00 |
| N1—C11—C10 | 122.37 (18) | ||
| C11—N1—N2—C12 | −177.23 (16) | C7—C4—C5—C10 | −1.2 (3) |
| C11—N1—N2—C14 | 9.1 (2) | C3—C4—C7—C8 | −179.6 (2) |
| N2—N1—C11—C10 | −179.57 (16) | C5—C4—C7—C8 | −0.2 (3) |
| N1—N2—C12—O1 | 2.8 (3) | C4—C5—C6—C1 | 2.3 (3) |
| N1—N2—C12—N3 | −177.61 (15) | C10—C5—C6—C1 | −178.07 (19) |
| C14—N2—C12—O1 | 177.26 (19) | C4—C5—C10—C9 | 2.3 (3) |
| C14—N2—C12—N3 | −3.1 (2) | C4—C5—C10—C11 | −175.93 (17) |
| N1—N2—C14—C13 | 176.87 (16) | C6—C5—C10—C9 | −177.34 (18) |
| C12—N2—C14—C13 | 2.7 (2) | C6—C5—C10—C11 | 4.4 (3) |
| C13—N3—C12—O1 | −178.02 (19) | C4—C7—C8—C9 | 0.6 (3) |
| C13—N3—C12—N2 | 2.4 (2) | C7—C8—C9—O3 | 179.31 (19) |
| C12—N3—C13—O2 | 179.8 (2) | C7—C8—C9—C10 | 0.6 (3) |
| C12—N3—C13—C14 | −0.7 (2) | O3—C9—C10—C5 | 179.36 (17) |
| C6—C1—C2—C3 | −0.7 (3) | O3—C9—C10—C11 | −2.4 (3) |
| C2—C1—C6—C5 | −0.9 (3) | C8—C9—C10—C5 | −2.1 (3) |
| C1—C2—C3—C4 | 0.8 (3) | C8—C9—C10—C11 | 176.19 (18) |
| C2—C3—C4—C5 | 0.7 (3) | C5—C10—C11—N1 | 179.33 (17) |
| C2—C3—C4—C7 | −179.9 (2) | C9—C10—C11—N1 | 1.1 (3) |
| C3—C4—C5—C6 | −2.2 (3) | O2—C13—C14—N2 | 178.40 (19) |
| C3—C4—C5—C10 | 178.15 (18) | N3—C13—C14—N2 | −1.1 (2) |
| C7—C4—C5—C6 | 178.47 (18) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3N···O1i | 0.890 (18) | 1.962 (18) | 2.851 (2) | 177.9 (19) |
| O3—H4···N1 | 0.82 | 1.91 | 2.622 (2) | 145 |
| C6—H6···O2ii | 0.93 | 2.55 | 3.469 (2) | 169 |
| C14—H14B···O2ii | 0.97 | 2.36 | 3.038 (2) | 127 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x+1, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2180).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Iijima, T., Momotake, A., Shinohara, Y., Sato, T., Nishimura, Y. & Arai, T. (2010). J. Phys. Chem. A, 114, 1603–1609. [DOI] [PubMed]
- Li, L., Dang, Y.-Q., Li, H.-W., Wang, B. & Wu, Y.-Q. (2010). Tetrahedron Lett.51, 618–621.
- Oxford Diffraction (2009). CrysAlis PRO Oxford Diffraction Ltd, Yarnton, Oxfordshire, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Xu, H.-J., Du, N.-N., Jiang, X.-Y., Sheng, L.-Q. & Tian, Y.-P. (2009). Acta Cryst. E65, o1047. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810020118/su2180sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810020118/su2180Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




