Abstract
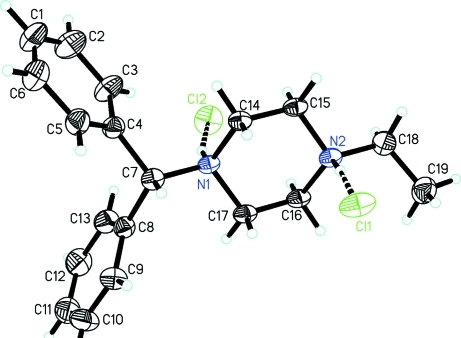
In the title compound, C19H26N2 2+·2Cl−, the piperazinediium ring exhibits a chair conformation. The dihedral angle between the two benzene ring planes is 76.45 (13)°. Both amine-group H atoms participate in hydrogen bonding with the two Cl atoms.
Related literature
The title compound was obtained in our search for a strong anti-Helicobacter pylori secondary metabolite. For general background to H. pylori, see: Gebert et al. (2003 ▶); Li et al. (2007 ▶); Moran & Upton (1986 ▶). For bond lengths and angles in related structures, see: Raves et al. (1992 ▶); Ilangovan et al. (2007 ▶).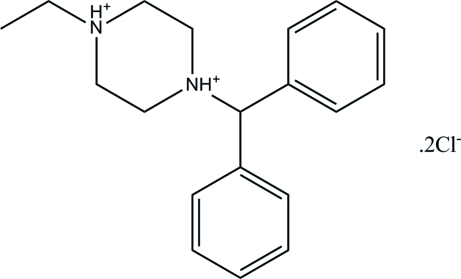
Experimental
Crystal data
C19H26N2 2+·2Cl−
M r = 353.32
Monoclinic,
a = 15.069 (3) Å
b = 7.2950 (15) Å
c = 18.565 (4) Å
β = 106.35 (3)°
V = 1958.3 (7) Å3
Z = 4
Mo Kα radiation
μ = 0.33 mm−1
T = 293 K
0.30 × 0.20 × 0.10 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.907, T max = 0.968
3684 measured reflections
3542 independent reflections
2101 reflections with I > 2σ(I)
R int = 0.028
200 standard reflections every 3 reflections intensity decay: 1%
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.157
S = 1.03
3542 reflections
216 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.38 e Å−3
Δρmin = −0.25 e Å−3
Data collection: CAD-4 Software (Enraf–Nonius, 1989 ▶); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810024530/zq2043sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810024530/zq2043Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1B⋯Cl2 | 0.96 (4) | 2.09 (4) | 3.028 (3) | 165 (3) |
| N2—H2B⋯Cl1 | 0.85 (4) | 2.16 (4) | 3.006 (3) | 174 (3) |
supplementary crystallographic information
Comment
The human pathogenic bacterium Helicobacter pylori has been ascertained to be an antiological agent for chronic active gastritis and a significant determinant in peptic and duodenal ulcer diseases (Gebert et al., 2003; Li et al., 2007). Sustained infection with this bacterium could lead to development of gastric cancer (Moran & Upton, 1986). Endophytic metabolites are recognized as a versatile arsenal of antimicrobial agents, since some endophytes have been shown to possess superior biosynthetic capabilities owing to their presumable gene recombination with the host, while residing and reproducing inside the healthy plant tissues. Our particular attention was extended to anti-Helicobacter pylori constituents. A detailed bioassay-guided fractionation of the culture extract of Fusarium sp., an endophytic fungus in Quercus variabilis Bl., was performed to afford a strong anti-H. pylori secondary metabolite. In this paper we report the structural information for the title compound, C19H26N22+.2Cl-, for which the asymmetric unit contains one 1-(diphenylmethyl)-4-ethylpiperazine-1,4-diium dication and two chloride anions. The bond lengths and angles of the title compound are in normal ranges when comparing with similar structures reported previously (Raves et al., 1992; Ilangovan et al., 2007). In the title compound, the piperazine fragment is in a chair conformation. The dihedral angle between the two benzene ring planes is 76.45 (13) °. Both amine-group H atoms participate in hydrogen bonding with the two Cl atoms.
Experimental
The cultivation of Fusarium sp. AMB-111, an endophytic fungus in Quercus variabilis, extraction and isolation were described in a preceding communication. A residue (149 g) from the dark brown tarry mass was obtained after depositing lipids, which was then subjected to column chromatography (CC) on silica gel (1300 g, 200–300 mesh), eluting with chloroform/methanol (1:0–0:1) to give seven fractions (F-1: 28.3 g, F-2: 12.2 g, F-3: 12.5 g, F-4: 14.0 g, F-5: 13.7 g, F-6: 12.3 g and F-7: 27.4 g). F-2, showing pronounced anti-Helicobacter pylori activity, was re-chromatographed over Si-gel column eluting with chloroform/acetone (50:1–4:1) to afford four subfractions (F-2–1: 4.5 g, F-2–2: 1.4 g, F-2–3: 2.3 g and F-2–4: 2.0 g). F-2–2 was subjected to gel filtration over Sephadex LH-20 with chloroform/methanol (1:1), followed by recrystalization repeatedly to give the title compound, a yellow crystal (300 mg).
Refinement
All H atoms were positioned geometrically (C—H = 0.93 Å for the aromatic H atoms and C—H = 0.96 Å for the aliphatic H atoms) and were refined as riding, with Uiso(H) = 1.2Ueq(C) and Uiso(H) = 1.2Ueq(N).
Figures
Fig. 1.
The structure of the title compound showing 30% probability displacement ellipsoids and the atom-numbering scheme.
Crystal data
| C19H26N22+·2Cl− | F(000) = 752 |
| Mr = 353.32 | Dx = 1.198 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 15.069 (3) Å | θ = 9–12° |
| b = 7.2950 (15) Å | µ = 0.33 mm−1 |
| c = 18.565 (4) Å | T = 293 K |
| β = 106.35 (3)° | Block, yellow |
| V = 1958.3 (7) Å3 | 0.30 × 0.20 × 0.10 mm |
| Z = 4 |
Data collection
| Enraf–Nonius CAD-4 diffractometer | 2101 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.028 |
| graphite | θmax = 25.3°, θmin = 1.4° |
| ω/2θ scan | h = 0→18 |
| Absorption correction: ψ scan (North et al., 1968) | k = 0→8 |
| Tmin = 0.907, Tmax = 0.968 | l = −22→21 |
| 3684 measured reflections | 200 standard reflections every 3 reflections |
| 3542 independent reflections | intensity decay: 1% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.157 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.065P)2 + 0.1129P] where P = (Fo2 + 2Fc2)/3 |
| 3542 reflections | (Δ/σ)max < 0.001 |
| 216 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.6307 (3) | 0.4082 (10) | 0.0245 (3) | 0.0936 (17) | |
| H1A | 0.6052 | 0.4038 | −0.0273 | 0.112* | |
| C2 | 0.6600 (4) | 0.2480 (8) | 0.0637 (3) | 0.0916 (16) | |
| H2A | 0.6549 | 0.1369 | 0.0383 | 0.110* | |
| C3 | 0.6968 (3) | 0.2534 (6) | 0.1407 (2) | 0.0710 (13) | |
| H3A | 0.7158 | 0.1459 | 0.1676 | 0.085* | |
| C4 | 0.7053 (2) | 0.4206 (5) | 0.17766 (19) | 0.0473 (9) | |
| C5 | 0.6766 (3) | 0.5794 (6) | 0.1372 (2) | 0.0576 (10) | |
| H5A | 0.6830 | 0.6915 | 0.1619 | 0.069* | |
| C6 | 0.6387 (3) | 0.5729 (8) | 0.0607 (3) | 0.0816 (14) | |
| H6A | 0.6187 | 0.6799 | 0.0338 | 0.098* | |
| C7 | 0.7408 (2) | 0.4406 (4) | 0.26239 (18) | 0.0404 (8) | |
| H7A | 0.7627 | 0.5672 | 0.2719 | 0.049* | |
| C8 | 0.6648 (2) | 0.4179 (5) | 0.30018 (19) | 0.0444 (9) | |
| C9 | 0.6408 (3) | 0.5678 (5) | 0.3367 (2) | 0.0572 (10) | |
| H9A | 0.6747 | 0.6758 | 0.3413 | 0.069* | |
| C10 | 0.5665 (3) | 0.5563 (7) | 0.3663 (2) | 0.0754 (13) | |
| H10A | 0.5506 | 0.6578 | 0.3903 | 0.090* | |
| C11 | 0.5164 (3) | 0.4010 (8) | 0.3610 (2) | 0.0766 (14) | |
| H11A | 0.4664 | 0.3957 | 0.3810 | 0.092* | |
| C12 | 0.5400 (3) | 0.2510 (7) | 0.3258 (2) | 0.0729 (13) | |
| H12A | 0.5056 | 0.1437 | 0.3222 | 0.087* | |
| C13 | 0.6142 (3) | 0.2571 (6) | 0.2956 (2) | 0.0606 (11) | |
| H13A | 0.6300 | 0.1541 | 0.2724 | 0.073* | |
| C14 | 0.9003 (2) | 0.3678 (4) | 0.26354 (17) | 0.0410 (8) | |
| H14A | 0.9148 | 0.4970 | 0.2721 | 0.049* | |
| H14B | 0.8801 | 0.3476 | 0.2097 | 0.049* | |
| C15 | 0.9859 (2) | 0.2566 (5) | 0.29714 (17) | 0.0432 (9) | |
| H15A | 0.9727 | 0.1281 | 0.2854 | 0.052* | |
| H15B | 1.0342 | 0.2941 | 0.2751 | 0.052* | |
| C16 | 0.9435 (2) | 0.2275 (5) | 0.41299 (18) | 0.0458 (9) | |
| H16A | 0.9638 | 0.2468 | 0.4669 | 0.055* | |
| H16B | 0.9295 | 0.0983 | 0.4039 | 0.055* | |
| C17 | 0.8575 (2) | 0.3387 (5) | 0.37957 (17) | 0.0464 (9) | |
| H17A | 0.8093 | 0.2996 | 0.4015 | 0.056* | |
| H17B | 0.8705 | 0.4668 | 0.3921 | 0.056* | |
| C18 | 1.1074 (3) | 0.1783 (5) | 0.4132 (2) | 0.0553 (10) | |
| H18A | 1.1505 | 0.2061 | 0.3845 | 0.066* | |
| H18B | 1.0950 | 0.0476 | 0.4091 | 0.066* | |
| C19 | 1.1516 (3) | 0.2259 (6) | 0.4946 (2) | 0.0720 (13) | |
| H19A | 1.2073 | 0.1561 | 0.5132 | 0.108* | |
| H19B | 1.1095 | 0.1975 | 0.5234 | 0.108* | |
| H19C | 1.1660 | 0.3543 | 0.4989 | 0.108* | |
| Cl1 | 1.05758 (8) | 0.68359 (12) | 0.40163 (5) | 0.0639 (4) | |
| Cl2 | 0.80657 (8) | −0.09301 (13) | 0.27873 (7) | 0.0756 (4) | |
| H1B | 0.807 (2) | 0.193 (5) | 0.2866 (18) | 0.059 (11)* | |
| H2B | 1.033 (2) | 0.392 (5) | 0.3885 (19) | 0.056 (11)* | |
| N1 | 0.82357 (19) | 0.3192 (4) | 0.29633 (14) | 0.0370 (7) | |
| N2 | 1.0189 (2) | 0.2800 (4) | 0.37993 (15) | 0.0401 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.082 (4) | 0.141 (5) | 0.045 (3) | −0.003 (4) | −0.003 (2) | −0.009 (3) |
| C2 | 0.105 (4) | 0.097 (4) | 0.064 (3) | −0.014 (3) | 0.010 (3) | −0.028 (3) |
| C3 | 0.095 (3) | 0.062 (3) | 0.047 (3) | −0.010 (3) | 0.005 (2) | −0.012 (2) |
| C4 | 0.044 (2) | 0.054 (2) | 0.044 (2) | −0.0073 (19) | 0.0123 (17) | −0.0052 (19) |
| C5 | 0.057 (3) | 0.061 (3) | 0.052 (2) | 0.004 (2) | 0.011 (2) | 0.006 (2) |
| C6 | 0.078 (3) | 0.100 (4) | 0.059 (3) | 0.013 (3) | 0.007 (3) | 0.012 (3) |
| C7 | 0.049 (2) | 0.0257 (17) | 0.047 (2) | −0.0090 (16) | 0.0137 (17) | −0.0047 (15) |
| C8 | 0.044 (2) | 0.045 (2) | 0.044 (2) | −0.0045 (18) | 0.0120 (17) | −0.0011 (17) |
| C9 | 0.067 (3) | 0.053 (2) | 0.058 (2) | −0.001 (2) | 0.027 (2) | −0.0058 (19) |
| C10 | 0.081 (3) | 0.085 (4) | 0.071 (3) | 0.010 (3) | 0.039 (3) | −0.010 (3) |
| C11 | 0.062 (3) | 0.108 (4) | 0.064 (3) | −0.001 (3) | 0.026 (2) | 0.009 (3) |
| C12 | 0.060 (3) | 0.086 (3) | 0.076 (3) | −0.028 (3) | 0.024 (2) | 0.003 (3) |
| C13 | 0.064 (3) | 0.062 (3) | 0.057 (2) | −0.012 (2) | 0.019 (2) | −0.005 (2) |
| C14 | 0.051 (2) | 0.0358 (19) | 0.0404 (19) | −0.0036 (17) | 0.0191 (17) | −0.0005 (15) |
| C15 | 0.060 (2) | 0.0354 (18) | 0.0395 (19) | −0.0084 (17) | 0.0220 (17) | −0.0077 (15) |
| C16 | 0.061 (2) | 0.043 (2) | 0.0389 (19) | −0.0094 (19) | 0.0220 (18) | −0.0034 (16) |
| C17 | 0.055 (2) | 0.050 (2) | 0.039 (2) | −0.0078 (19) | 0.0220 (17) | −0.0059 (17) |
| C18 | 0.059 (2) | 0.040 (2) | 0.063 (2) | 0.008 (2) | 0.012 (2) | −0.0043 (19) |
| C19 | 0.068 (3) | 0.077 (3) | 0.064 (3) | 0.011 (2) | 0.005 (2) | −0.001 (2) |
| Cl1 | 0.1106 (9) | 0.0332 (5) | 0.0560 (6) | −0.0187 (5) | 0.0369 (6) | −0.0078 (4) |
| Cl2 | 0.0951 (9) | 0.0287 (5) | 0.0988 (9) | −0.0145 (5) | 0.0205 (7) | −0.0052 (5) |
| N1 | 0.0494 (18) | 0.0276 (14) | 0.0358 (15) | −0.0083 (14) | 0.0151 (13) | −0.0055 (12) |
| N2 | 0.0539 (19) | 0.0269 (16) | 0.0408 (17) | −0.0027 (14) | 0.0155 (14) | −0.0052 (13) |
Geometric parameters (Å, °)
| C1—C6 | 1.366 (7) | C13—H13A | 0.9300 |
| C1—C2 | 1.382 (7) | C14—N1 | 1.493 (4) |
| C1—H1A | 0.9300 | C14—C15 | 1.502 (4) |
| C2—C3 | 1.381 (6) | C14—H14A | 0.9700 |
| C2—H2A | 0.9300 | C14—H14B | 0.9700 |
| C3—C4 | 1.388 (5) | C15—N2 | 1.486 (4) |
| C3—H3A | 0.9300 | C15—H15A | 0.9700 |
| C4—C5 | 1.382 (5) | C15—H15B | 0.9700 |
| C4—C7 | 1.519 (4) | C16—N2 | 1.486 (4) |
| C5—C6 | 1.373 (5) | C16—C17 | 1.506 (5) |
| C5—H5A | 0.9300 | C16—H16A | 0.9700 |
| C6—H6A | 0.9300 | C16—H16B | 0.9700 |
| C7—C8 | 1.512 (5) | C17—N1 | 1.492 (4) |
| C7—N1 | 1.514 (4) | C17—H17A | 0.9700 |
| C7—H7A | 0.9800 | C17—H17B | 0.9700 |
| C8—C9 | 1.387 (5) | C18—N2 | 1.499 (4) |
| C8—C13 | 1.388 (5) | C18—C19 | 1.510 (5) |
| C9—C10 | 1.382 (5) | C18—H18A | 0.9700 |
| C9—H9A | 0.9300 | C18—H18B | 0.9700 |
| C10—C11 | 1.350 (6) | C19—H19A | 0.9600 |
| C10—H10A | 0.9300 | C19—H19B | 0.9600 |
| C11—C12 | 1.371 (6) | C19—H19C | 0.9600 |
| C11—H11A | 0.9300 | N1—H1B | 0.96 (4) |
| C12—C13 | 1.386 (5) | N2—H2B | 0.85 (4) |
| C12—H12A | 0.9300 | ||
| C6—C1—C2 | 120.9 (4) | N1—C14—H14B | 109.2 |
| C6—C1—H1A | 119.6 | C15—C14—H14B | 109.2 |
| C2—C1—H1A | 119.6 | H14A—C14—H14B | 107.9 |
| C3—C2—C1 | 119.8 (5) | N2—C15—C14 | 111.4 (3) |
| C3—C2—H2A | 120.1 | N2—C15—H15A | 109.3 |
| C1—C2—H2A | 120.1 | C14—C15—H15A | 109.3 |
| C2—C3—C4 | 119.4 (4) | N2—C15—H15B | 109.3 |
| C2—C3—H3A | 120.3 | C14—C15—H15B | 109.3 |
| C4—C3—H3A | 120.3 | H15A—C15—H15B | 108.0 |
| C5—C4—C3 | 119.8 (3) | N2—C16—C17 | 111.1 (3) |
| C5—C4—C7 | 116.6 (3) | N2—C16—H16A | 109.4 |
| C3—C4—C7 | 123.5 (3) | C17—C16—H16A | 109.4 |
| C6—C5—C4 | 120.6 (4) | N2—C16—H16B | 109.4 |
| C6—C5—H5A | 119.7 | C17—C16—H16B | 109.4 |
| C4—C5—H5A | 119.7 | H16A—C16—H16B | 108.0 |
| C1—C6—C5 | 119.5 (5) | N1—C17—C16 | 112.3 (3) |
| C1—C6—H6A | 120.2 | N1—C17—H17A | 109.1 |
| C5—C6—H6A | 120.2 | C16—C17—H17A | 109.1 |
| C8—C7—N1 | 112.7 (3) | N1—C17—H17B | 109.1 |
| C8—C7—C4 | 112.2 (3) | C16—C17—H17B | 109.1 |
| N1—C7—C4 | 112.6 (3) | H17A—C17—H17B | 107.9 |
| C8—C7—H7A | 106.3 | N2—C18—C19 | 112.9 (3) |
| N1—C7—H7A | 106.3 | N2—C18—H18A | 109.0 |
| C4—C7—H7A | 106.3 | C19—C18—H18A | 109.0 |
| C9—C8—C13 | 118.8 (3) | N2—C18—H18B | 109.0 |
| C9—C8—C7 | 118.4 (3) | C19—C18—H18B | 109.0 |
| C13—C8—C7 | 122.6 (3) | H18A—C18—H18B | 107.8 |
| C10—C9—C8 | 119.9 (4) | C18—C19—H19A | 109.5 |
| C10—C9—H9A | 120.1 | C18—C19—H19B | 109.5 |
| C8—C9—H9A | 120.1 | H19A—C19—H19B | 109.5 |
| C11—C10—C9 | 121.4 (4) | C18—C19—H19C | 109.5 |
| C11—C10—H10A | 119.3 | H19A—C19—H19C | 109.5 |
| C9—C10—H10A | 119.3 | H19B—C19—H19C | 109.5 |
| C10—C11—C12 | 119.4 (4) | C17—N1—C14 | 108.7 (3) |
| C10—C11—H11A | 120.3 | C17—N1—C7 | 112.2 (3) |
| C12—C11—H11A | 120.3 | C14—N1—C7 | 109.4 (2) |
| C11—C12—C13 | 120.9 (4) | C17—N1—H1B | 106 (2) |
| C11—C12—H12A | 119.5 | C14—N1—H1B | 110 (2) |
| C13—C12—H12A | 119.5 | C7—N1—H1B | 110 (2) |
| C12—C13—C8 | 119.6 (4) | C16—N2—C15 | 109.0 (3) |
| C12—C13—H13A | 120.2 | C16—N2—C18 | 113.4 (3) |
| C8—C13—H13A | 120.2 | C15—N2—C18 | 111.7 (3) |
| N1—C14—C15 | 112.0 (3) | C16—N2—H2B | 111 (2) |
| N1—C14—H14A | 109.2 | C15—N2—H2B | 107 (2) |
| C15—C14—H14A | 109.2 | C18—N2—H2B | 104 (2) |
| C6—C1—C2—C3 | 0.9 (8) | C10—C11—C12—C13 | −0.1 (7) |
| C1—C2—C3—C4 | −1.0 (8) | C11—C12—C13—C8 | −0.8 (6) |
| C2—C3—C4—C5 | 0.2 (6) | C9—C8—C13—C12 | 1.5 (6) |
| C2—C3—C4—C7 | 177.2 (4) | C7—C8—C13—C12 | −174.2 (3) |
| C3—C4—C5—C6 | 0.8 (6) | N1—C14—C15—N2 | −57.9 (4) |
| C7—C4—C5—C6 | −176.4 (4) | N2—C16—C17—N1 | 57.7 (4) |
| C2—C1—C6—C5 | 0.1 (8) | C16—C17—N1—C14 | −55.1 (4) |
| C4—C5—C6—C1 | −0.9 (7) | C16—C17—N1—C7 | −176.3 (3) |
| C5—C4—C7—C8 | 90.9 (4) | C15—C14—N1—C17 | 55.1 (3) |
| C3—C4—C7—C8 | −86.2 (4) | C15—C14—N1—C7 | 177.9 (2) |
| C5—C4—C7—N1 | −140.7 (3) | C8—C7—N1—C17 | −51.4 (3) |
| C3—C4—C7—N1 | 42.2 (5) | C4—C7—N1—C17 | −179.6 (3) |
| N1—C7—C8—C9 | 117.2 (4) | C8—C7—N1—C14 | −172.2 (3) |
| C4—C7—C8—C9 | −114.5 (4) | C4—C7—N1—C14 | 59.7 (3) |
| N1—C7—C8—C13 | −67.0 (4) | C17—C16—N2—C15 | −57.0 (4) |
| C4—C7—C8—C13 | 61.3 (4) | C17—C16—N2—C18 | 177.9 (3) |
| C13—C8—C9—C10 | −1.4 (6) | C14—C15—N2—C16 | 57.4 (3) |
| C7—C8—C9—C10 | 174.5 (3) | C14—C15—N2—C18 | −176.6 (3) |
| C8—C9—C10—C11 | 0.5 (7) | C19—C18—N2—C16 | −66.6 (4) |
| C9—C10—C11—C12 | 0.3 (7) | C19—C18—N2—C15 | 169.8 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···Cl2 | 0.96 (4) | 2.09 (4) | 3.028 (3) | 165 (3) |
| N2—H2B···Cl1 | 0.85 (4) | 2.16 (4) | 3.006 (3) | 174 (3) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZQ2043).
References
- Enraf–Nonius (1989). CAD-4 Software Enraf–Nonius, Delft, The Netherlands.
- Gebert, B., Fischer, W., Weiss, E., Hoffmann, R. & Haas, R. (2003). Science, 301, 1099–1102. [DOI] [PubMed]
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- Ilangovan, A., Kumar, R. G., Liang, H., Balasubramani, K. & Muthiah, P. T. (2007). Acta Cryst. E63, o4087.
- Li, H.-Q., Xu, C., Li, H.-S., Xiao, Z.-P., Shi, L. & Zhu, H.-L. (2007). ChemMedChem, 2, 1361–1369. [DOI] [PubMed]
- Moran, A. P. & Upton, M. E. (1986). J. Appl. Bacteriol.60, 103–110. [DOI] [PubMed]
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Raves, M. L., Kanters, J. A. & Tollenaere, J. P. (1992). Acta Cryst. C48, 1712–1713.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810024530/zq2043sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810024530/zq2043Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report